Abstract
Self-assembled monolayers (SAMs) of alkanethiolates on gold have become an important tool for probing cell-material interactions. Emerging studies in stem cell biology are particularly reliant on well-defined model substrates, and rapid and highly controllable fabrication methods may be necessary to characterize the wide array of stem cell-material interactions. Therefore, this study describes a rapid method to create SAM cell culture substrates with multiple discrete regions of controlled peptide identity and density. The approach uses an NaBH4 solution to selectively remove regions of bio-inert, hydroxyl-terminated oligo(ethylene glycol) alkanethiolate SAM, then locally replace them with mixed SAMs of hydroxyl- and carboxylic acid-terminated oligo(ethylene glycol) alkanethiolates. The cell adhesion peptide Arg-Gly-Asp-Ser-Pro (RGDSP) was then covalently linked to carboxylic acid-terminated mixed SAM regions to create cell adhesive environments within a bio-inert background. SAM preparation and peptide immobilization were characterized using polarization modulation–infrared reflection-absorption spectroscopy (PMIRRAS), as well as assays to monitor conjugation of a fluorescently-labeled peptide. This “localized SAM replacement” method was achieved using an array of microchannels, which facilitated rapid and simple processing. Results indicate that immobilized RGDSP promoted spatially localized attachment of human mesenchymal stem cells (hMSCs) within specified regions, while maintaining a stable, bio-inert background in serum-containing cell culture conditions for up to 14 days. Cell attachment to patterned regions presenting a range of cell adhesion peptide densities demonstrated that peptide identity and density strongly influence hMSC spreading and focal adhesion density. These substrates contain discrete, well-defined microenvironments for stem cell culture, which could ultimately enable high-throughput screening for the effects of immobilized signals on stem cell phenotype.
Introduction
Self-assembled monolayers (SAMs) of alkanethiolates on gold can be used to design biological interfaces that regulate protein-surface interactions and, consequently, cell-surface interactions. Specifically, this can be achieved by terminating SAMs with two components: (1) oligo(ethylene glycol) (OEG) moieties that prevent non-specific protein interactions; and (2) controlled densities of extracellular matrix (ECM)-derived peptide ligands that promote integrin-mediated cell adhesion. In this manner, any downstream changes in cell phenotype due to regulation of cell adhesion can be directly correlated to the density or type of cell adhesion ligand displayed on the surface. This offers significant advantages over canonical cell culture, in which the type and density of cell-binding motifs present in an adsorbed protein layer are difficult to determine or predict,1 and thus difficult to correlate to downstream changes in cell phenotype.
In view of their advantageous characteristics, well-defined SAM substrates have been used to study cell attachment, spreading, proliferation, and migration.2, 3, 4, 5 For example, Roberts et al. demonstrated that the density of the cell adhesion ligand Arg-Gly-Asp (RGD) presented on an otherwise bio-inert SAM substrate dictated attachment and spreading of bovine endothelial cells.2 In addition, SAMs have been used as biological tools to investigate more detailed cell signaling mechanisms associated with cell adhesion. For example, Kato et al. used well-defined SAM substrates to examine the effects of linear versus cyclic versions of RGD on integrin-mediated activation of the intracellular enzyme focal adhesion kinase (FAK). Immobilized cyclic RGD promoted increased FAK activation as well as increased downstream mitogen-activated protein (MAP) kinase activation when compared to linear RGD.6 Taken together, these studies demonstrate that SAMs can be used as well-defined platforms to probe the influence of specific signals on cell adhesion and signal transduction. Therefore, SAM substrates can potentially be used as a broad platform to study cell interactions with their microenvironment. However, the experimental scope of cell culture on SAMs has been limited due to the low-throughput of standard SAM fabrication, and the difficulties in applying standard soft lithography methods to oligo(ethylene glycol)-terminated SAMs.
Well-defined substrates capable of controlling the cell adhesion microenvironment may be particularly important in stem cell biology. Stem cells reside in complex microenvironments in vivo that govern cell behavior through a plethora of pathways, including signaling via cell adhesion to the ECM.7 In addition, recent in vitro studies indicate that the behavior of human mesenchymal stem cells (hMSCs), including osteogenic differentiation8, 9, 10, 11 and maintenance of differentiation potential,12, 13 can be regulated through adhesion to ECM-derived molecules. In particular, vitronectin, collagen I,8, 9 laminin 5,10 laminin-332,11 and mixtures of ECM proteins12, 13 have each been shown to influence MSC phenotype. Due to the large number of ECM-derived molecules that can influence hMSC phenotype, and the multivalency of many ECM proteins, a mechanistic understanding of hMSC adhesion is likely to require a high-throughput approach capable of exploring a wide range of experimental conditions simultaneously. Therefore we aimed to develop a rapid SAM fabrication approach that could ultimately be compatible with high-throughput experimental designs for screening of stem cell-material interactions.
We aimed to develop a facile, rapid fabrication approach to create arrays of spatially distinct cell culture environments on otherwise bioinert SAM substrates. This was achieved using microfluidics in combination with aqueous chemistries to selectively remove regions of hydroxyl-terminated oligo(ethylene glycol) alkanethiolate SAMs and locally replace them with a mixture of carboxylic acid- and hydroxyl-terminated oligo(ethylene glycol) alkanethiolate SAMs. A carbodiimide condensation reaction was then used to covalently immobilize the cell adhesion peptide Arg-Gly-Asp-Ser-Pro (RGDSP), which is derived from the 10th domain of the cell adhesion protein fibronectin type III.14 The resulting substrates contained multiple, discrete cell culture environments, with varied densities of a cell adhesion peptide. Importantly, the use of a microfluidics approach facilitated rapid substrate processing to yield arrays of discrete cell culture environments, as well as adaptability to generate multiple array geometries. The resulting approach may provide adaptable, broadly applicable substrates to investigate cell-material interactions.
