Abstract
Characterization of the functional components in mammalian genomes depends on our ability to completely elucidate the genetic and epigenetic regulatory networks of chromatin states and nuclear architecture. Such endeavors demand the availability of robust and effective approaches to characterizing protein-DNA associations in their native chromatin environments. Consider able progress has been made through the applica tion of chromatin immunoprecipitation (ChIP) to study chromatin biology in cells. Coupled with genome-wide analyses, ChIP-based assays enable us to take a global, unbiased and comprehensive view of transcriptional control, epigenetic regulation and chromatin structures, with high precision and versatility. The integrated knowledge derived from these studies is used to decipher gene regulatory networks and define genome organization. In this review, we discuss this powerful approach and its current advances. We also explore the possible future developments of ChIP-based approaches to interrogating long-range chromatin interactions and their impact on the mechanisms regulating gene expression.
Introduction
Now that the complete human genome sequence is available [1], the current challenges are to identify all the functional genetic elements it encodes and to elucidate the complex regulatory networks that coordinate the function of all genetic and epigenetic elements that are crucial for cellular homeostasis, development and disease progression [2,3]. Hence, research focus has turned to the annotation of the genome for functional properties and the characterization of regulatory elements involved in controlling gene expression, gene function and genome stability.
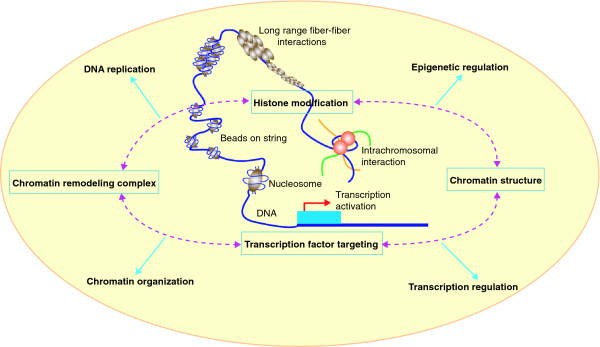
Among all the functional features of genome activities, dissecting the complex regulatory mechanisms controlling the precise spatial and temporal patterns of gene expression is critical for understanding developmental and cellular processes. The regulation of genome functions is largely mediated through highly controllable, dynamic and transient protein-chromatin interactions. In eukaryotes, genomic DNA is packaged by an octamer of four core histones into a nucleosome, the basic building block of chromatins. The intimate associations between DNA, histone and regulatory protein complexes within nucleosomes are critical for many nuclear activities such as transcription, DNA repair and replication (Figure 1) [4]. A detailed characterization of chromatin-DNA interactions is therefore required for understanding the molecular mechanisms behind gene regulation.
Figure 1.
Overview of activities in the nucleus. Regulation of genome functions involved in complex interactions between DNA, histone and protein complexes within the nucleosomes to bring about highly controlled and organized nucleus activities, such as transcription, DNA repair and replication, critical for cellular development.
Significant efforts have been dedicated to deciphering global chromatin structures, modifications and chromatin-protein interactions. Due to the dynamic and transient nature of such interactions, early attempts using biochemical fractionation were problematic [5]. Thanks to a powerful approach called chromatin immunoprecipitation (ChIP) [4], our understanding of protein-DNA interactions within their native chromatin context, in relation to different nuclear activities, has been greatly advanced. ChIP captures snapshots of these interactions in living cells by employing efficient cross-linking agents. The chromatin is disrupted by sonication and the DNA fragments cross-linked to the proteins of interest are then selectively enriched by immunoprecipitation with specific antibodies. After reversal of the cross-links, the enriched DNA can be subjected to further characterization. The ChIP method has been applied successfully in different areas, with focus on the analysis of chromatin structures and transcriptional dynamics. These areas include transcription factor (TF) binding [6], structural components of chromatin complexes [7,8], histone modifications [9-11] and enzyme function in histone modifications [12,13] across a wide range of organisms. Here, we summarize the developments of ChIP-based assays, their technical specifications and how they are applied to reveal insights into the molecular mechanisms during transcriptional and epigenetic regulation.
Technical considerations in conducting ChIP analysis
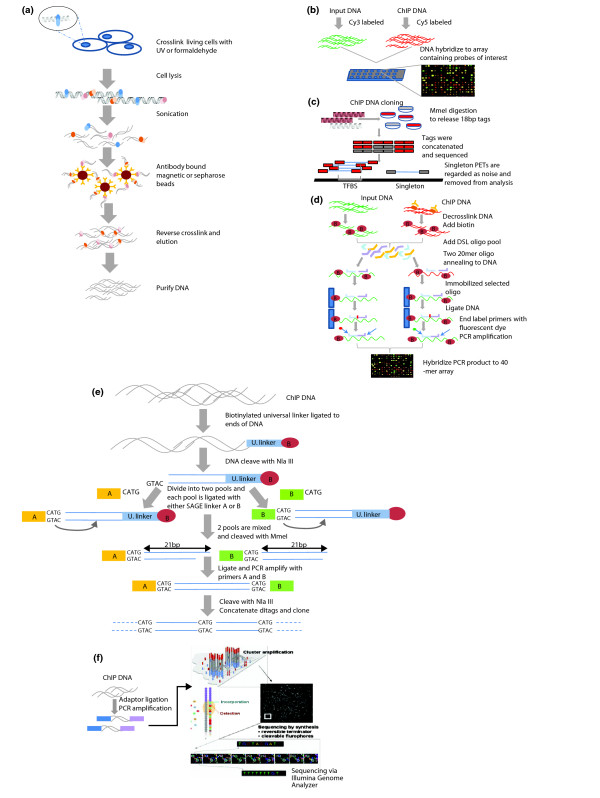
The basic principle of ChIP is schematically illustrated in Figure 2a. In this process, intact cells are subjected to cross-linking and nuclear extracts are prepared from the cross-linked cells, which are then sonicated to shear chromatin fragments into fragments of manageable size [14]. The methods used to covalently link the protein to DNA in vivo include ultraviolet (UV) and formaldehyde [15]. Formaldehyde, which cross-links DNA (primarily dA and dC) to the α-amino group of all amino acids [16], produces both protein-nucleic acid and protein-protein cross-links in vivo, making it a simple, fast and highly efficient agent for cross-linking. Experiments have suggested that different proteins are cross-linked with their interacting DNA with different efficiency [17], and excess exposure to formaldehyde can cause resistance to sonication and loss of material and low recovery. Therefore, small-scale trials with different cross-linking stringencies are recommended to evaluate the optimal condition. A key feature of formaldehyde-based cross-linking is that the cross-links are fully reversible through extensive proteinase K digestion and heat treatment. Thus, the proteins and DNA can be purified separately to enable subsequent analyses [18]. As a result, formaldehyde has been the preferred and general strategy for cross-linking.
Figure 2.
Overview of ChIP-based analysis. (a) Schematic view of the ChIP method. (b) Outline of ChIP-chip method. Input DNA and ChIP DNA are fluorescently labeled and hybridized to an array of probes where intensities from individual probes are recorded. A high probe intensity from the ChIP DNA over the input DNA will represent a potential binding site. Here a red dot represents a potential protein-binding site. (c) ChIP-PET overview. ChIP DNA is cloned into a plasmid vector and converted into PETs via MmeI digestion. The released PETs are concatenated, cloned and sequenced. PET sequences are mapped back to the genome to locate the TF binding site. Sites with multiple overlapping PETs are regarded as potential TF binding sites, while singleton PETs are regarded as background and removed from the analysis. (d) ChIP-DSL scheme. The technique avoids direct amplification of ChIP DNA for hybridization, hence providing an unbiased amplification with increased sensitivity over ChIP-chip. (e) ChIP-SAGE. Summary of the ChIP-SAGE experimental procedure. (f) Illustration of ChIP-Seq. ChIP DNA is ligated with specific sequencing adaptors and PCR amplified. The PCR product is hybridized onto the surface of a flowcell for sequencing.