Design rationale
The rapid lateral diffusion15, 16 and solution exchange17 of alkanethiolates that occur during SAM formation present challenges to conventional soft lithography approaches (e.g. stamping, microcontact printing, “lift-off” techniques).18, 19 In addition, the inherent instability of alkanethiolate SAMs when exposed to ambient air, perhaps due to the susceptibility of the thiol moiety to oxidation,20, 21 presents a significant practical processing challenge. Previous SAM patterning techniques have attempted to address these issues by using serum-free cell seeding conditions to limit the confounding effects of non-specific protein adsorption, or using novel alkanethiolate chemistries to stabilize patterned regions of a SAM and maintain background inertness.22, 23 Here we use a “localized SAM replacement” approach (Fig. 1) that involves the following steps: (1) An inert, hydroxyl terminated oligo(ethylene glycol) alkanethiolate SAM was pre-formed on a gold substrate. (2) A polydimethylsiloxane (PDMS) microfluidic device containing an array of microchannels was placed on the pre-formed SAM. (3) An aqueous solution of NaBH4 was flowed through microchannels to locally remove regions of bioinert, pre-formed SAM. (4) Aqueous alkanethiolate solutions were flowed through the same microchannels to locally form new SAMs with varied ratios of carboxylic acid- and hydroxyl-terminated oligo(ethylene glycol) alkanethiolates. (5) A carbodiimide “activating” solution and a peptide solution were sequentially flowed through microchannels to covalently immobilize an amine-terminated cell adhesion peptide via carbodiimide condensation. (6) The PDMS device was removed, and stem cells were seeded on the resulting patterned SAM substrate in serum-containing medium. The use of PDMS materials for microchannel fabrication offered rapid microchannel device fabrication and easily adaptable pattern geometries (Fig. 2).24 Additionally, microchannels were designed to utilize “passive pumping” in which differences in surface tension across the microchannel were used to drive fluid transport.25, 26 This approach allowed for rapid liquid transport - and rapid local substrate modification - by simply maintaining a difference in inlet and outlet port droplet volumes. Microfluidic devices with arrays of microchannels also allowed for addressable control over local substrate modifications from channel-to-channel, including alkanethiolate composition and immobilized peptide type and density. Furthermore, aqueous SAM formation conditions in step (3) were chosen rather than traditional ethanolic solutions to prevent solvent leaking from microchannels and to minimize PDMS swelling during substrate processing.
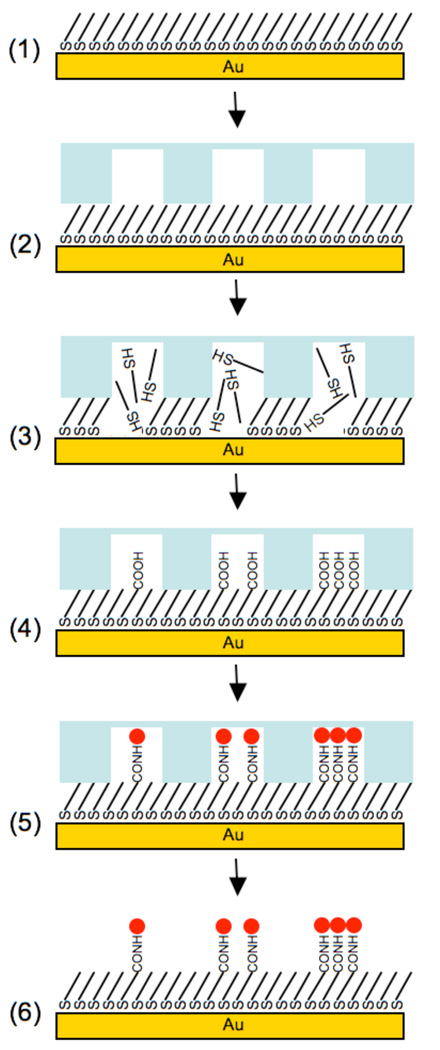
Figure 1.
Schematic representation of localized SAM replacement: (1) 100% EG3—OH terminated alkanethiolate SAM; (2) Apply microfluidic device (3) Flow aqueous 0.5 M NaBH4 through channels to locally remove SAM; (4) Locally form a new SAM with a mixture of aqueous EG3—OH and EG6—COOH alkanethiolates to create features with varied carboxylic acid densities; (5) Covalently conjugate primary amine-bearing peptide to COOH-terminated SAMs via carbodiimide chemistry; (6) Remove microfluidic device.
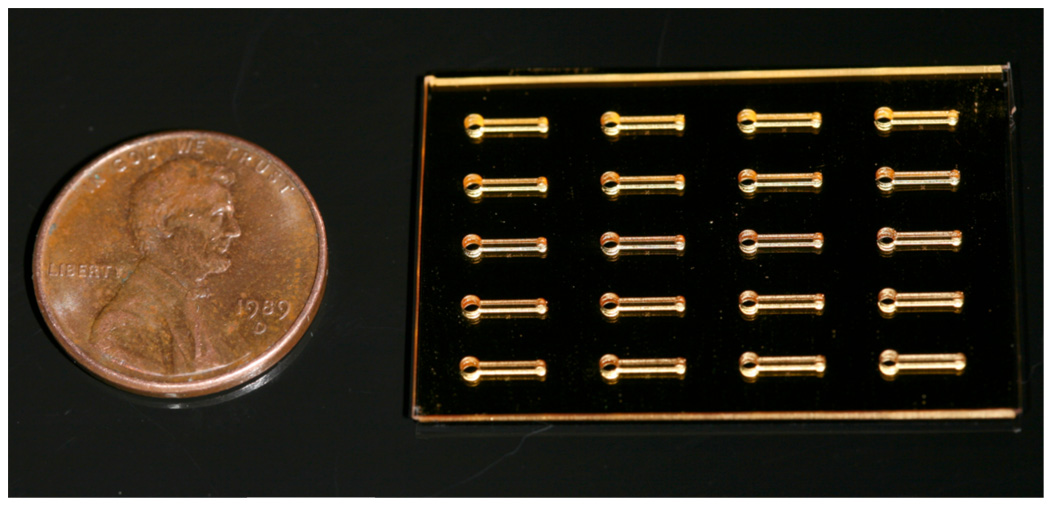
Figure 2.
Photograph of a PDMS microfluidic device with a 5 × 4 array of microchannels adhered to a 100 % HS—EG3 SAM on a gold-coated glass substrate.