Other important factors to be considered while doing ChIP include the antibody specificity and the fine balance between the cross-linking stringency and sonication conditions. The robustness of ChIP to differentially select target regions versus random genomic DNA is highly dependent on the availability of high-quality and high-affinity antibodies against the protein of interest. Community and industrial efforts have been initiated to characterize and catalog ChIP-grade antibodies against nuclear proteins of interest. Furthermore, ChIP with an antibody of different isotype is commonly used to validate the binding events found. To further improve the efficiency of the ChIP process, a sequential ChIP can be attempted. In this method, two rounds of ChIP are performed sequentially using different subtypes of antibodies against the same proteins but different epitopes. Although highly accurate, sequential ChIP is technically challenging and suffers from low yield, which limits its applications.
Readout methods for ChIP-based analysis
It is important to note that the ChIP process differentially enriches the targeted protein-DNA interactions from the entire nuclear cross-linked chromatin-protein complexes through antibody selection; however, it is not a purification step. Therefore, once the ChIP material is available, additional steps are required to characterize the material pulled down and determine their relative enrichments (Figure 2b-f). In a conventional ChIP assay, the enriched regions are initially analyzed using small-scale assays such as traditional cloning followed by a sequencing-based approach [19], Southern blot hybridization analysis [20] or quantitative real-time polymerase chain reaction (PCR) (ChIP-qPCR) [21]. The availability of the complete genome sequences of many complex organisms offers the opportunity to carry out genome-wide detection of protein-chromatin interactions. Two major approaches have been commonly adopted as readouts to determine the identity of these ChIP-enriched DNA fragments at the whole-genome scale: hybridization-based or sequencing-based methods.
Hybridization-based whole-genome ChIP analysis: ChIP-on-chip
To characterize the protein-DNA interaction profiles across different regions on the genome landscape, high-density microarrays are created and hybridization is used for the analysis of ChIP DNA (referred to as ChIP-on-chip). In brief, after reversal of the cross-links, ChIP-enriched DNA and control DNA will be amplified by PCR and fluorescently labeled with the cyanine dyes Cy5 and Cy3 for hybridization to the DNA microarrays containing probes that correspond to the genomic sequences of interest (Figure 2b). The ratio of the Cy5 to Cy3 fluorescence intensities measured for each DNA element provides a measure of the extent of the binding across the entire genomic regions covered in the array. Genomic loci with higher fluorescent intensity in the ChIP DNA than the control DNA will be considered enriched as the potential binding sites. Using this technique, the non-repeat sequences in the genome can be interrogated and many novel binding sites uncovered. For example, genes regulated by many TFs such as STE12 and GAL4p were characterized in detail in yeast systems and revealed new functional pathways regulated through multiple TF bindings [6,22].
Initially, array studies were limited to promoter regions amplified through PCR [23]. Over the years, significant improvements have been made to the ChIP-on-chip procedures as well as the array designs. High-density oligonucleotide tiling arrays that represent the entire genome are now available and enable comprehensive mapping of protein-DNA interactions [6,22,24].
Limitations of the ChIP-on-chip approach
Despite considerable success, array-based readout of ChIP signals does suffer from several limitations. Firstly, the hybridization-based platform is unable to detect signals in repeat regions. Due to the large size and complexity of mammalian genomes, the DNA microarrays available often only contain partial genomic content or promoter regions of well-characterized genes. Therefore, many of the ChIP-chip analyses provide incomplete information, as any biologically significant binding occurring within the non-interrogated regions cannot be captured. Nevertheless, the repetitive regions are important areas to examine, based on what we know about TF binding [25]. Secondly, PCR is generally used to amplify the ChIP material for hybridization, which can result in potential hybridization noise signals from biased amplification. To overcome non-specific amplification from direct PCR and cross-hybridization noise, an improved method called ChIP-DSL (DNA selection and ligation) was developed [26]. In ChIP-DSL, paired oligonucleotides corresponding to regions of interest are designed as signatures and selected by ChIP DNA. The annealed paired oligos are then ligated and PCR used for the array-based detection (Figure 2d). ChIP-DSL avoids direct amplification of ChIP fragments and the amplicons are uniform in size to minimize PCR bias. Thirdly, as many different array designs and genome assemblies exist, the results from different groups could be difficult to compare. Lastly, the global ChIP-chip approach is dependent on the construction of whole-genome arrays. For certain complex genomes, these are not commercially available or economically practical. Due to these limitations, the whole-genome tiling array approach has not yet been adopted by the entire research community and has only been used in several large projects studying the genomes of human and mouse.
Sequencing-based whole-genome ChIP analyses
Sequencing-based methods emerged as an alternative to genome-wide readouts of ChIP analysis, particularly for complex genomes. To determine the identities of ChIP DNA by sequencing methods, large numbers of sequence reads are required. As ChIP assay is only a process of enrichment, a significant amount of non-enriched background DNA will still be present in the ChIP DNA material. With a limited survey of the ChIP DNA pool, it is difficult to distinguish between genuine signal and noise. However, if the sampling of the DNA pool can be increased, the genuine ChIP-enriched sites can be defined by multiple overlapping ChIP fragments, whereas the non-specific regions will only be covered by random ChIP singletons. The bona fide sites can then be inferred by multiple mapped sequenced fragments.
ChIP-SAGE
To overcome the depth of sequencing coverage, short-tag-based sequencing strategies like serial analysis of gene expression (SAGE) have been adopted. SAGE was originally developed for counting transcript levels [27] and later applied to genome scanning for transcription factor binding site and histone modifications [28,29]. In ChIP-SAGE, the ChIP-enriched DNA fragments are end-ligated with a universal biotinylated linker, and 21-bp tags are generated by type II restriction enzyme digestion for sequencing (Figure 2e). Compared with the ChIP-on-chip hybridization approach, ChIP-SAGE increases the coverage and resolution to the entire genome [28]. However, this monotag approach suffers from mapping ambiguity and is unable to differentiate amplification bias, and thus has a lower accuracy.
ChIP-PET
In order to enhance the mapping accuracy of short-tags and increase the information content while still exploiting the short-tag sequencing efficiency, a paired-end-ditag (PET) method has been developed (ChIP-PET). Like SAGE, the PET approach was initially used for transcriptome analysis [30]. In ChIP-PET, the ChIP DNA is converted into PETs for ultra-high-throughput sequencing. Each PET sequence is mapped onto the genome and the locations of binding sites can be inferred by overlapping PET-defined clusters (Figure 2c). Over 90% of the sites identified can be validated by ChIP-qPCR, and de novo consensus binding motifs can be predicted from the overlapping regions [31]. The ChIP-PET approach has been demonstrated to map whole-genome TF binding sites and epigenetic modifications in both cancer and embryonic stem cells (ESCs) with high specificity and resolution [9,31,32]. Compared to ChIP-on-chip, the ChIP-PET approach is an unbiased and open system for identifying all DNA segments enriched by ChIP. This method is not restricted by the array coverage or probe performance and thus allows a real genome-wide analysis. Its only limitation is the upfront requirement for large sequencing capacity.
ChIP-Seq
Recently, the development of robust and advanced sequencing technologies, particularly the ability to rapidly decode millions of DNA fragments simultaneously with high efficiency and relative low cost, has facilitated our ability to characterize ChIP DNA by direct sequencing (ChIP-Seq) [11,33]. ChIP-Seq has proved to be a simple and robust method for global, unbiased interrogation of the TF binding sites and epigenetic modifications. In ChIP-Seq, the ChIP DNA is end polished and ligated with the sequencing adaptors, followed by limited PCR amplifications. Size selections of DNA fragments are subjected to cluster amplification and sequencing (Figure 2f). Between 25 and 36 nucleotides from either end of ChIP DNA fragments can be determined with high accuracy, and millions of high-quality reads can be generated within days. Based on their mapping locations, regions with a high number of clusters of ChIP tag sequences are defined as ChIP enrichment sites. To further distinguish the true binding sites from the non-specific sites, control DNA (input) is sequenced to determine the noise, which can then be removed. ChIP-Seq enables the performance of deep sequencing at high resolution and low cost.