Experimental Section
Materials and reagents
Carboxylic acid-capped hexa(ethylene glycol) undecanethiol (HS-C11-(O-CH2-CH2)6-O-CH2-COOH) (referred to herein as “HS—EG6—COOH”) was purchased from Prochimia (Sopot, Poland). 5(6)-carboxyfluorescein, Fmoc-protected amino acids and Rink amide MBHA peptide synthesis resin were purchased from NovaBiochem (San Diego, CA). Hydroxybenzotriazol (HOBt) was purchased from Advanced Chemtech (Louisville, KY). Diisopropylcarbodiimide (DIC) was purchased from Anaspec (San Jose, CA). Sodium borohydride (NaBH4), n-hydroxysuccinimide (NHS), n-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), sodium dodecyl sulfate (SDS), trifluoroacetic acid (TFA), diethyl ether, and deionized ultrafiltered water (DIUF) were purchased from Fisher Scientific (Fairlawn, NJ). Triisopropylsilane (TIPS), piperidine, dimethylformamide (DMF), dichloromethane (DCM), triethylamine (TEA), acetonitrile, and acetone were purchased from Sigma-Aldrich (St. Louis, MO). Absolute ethanol (EtOH) was purchased from AAPER Alcohol and Chemical Co. (Shelbyville, KY). All purchased items were of analytical grade and used as received. 11-tri(ethylene glycol)-undecane-1-thiol (HS-C11-(O-CH2-CH2)3-OH) (referred to herein as “HS—EG3”) was synthesized as described elsewhere.27 Thin films of 1000 Å Au <111>, 50 Å Cr on 1” × 3” × 0.040” glass slides were purchased from Evaporated Metal Films, Inc. (Ithaca, NY).
Peptide synthesis
Standard solid phase Fmoc-peptide synthesis (Fmoc SPPS) was performed using a 316c automated peptide synthesizer (C S Bio, Menlo Park, Ca). Rink amide MBHA resin was used as the solid phase, and HOBt and DIC were used for amino acid activation and coupling. After coupling the final amino acid, a 4-hour incubation in TFA, TIPS, and DIUF (95:2.5:2.5) released the peptide from resin and removed protecting groups. Released peptide was extracted from the TFA/TIPS/DIUF cocktail via precipitation in cold diethyl ether. Lyophilized peptides were analyzed using matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry with a Bruker Reflex II (Billerica, MA). The purity of synthesized peptides was evaluated via HPLC using a C18 analytical column (Shimadzu, Kyoto, Japan) with a gradient of 0–90% H2O + 0.1% % TFA/acetonitrile and a flow rate of 0.9 mL/minute (Supporting Information).
Peptide fluorescent labeling
Fluorescently labeled GGRGDSPK was synthesized using a lysine residue with methyltrityl (Mtt) epsilon primary amine protection and Fmoc alpha primary amine protection (N-a-Fmoc-N-e-4-methyltrityl-L-lysine, NovaBiochem, San Diego, Ca). In this orthogonal approach, the epsilon primary amine maintained Mtt protection while standard Fmoc SPPS was used to grow the peptide chain. Before removal of the final N-terminal Fmoc, the Mtt group was removed by incubating the resin and bound peptide chain in 5 mL of TFA, TIPS, and DCM (2:5:93) for ~30 seconds, five times. 5(6)carboxyfluorescein was then coupled to the deprotected lysine epsilon primary amine by incubating the peptide and resin in 5 mL DMF solution containing 3 X molar excess of 5(6)carboxyfluorescein, HOBt, DIC, and 20 X molar excess of TEA for 24 hours. The resin was then rinsed using copious amounts of DMF to remove any remaining 5(6)carboxyfluorescein and the final Fmoc was removed using 20% piperidine in DMF. The remaining peptide work-up was performed as previously described above.
Surface preparation
Gold slides were cut using a diamond scribe and placed into a 150 mm glass petri dish, covered with EtOH, and sonicated for ~3 mins using an ultrasonic bath (Bransonic 1510, Branson, Danbury, CT). Sonicated gold chips were then rinsed with EtOH, blown dry with nitrogen, and immersed in a 2 mM ethanolic solution of HS—EG3 for a minimum of 8 hours.
Fabrication of microfluidic devices
Microfluidic devices with arrays of microchannels were created using soft lithography and designed to facilitate passive pumping.25, 28 Briefly, master molds were fabricated from SU-8 (Microchem, Newton, MA) spin-coated silicon wafers using conventional photolithography techniques. Multiple layers of photoresist were used to create channels geometries of 4500 × 670 × 250 µm (l × w × h) and ports with radii of 603 µm and 1200 µm. Polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning, Midland, MI) was prepared by mixing a 10:1 ratio of base/curing agent (w/w) followed by degassing for ~30 mins. The degassed mixture was cast over the mold and cured for 4 hrs at 85 °C. Following curing, PDMS devices were removed from molds and cleaned in EtOH using an overnight Soxhlet extraction.29 After cleaning, PDMS devices were placed in vacuo to remove residual ethanol from the Soxhlet extraction process.