Insights from genome-wide ChIP analysis
With the availability of whole-genome and unbiased approaches to characterizing chromatin-DNA interactions, our knowledge of the genomic features, landscape, target genes and gene expression activity has drastically advanced in recent years. Here, we summarize what we have learnt collectively on the critical links between chromatin modifications and transcriptional outputs.
Identification of transcription factor binding sites
Applying ChIP-based assays for components in the transcription machinery or TFs, their genomic targets and regulatory circuitries can be reconstructed [33-35]. One of the unique and intriguing findings from these genome-wide studies indicates that there are large numbers of identified target binding sites located outside of the previously annotated promoters and suggests that the functional regulatory elements of the genome are larger than previously envisioned. For example, over 30% of the estrogen receptor binding sites were found in the inter-genic regions at least 50 kb away from the neighbor genes [36]. Such an observation raises interesting questions about the functional nature of these binding sites and about how to accurately correlate the genes and their corresponding regulatory regions. The genome-wide ChIP assay can also be used to uncover the sequences bound by specific TFs and characterize their binding site selection. Through the putative in vivo binding sites identified, the ab initio binding consensus sequences associated with the protein of interest can be efficiently derived [33]. We have also gained insights into how TFs have evolved different mechanisms to elicit target gene responses. Some individual TFs can elicit multiple transcriptional responses, while different TFs can be recruited to the same target regions to trigger transcriptional activation leading to cell differentiation [33]. In ESCs, key reprogramming factors and TFs involved in signaling pathways as well as self-renewal have been analyzed. Specifically, two clusters of genomic loci were found that were extensively targeted by multiple transcription factors in the ESC genome. The first cluster includes NANOG, OCT4, SOX2, SMAD1 and STAT3. The second cluster consists of c-Myc (MYC), n-Myc (MYCN), ZFX and E2F1. STAT3 and SMAD1 are major signaling components modulating the leukemia inhibitory factor (LIF) and bone morphogenetic protein (BMP) pathways. LIF and BMPs are protein factors required for the maintenance of the pluripotency state of ESCs. These results have shown that LIF and BMP signaling pathways are integrated into the ESC pluripotency maintenance TF cluster (OCT4, SOX2 and NANOG) through SMAD1 and STAT3; and multiple transcription factor clustering is the mechanism to recruit cell-specific enhancer targeting for lineage-specific transcription regulation.
Profiling chromatin modifications
In addition to TF binding, the ChIP assay can also be used to profile the distribution of the chromatin modification components, histone variants and modifications [10]. One of the pioneering efforts was to understand the mechanisms by which histone modifications regulate transcription and chromatin organization. Starting in the yeast system, the application of ChIP assays demonstrated that histone acetylation was a critical link between chromatin structure and transcriptional activation [37]. In mammalian genomes, Barski et al. have characterized the histone codes through profiling 20 lysine and arginine methylation modification patterns in histones, and identified the signatures for histone methylation patterns surrounding promoters, enhancers, insulators and transcribed regions [11]. Among them, monomethylations of H3K27, H3K9, H4K20, H3K79 and H2BK5 were found to be associated with gene activation, while trimethylation of H3K27, H3K9 and H3K79 was linked to gene repression. In a study to investigate the types of histone modifications that underlie the chromatin properties to maintain the pluripotent nature of the ESC genome, Lander and colleagues un covered 109 domains showing overlapping opposing histone modification marks, termed 'bivalent domains', where large regions of H3K27me3 harbor smaller regions of H3K4me3 [10]. Following further characterization using a genome-wide ChIP-PET approach in human ESCs [9], H3K4me3 was found to be prevalent and occurred in nearly 70% of promoters in annotated genes, while H3K27me3 appears less occupied in promoter regions and forms a 'bivalent domain' by co-marking 10% of genes with H3K4me3. A large portion of genes that are important for mesoderm development, neuroectoderm and other developmental processes are among the genes co-modified by H3K4me3 and H3K27me3 [9].