Polarization modulation-infrared reflection-adsorption spectroscopy
Polarization modulation-infrared reflection-adsorption spectroscopy (PM-IRRAS) was used to evaluate key steps in the patterning process. The formation and removal of inert hydroxyl-terminated oligo(ethylene glycol) alkanethiolate (HS—EG3) SAMs, as well as subsequent replacement with SAMs from aqueous solutions of mixed HS—EG3 and COOH-terminated oligo(ethylene glycol) alkanethiolates (HS—EG6—COOH) were characterized via PM-IRRAS. Additionally, PM-IRRAS was used to confirm peptide conjugation to carboxylate presenting SAMs. Due to constraints of the PM-IRRAS apparatus, bulk SAM processing, as opposed to localized SAM removal and replacement in microchannels, was used on gold chips with dimensions of 0.5” × 1.5” for infrared analysis. Bulk SAM processing was carefully performed to recapitulate all of the processing steps used during localized SAM removal and replacement in microchannels. IR spectra of alkanethiolate SAMs on gold were obtained using a Nicolet Manga-IR 860 FT-IR spectrometer with a photoelastic modulator (PEM-90, Hinds Instruments, Hillsboro, OR), synchronous sampling demodulator (SSD-100, GWC Technologies, Madison, WI), and a liquid-N2-cooled mercury telluride (MCT) detector. All spectra were obtained at an incident angle of 83° with modulation centered at 1500 cm−1 and 2500 cm−1 to produce spectrum with a range of 1000–3000 cm−1. For each sample, 1000 scans were taken using a resolution of 4 cm−1 per modulation center. Data were acquired as differential reflectance (%ΔR/R) versus wavenumber, and spectra were normalized and converted to absorbance units versus wavenumber via methods outlined by Frey et al.30
Patterning via localized SAM replacement
The method used to create patterned SAM features is shown schematically in Figure 1. First, gold chips measuring 1” × 1.5” were removed from a 2 mM ethanolic HS—EG3 solution, rinsed with EtOH, blown dry with nitrogen, and placed in a polystyrene petri dish. Approximately 15 µL of EtOH was applied to the center of the chip, followed by application of the PDMS microfluidic device. The SAM chip and device were then placed in vacuo for ~30 minutes to remove the EtOH and create a tight seal between the microfluidic device and SAM surface (Figure 2).
Localized SAM removal was achieved by filling all channels with 0.5 M aqueous NaBH4 for ~ 1 min.31 Channels were then aspirated, and rinsed with DIUF H2O. In all rinsing steps, microchannel arrays were filled with fluid, and then flushed three times with the same volume to ensure that reagents from previous processing steps were completely removed. Mixed 100 µM aqueous solutions of HS—EG6—COOH and HS—EG3 alkanethiolates at varied mol percentages were then added to channels to locally regenerate mixed SAMs presenting varied densities of carboxylic acid functional groups. Aqueous alkanethiolate solutions were incubated in channels for 15 minutes, then channels were again rinsed with DIUF H2O. Channels were then exposed to an aqueous 250 mM EDC, 100 mM NHS solution for 10 min. to yield NHS-activated surfaces in each channel. After an additional DIUF H2O rinse, solutions of 1 mM peptide in 1 X PBS (pH 7.4) were added to channels and incubated for 45 minutes. Channels were then rinsed with DIUF H2O, followed by immersion of the gold chip and microfluidic device in 0.1% w/v SDS in water. The device was then carefully removed from the SAM substrate while still immersed in the SDS solution. The patterned substrate was then transferred to a 50 mL conical tube (BD Falcon, San Jose, CA) containing ~30 mL of SDS solution and sonicated for 3 minutes. Following sonication, the patterned substrates were finished with 5-second rinses of additional SDS solution, H2O, EtOH, and then dried with nitrogen. Patterned substrates for cell adhesion studies were transferred to 60 mm petri dishes containing sterile 1 X PBS while substrates for fluorescent surface scanning were imaged immediately.
Fluorescence surface scanning
A GE Healthcare Typhoon Trio Variable Mode Imager was used to scan patterned SAM surfaces containing fluorescently-labeled peptide. In addition to conjugating fluorescently-labeled GGRGDSPK to a range of HS—EG6—COOH mol percent conditions, a control “non-replace” condition was included in which a channel was exposed to DIFU H2O and H2O pH 2 instead of NaBH4 and alkanethiol solutions, respectively. The highest scan quality was achieved by bridging a patterned gold chip across two glass slides resting on the scanning surface and filling the region between the chip and scanner surface with 1 X PBS (pH 7.4). The patterned surface was then scanned with 25 µm resolution using the blue laser (488 nm) in combination with the 526 nm emission filter and a PMT intensity of 600. Scanned images were colored and analyzed using ImageJ (ImageJ, Freeware, NIH, Bethesda, MD).
hMSC attachment to patterned surfaces
Human mesenchymal stem cells (Lonza, Walkersville, MD) were expanded at low density on tissue culture polystyrene plates to maintain pluripotency as described by Sotiropoulou et al.32 At passage 7, cells were removed from the plate using a 0.05% trypsin solution, resuspended at a concentration of 26,000 cells/mL. Sterile 1 X PBS was aspirated from 60 mm petri dishing containing the patterned SAMs substrates fabricated as previously described, and 5 mL of cell suspension was added to the dish. Cells were seeded in minimum essential medium, alpha (Mediatech, Manassas, VA) containing 10% MSC qualified fetal bovine serum (Invitrogen, Carlsbad, Ca) and 1% penicillin/streptomycin (Hyclone, Logan, UT). Patterned substrates were then incubated in a humid environment at 37 °C and 5% CO2 until cell adhesion was examined. During extended cell culture on patterned substrates medium was replenished every 48 hours. To visualize cell attachment, patterned surfaces were carefully rinsed with sterile 1 X PBS, and incubated in a 4 µM solution of Calcein AM (Invitrogen, Carlsbad, Ca) for ~10 minutes. Living cells could be visualized via green fluorescence and were imaged using an Olympus IX51 inverted epifluorescent microscope equipped with a FITC filter cube set. To achieve this, stained substrates were removed from the staining solutions and inverted onto a glass microscope slide with ~500 µL of PBS to maintain sample hydration during imaging.