Through the applications of genome-wide ChIP analyses across different organisms, we learnt that TF binding sites are not necessarily conserved among species [34,38] and that not all TF-chromatin interactions are functional [25]. Using the binding regions of seven mammalian TFs (ESR1, TP53, MYC, RELA, POU5F1, SOX2 and CTCF) identified on a genome-wide scale, we found only a minority of sites appeared to be conserved at the sequence level, suggesting that evolution has adapted factor binding sites to aid the dynamic regulation of mammalian genomes.
New advances in ChIP technology
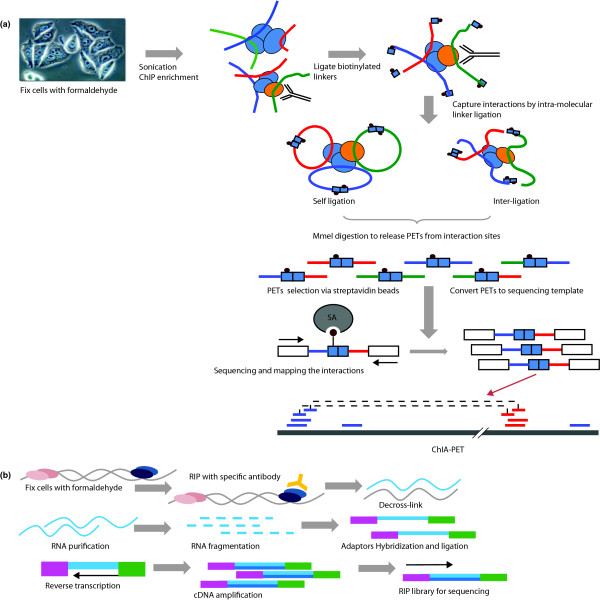
Up to now, most studies using the ChIP assay have been focused on characterization of the DNA portions associated with the pulled-down ChIP material. Analysis of the proteins in their in vivo chromosomal context recovered from ChIP has only been reported recently [39]. In addition to protein-DNA interactions, ChIP can also be used to study RNA-protein interactions, especially non-coding, nuclear RNA-directed epigenetic control [40,41]. Applying RNA immunoprecipitation followed by PCR (RIP-PCR), non-coding RNAs (ncRNAs), such as HOTAIR and Kcnq1ot1, have been shown to associate with Suz12 and G9a in primary human fibroblasts and mouse fetus, and these associations affect Hox genes as well as the expression of imprinting genes [42,43]. Although RIP has only been carried out in selected cells and at a limited scale, it is intriguing to suggest that there is a specific population of ncRNAs that acts in coordination with different components of histone and DNA modification machineries to achieve gene-expression control. Through further advancement in RIP-based analysis (Figure 3b), it will be interesting to determine their identity, specificity and impact on cell differentiation.
Figure 3.
Future advances of ChIP-based approaches. (a) ChIA-PET represents a new approach to interrogate long-range inter- and intra-chromosomal interactions that are mediated by TFs or chromatin-modifying complexes to drive gene expression. (b) RNA immunoprecipitation method coupled with ultra-high-throughput sequencing (RIP-Seq).
The recent expansion of ChIP technologies has enabled a better understanding of the interactions between TFs and the regulatory networks contributing to gene regulation. Surprisingly, these analyses have demonstrated that many TFs rarely bind to promoter regions compared with intergenic regions [36], suggesting critical roles for long-distance, promoter-enhancer interactions in regulating gene expression in mammalian cells [44]. In some cases, it was found that the transcriptional activation involved distal control elements located hundreds of kilobases away, which are brought together through connecting DNA loops that allow physical interactions between the regulatory elements for gene expression [45]. However, methods like ChIP-Seq can only reveal the functional genome in a linear fashion. Information on long-range interactions harnessed within the chromatin-protein complexes and how they impact transcriptional regulation is still lacking.
Initial efforts to characterize the distant interactions have been technically challenging and mostly limited to microscopy techniques, which are laborious and of poor resolution. Through formaldehyde cross-linking followed by proximity-based ligation, long-range chromosomal interactions can be captured and detected by PCR (chromatin conformation capture, 3C), microarray analysis or high-throughput sequencing (4C or 5C), with limited scale and selective bias [46-48]. Applying 3C in the human β-globin loci, various specific interactions between the genes and the regulatory elements were demonstrated [49]. Although 3C and its variants are excellent tools to study complex interactions, these methods require prior knowledge of interacting candidates, hence cannot be used for genome-wide profiling for all chromatin interactions. As such, there is a need for approaches that reveal global chromatin interactions at the whole-genome scale in an unbiased and de novo manner. With the pair end ditag concept, we further explored the ability of PET to connect two ends of DNA and delineate their relationships to characterize interacting chromatins (chromatin interaction analysis by pair-end-ditagging; ChIA-PET) [50]. In this approach, ChIP was performed with antibodies specific to the TF of interest. Specially designed short oligonucleotide linkers were ligated to the ends of each interacting DNA fragment, followed by second intra-molecular ligations to connect two interacting DNA fragments together. PETs from ligated DNA are extracted and analyzed by pair end ditag sequencing. The linear binding sites along genomic DNA can be revealed from self-ligation PETs and the interactions between the binding sites can be determined from inter/intra-chromatin ligating PETs (Figure 3a). Therefore, a single ChIA-PET experiment can generate two interrelated datasets, depending on the step at which the ligation occurs (before or after the de-cross-link). Such a feature, when supported by ultra-high-throughput sequencing, can reveal interactomes mediated by TFs or chromatin-modifying complexes. We expect that the mapping of the whole-genome interactome mediated by pertinent TFs or chromatin modifications will translate into knowledge that is critical for understanding the fundamental transcriptional regulation programs.
Future prospects
As described in this paper, combination of the ChIP assay with robust readout methods is extremely powerful for a variety of whole-genome analyses in order to define the functional components within mammalian genomes. The wide range of interactions and diverse organisms it has been applied to have already demonstrated the power of this approach. Considerable progress has been made in our understanding of transcriptional and epigenetic regulation, as well as in the elucidation of transcriptional regulatory networks and chromatin organization. Ultimately, with further improvement of the ChIP-based assays, particularly in the robustness of the enrichment and expansion of their applications, we foresee that ChIP will continue to be the critical approach to study chromatin biology and genome regulation. If successfully implemented, particularly for individual and personal human genome interrogations, such applications will further our understanding of how genetic and epigenetic regulation coordinates eukaryotic development. This knowledge has the potential to translate into a better understanding of the fundamental transcriptional regulation programs, and lead to biomarker discovery or therapeutic target stratifications, which ultimately guide the development of strategies for personalized medicine.
Abbreviations
BMP: bone morphogenetic protein; 3C: chromatin conformation capture; ChIA-PET: chromatin interaction analysis followed by pair-end-ditagging; ChIP: chromatin immunoprecipitation; ChIP-chip: chromatin immunoprecipitation followed by DNA microarray hybridization; ChIP-DSL: chromatin immunoprecipitation followed by DNA selection and ligation; ChIP-PET: chromatin immunoprecipitation followed by pair-end-ditagging sequencing; ChIP-qPCR: chromatin immunoprecipitation followed by quantitative PCR; ChIP-SAGE: chromatin immunoprecipitation coupled with serial analysis of gene expression; ChIP-Seq: chromatin immunoprecipitation followed by high-throughput sequencing; ESC: embryonic stem cell; LIF: leukemia inhibitory factor; ncRNA: non-coding RNA; RIP-PCR: RNA immunoprecipitation followed by PCR; TF: transcription factor; UV: ultraviolet.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CLW provided the overall direction and outline. EW generated the figures and produced the first draft. EW and CLW jointly wrote the manuscript.