Immunocytochemistry
hMSCs were seeded onto a patterned SAM as previously described, and allowed to attach overnight. After a gentle rinse with 1 X PBS, hMSC immunocytochemical staining for the actin cytoskeleton and focal adhesion markers was performed as directed by the manufacturer (Catalogue No. FAK100, Millipore, Billerica, MA). Briefly, cells were fixed using 4% formaldehyde in 1 X PBS for 15 minutes. Following fixing, the patterned substrate was then washed gently twice with wash buffer containing 0.05% Tween 20 (Fisher Scientific, Fairlawn, NJ) in 1 X PBS. Cells were then permeabilized using 0.1 % Triton X-100 (MP Biomedicals, Aurora, OH) in 1 X PBS for 5 minutes. The substrate was again rinsed twice with wash buffer and then blocked to prevent non-specific antibody binding using 1% (w/v) bovine serum albumin (Fisher Scientific, Fairlawn, NJ) in 1 X PBS for 30 minutes. After blocking, the patterned substrate was exposed to a 1 X PBS solution containing an anti-vinculin primary antibody and incubated at room temperature for 1 hour. Following another rinse, three times with wash buffer, the patterned substrate was exposed to a 1 X PBS solution containing a FITC-conjugated secondary antibody and TRITC-conjugated Phalloidin for 60 minutes at room temperature. The substrate was again rinsed with wash buffer, and then incubated in a 1 X PBS solution of DAPI nuclear counterstain for 5 minutes. The substrate was then rinsed an additional time and imaged using Olympus IX51 inverted epifluorescent microscope equipped with FITC, TRITC, and DAPI filter cube sets. Exposure times and gain settings for each filter set were held constant during imaging across all conditions. Analysis of projected cell area and focal adhesion density was achieved using ImageJ (ImageJ, Freeware, NIH, Bethesda, MD) imaging software. Briefly, images from all conditions were collected into a single stack and, in this manner, any image processing procedures were consistently applied to the entire image stack. Projected cell area quantitation was achieved by thresholding and measuring the red-labled actin component of the stacked images while focal adhesion quantitation was achieved by thresholding and measuring the green-labeled vincullin component of the stacked images (Supporting Information). In both cases, the “analyze particles” function of ImageJ was used to measure cell area or count focal adhesions in thresholded red and green channels, respectively.
Results and discussion
SAM removal, replacement, and peptide coupling
To simplify the description of this work, SAMs that were formed from solutions containing a specific mol percentage of HS—EG6—COOH mixed with HS—EG3 will simply be referred to as “X% HS—EG6—COOH SAMs”. The authors emphasize that this terminology refers strictly to mol percentage of alkanethiol species in formation solutions, and does not necessarily indicate the mol percentage of each component in the formed SAM.33 Furthermore, SAMs formed in ethanol or aqueous solution will be referred to as “ethanolic SAMs” or “aqueous SAMs”, respectively.
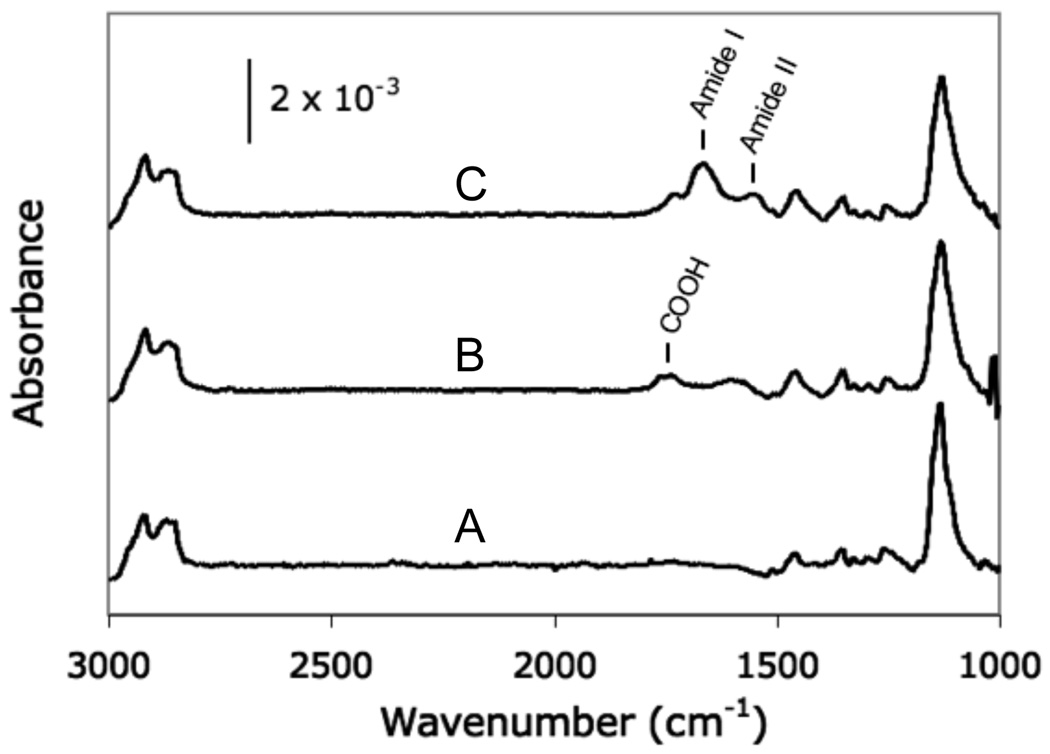
A PM-IRRAS spectrum of an aqueous 100% HS—EG3 SAM (Figure 3A) was similar to a spectrum of a 100% HS—EG3 SAM formed from an overnight incubation in an ethanolic solution (Figure 4A). Spectra exhibited the characteristics of well-packed SAMs, including the symmetric and asymmetric alkyl (CH) stretching bands of C11 methylene units at 2850 cm−1 and 2920 cm−1, and the OEG (C-O-C) stretching band centered around 1130 cm−1.34 Additionally, aqueous 25% HS—EG6—COOH SAMs spectra (Figure 3B) exhibited the carboxylic acid stretch band centered around 1740 cm−1, which verified the incorporation of HS—EG6—COOH. Furthermore, the emergence of the amide I (C=O) and amide II (NH) stretch bands centered around 1675 cm−1 and 1550 cm−1 following peptide conjugation (Figure 3C) verified covalent peptide incorporation.35, 36, 37, 38
Figure 3.