Contributor Information
Eleanor Wong, Email: wonghe@gis.a-star.edu.sg.
Chia-Lin Wei, Email: weicl@gis.a-star.edu.sg.
Acknowledgements
The authors would like to acknowledge Genome Technology and Biology Group at the Genome Institute of Singapore for technical details on sequencing, particularly the GIS sequencing team. The authors are supported by the Agency for Science, Technology and Research (A*STAR) of Singapore and the NIH ENCODE grant 1R01HG003521-01.
References
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. In vivo cross-linking and immunoprecipitation for studying dynamic Protein:DNA associations in a chromatin environment. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- Dedon PC, Soults JA, Allis CD, Gorovsky MA. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem. 1991;197:83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber SA, Gerton JL, Polancic JE, DeRisi JL, Koshland D, Megee PC. The kinetochore is an enhancer of pericentric cohesin binding. PLoS Biol. 2004;2:E260. doi: 10.1371/journal.pbio.0020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/S1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Jackson V, Chalkley R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell. 1981;23:121–134. doi: 10.1016/0092-8674(81)90277-4. [DOI] [PubMed] [Google Scholar]
- Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci USA. 1984;81:4275–4279. doi: 10.1073/pnas.81.14.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaw YF, Crane LE, Lange P, Shapiro R. Isolation and identification of cross-links from formaldehyde-treated nucleic acids. Biochemistry. 1980;19:5525–5531. doi: 10.1021/bi00565a010. [DOI] [PubMed] [Google Scholar]
- Solomon MJ, Varshavsky A. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc Natl Acad Sci USA. 1985;82:6470–6474. doi: 10.1073/pnas.82.19.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem Sci. 2000;25:99–104. doi: 10.1016/S0968-0004(99)01535-2. [DOI] [PubMed] [Google Scholar]
- Grandori C, Mac J, Siebelt F, Ayer DE, Eisenman RN. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- Guccione E, Martinato F, Finocchiaro G, Luzi L, Tizzoni L, Dall' Olio V, Zardo G, Nervi C, Bernard L, Amati B. Myc-binding-site recognition in the human genome is determined by chromatin context. Nat Cell Biol. 2006;8:764–770. doi: 10.1038/ncb1434. [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Hanlon SE, Lieb JD. Progress and challenges in profiling the dynamics of chromatin and transcription factor binding with DNA microarrays. Curr Opin Genet Dev. 2004;14:697–705. doi: 10.1016/j.gde.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, Liu ET. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, Fan JB, Duan L, Glass CK, Rosenfeld MG, Fu XD. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci USA. 2007;104:4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- Ng P, Wei CL, Sung WK, Chiu KP, Lipovich L, Ang CC, Gupta S, Shahab A, Ridwan A, Wong CH, Liu ET, Ruan Y. Gene identification signature (GIS) analysis for transcriptome characterization and genome annotation. Nat Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, Emanuelsson O, Stolc V, Weissman S, Gerstein MB, Ruan Y, Snyder M. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, Bourque G, Wei CL, Liu ET. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3:e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin J, Kingston RE. Purification of proteins associated with specific genomic loci. Cell. 2009;136:175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988;335:683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F. The effect of distance on long-range chromatin interactions. Mol Cell. 1997;1:131–139. doi: 10.1016/S1097-2765(00)80014-3. [DOI] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Han X, Mohamed YB, Orlov YL, Velkov S, Ho A, Poh HM, Chew EGY, Huang PYH, Welboren WJ, Han YY, Ooi HS, Ariyaratna PN, Vega BV, Luo YQ, Tan PY, Choy PY, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li HX, Desai KV, Thomsen JS, Lee KY. An oestrogen receptor α-bound human chromatain interactome. Nature. in press . [DOI] [PMC free article] [PubMed]