PM-IRRAS analysis of SAMs formed from aqueous solutions of: (A) 100 mol % HS—EG3; (B) 25 mol % HS—EG6—COOH; and (C) 25 mol % HS—EG6—COOH following conjugation of RGDSP via carbodiimide condensation
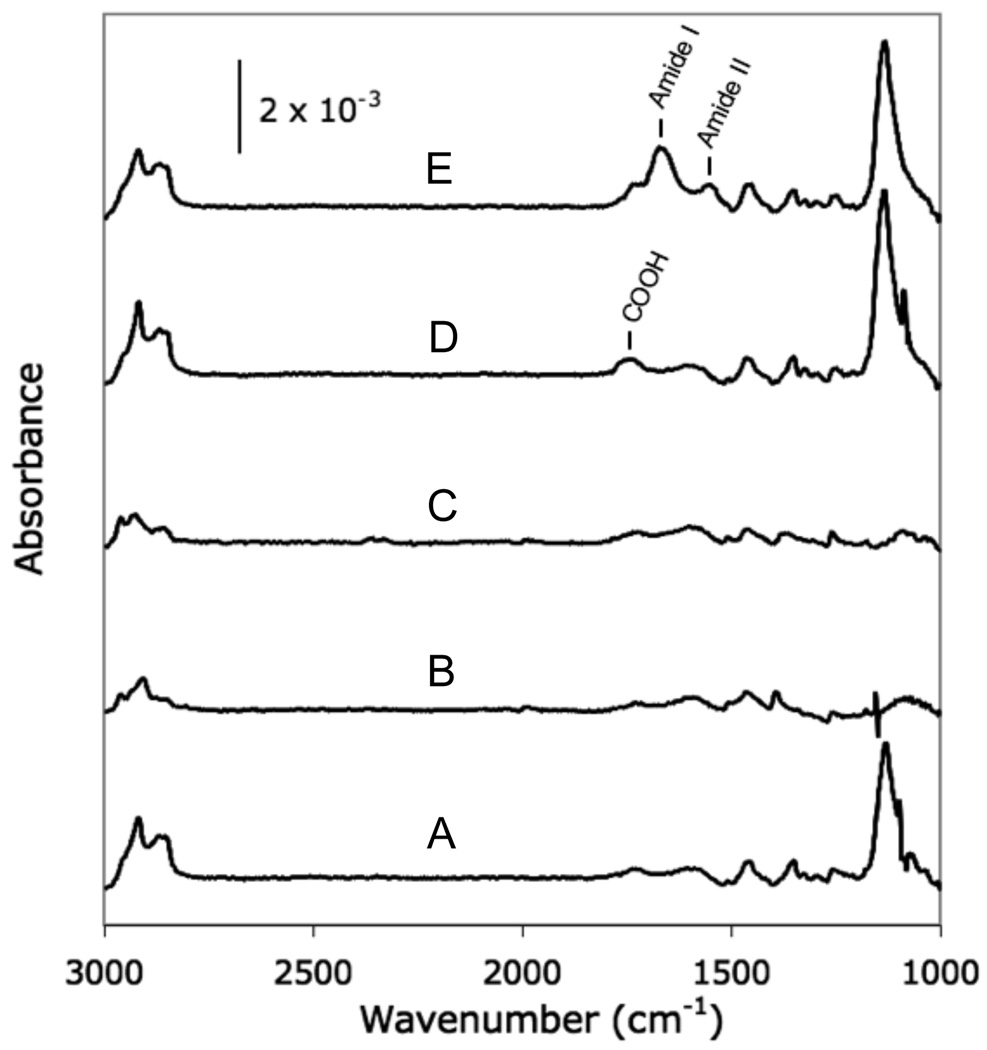
Figure 4.
PM-IRRAS analysis of each step of the localized SAM replacement process. (A) 100% HS—EG3—OH terminated SAM, (B) following removal of SAM via aqueous NaBH4, (C) bare gold for comparison, (D) following formation of a new SAM using an aqueous 25 mol % HS—EG6—COOH alkanethiolate solution, and (E) following conjugation of RGDSP via carbodiimide condensation.
Alkanethiolate SAMs could be rapidly and efficiently removed from a gold substrate via treatment with NaBH4. Exposure of a 100% HS—EG3 SAM to 0.5 M NaBH4 for 1 minute resulted in an IR spectrum (Figure 4B) lacking the characteristic stretching bands of well-packed SAMs, and was similar to a spectrum of bare gold (Figure 4C), indicating complete alkanethiolate removal. These results are generally consistent with the NaBH4-facilitated SAM removal process described by Yuan and co-workers.31, in which complete removal of a 1-dodecanethiolate SAM was achieved after a 10 minute exposure to 0.5 M NaBH4 in a 1:1 EtOH:H2O solution. The shorter timescale required for 100% HS—EG3 SAM removal observed here when compared to the work by Yuan and co-workers is most likely due to differences in 1-dodecanethiolate and OEG alkanethiolate solubility, as well as the use of fully aqueous NaBH4 removal conditions in our study. In general, aqueous NaBH4 solutions offered a rapid and convenient method for removing HS—EG3 SAMs to leave behind a bare gold substrate ready for SAM re-formation.
Alkanethiolate SAMs could be re-formed on the same gold surface after removal of an initial SAM layer. Following SAM removal via NaBH4, aqueous re-formation conditions were used to create 25% HS—EG6—COOH SAMs (Figure 4D), which allowed for subsequent peptide conjugation (Figure 4E, Supporting Information). Importantly, re-formed aqueous SAMs spectra (Figures 4D, 4E) were similar to aqueous SAMs formed on freshly cleaned gold substrates (Figures 3B, 3C), indicating that incorporation of HS—EG6—COOH and subsequent peptide conjugation were unaffected by the NaBH4-mediated SAM removal process. These results are consistent with findings by Yuan and co-workers, in which 1-dodecanethiolate SAMs were regenerated following SAM removal by immersing gold substrates in ethanolic alkanethiolate solutions for 24 hours.31 However, our current work further demonstrates that SAMs can be re-formed in aqueous conditions to create monolayers with mixed HS—EG3 and HS—EG6—COOH compositions. Kiessling and co-workers have previously removed inert perfluorinated alkanethiolate SAMs via exposure to UV-irradiation and replaced with mixed SAMs of glucamine-terminated and peptide-terminated alkanethiolates.22 Our results offer an alternative strategy, in which SAMs can be rapidly removed in aqueous solution to facilitate subsequent SAM re-formation with different, commercially available alkanethiolate compositions. Taken together, the relatively short timescales required for both SAM removal and re-formation facilitate rapid processing, which allows for creation of a substrate patterning approach via localized SAM replacement.
Analysis of aqueous SAM bio-inertness
Aqueous 100% HS—EG3 SAMs formed over various timeframes were each bioinert, as they prevented cell attachment in serum-containing cell culture medium. Aqueous 100% HS—EG3 SAMs were allowed to form for various durations, seeded with hMSCs in media containing 10 % fetal bovine serum, and incubated for 12 hours. Each SAM formed from 1 minute incubation or longer was capable of completely preventing cell attachment (blank images devoid of cells not shown). Similarly, a previous study by Li et al.39 also demonstrated that OEG alkanethiolate SAMs formed on gold from aqueous solutions resisted protein adsorption, as detected by surface plasmon resonance (SPR). However, this previous study did not examine SAM formation times less than 24 hours. Importantly, our results demonstrate that bioinert SAMs can be formed in aqueous solution over as little as 1 minute. Interestingly, our result is consistent with previous results from ethanolic SAM formation, as Harder and co-workers34 have demonstrated that protein-resistant SAMs could be formed via 15-second immersions in ethanolic alkanethiolate solutions. The ability to rapidly form bioinert SAM layers is important for a variety of SAM patterning approaches, including the localized SAM replacement approach described in this manuscript.
Spatial control over peptide immobilization
The local density of fluorescently-labeled peptide could be controlled using localized SAM replacement. 5(6)carboxyfluorescein-labeled RGDSPK was coupled to a patterned SAM substrate with locally re-formed aqueous SAMs from various mol percent HS—EG6—COOH solutions (Figure 5A). The clear interface between each patterned region and the surrounding background suggested that labeled peptide was strictly confined to regions locally replaced with HS—EG6—COOH containing SAM. The small amount of fluorescence observed in both the non-replace (NR) condition, as well as the 0% HS—EG6—COOH condition was not unexpected (Figure 5B, 5C), as previous work by Clare and coworkers demonstrated low levels of non-specific fluorescein-labeled protein adsorption on HS—EG3 SAMs.40 Interestingly, the NR condition exhibited slightly less fluorescence than 0% HS—EG6—COOH condition suggesting that aqueous HS—EG3 SAMS may be more susceptible to non-specific adsorption of fluorescein-labeled peptides. This may be due to differences in aqueous and ethanolic HS-EG3 SAMs as described by Li and co-workers.39 Regardless, as indicated by cell attachment results described above, aqueous 100% HS—EG3 SAMs were still capable of preventing cell attachment in serum-containing cell seeding conditions.
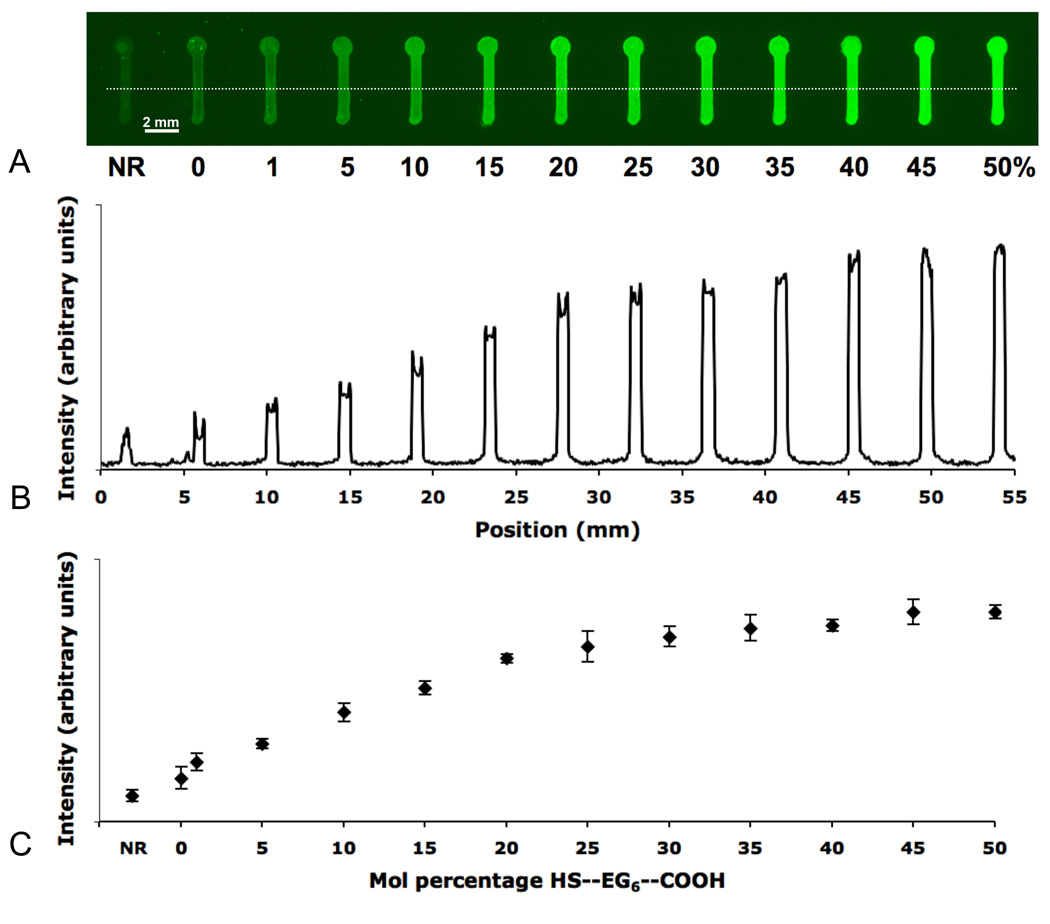
Figure 5.
Conjugation of GGRGDSPK-fluorescein to a patterned SAM containing discrete features with varied mol % HS—EG6—COOH alkanethiolate and non-replacement (NR) conditions. (A) Fluorescence surface scan of the pattern, (B) linear fluorescence intensity profile, and (C) average fluorescent intensity profile. (Average obtained from two different patterned samples, three intensity profiles each; Error bars represent standard deviation).
As the mol percentage of HS—EG6—COOH present during SAM re-formation increased, there was a concomitant increase in fluorescence intensity associated with immobilized, fluorescently-labeled peptide (Figures 5B, 5C). Additionally, this trend appeared to be linear from 0 % to ~15% HS—EG6—COOH, followed by a less prominent increase in fluorescence intensity up to 50 %. This corresponds well with theoretical calculations that would predict surface peptide saturation at ~ 13% HS—EG6—COOH, assuming an alkanethiolate spacing of 4.97 Å41, and estimating each labeled peptide molecule to be hard sphere with an approximate volume of 1350 Å3. Although the explicit surface densities of HS—EG6—COOH that resulted from changes in HS—EG6—COOH mol percentage during SAM re-formation were not measured in this work, changes in HS—EG6—COOH mol fraction during re-formation correlated to similar changes in immobilized fluorescent peptide density. Thus, localized SAM replacement not only afforded spatial control over peptide conjugation, but also afforded control over immobilized peptide densities over a broad range.
Spatial localization of human mesenchymal stem cell attachment
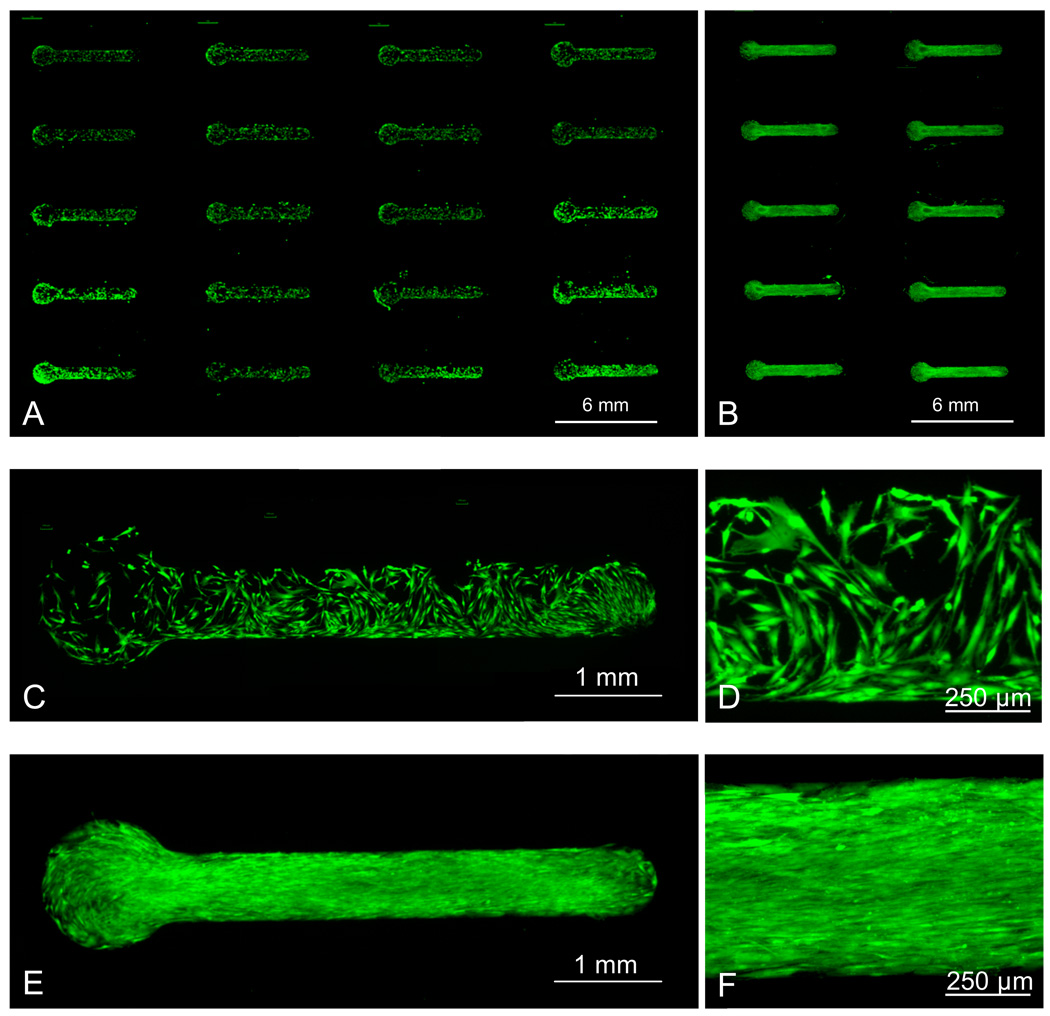
hMSC attachment could be confined to regions presenting cell adhesion peptides, which were patterned via localized SAM replacement. First, localized SAM replacement was used to pattern an array of features presenting a single density of cell adhesion ligand. hMSCs were then seeded and incubated in serum-containing medium for 12 hours on a 5 × 4 array of RGDSP conjugated to 5% HS— EG6—COOH SAM features (Figure 6A). hMSC attachment was confined within patterned features (Figures 6C, 6D) and exhibited cell morphology similar to hMSCs on tissue culture polystyrene surfaces.42 Importantly, even after 14 days of extended culture in serum-supplemented medium, hMSCs proliferated and remained confined to patterned regions (Figures 6B, 6E, 6F).
Figure 6.
Attachment of human mesenchymal stem cells to patterns presenting RGDSP conjugated to 5% HS—EG6—COOH. Cells were stained with Calcein AM (A) 12 hours after seeding, or (B) 14 days after seeding, with media replacement every 2 days. (C, D) High magnification images of 12 hour time point. (E, F) High magnification images of 14 day time point.
The localized replacement approach maintained a bio-inert background between array features. This conclusion is supported by the qualitative observation that no significant feature “broadening” was detected in any fluorescent peptide immobilization (Figure 5) or cell adhesion experiments (Figure 6). Therefore, the use of PDMS materials for microchannel fabrication not only offered rapid microchannel device fabrication and easily adaptable pattern geometries, but also served to maintain a bio-inert background in several ways. First, we observed that contact between PDMS materials and a 100% HS—EG3 SAM during localized SAM replacement maintained the HS—EG3 background inertness even after extended cell culture. We hypothesize that by limiting SAM exposure to ambient air, PDMS served to retard thiol oxidation and maintain the thiol-gold coordination bonds of the inert pre-formed HS—EG3 SAM during substrate processing. This hypothesis is supported by a study from Willey and co-workers, in which only minor SAM thiol oxidation was detected by Near-edge X-ray absorption fine structure spectroscopy (NEXAFS) when SAM exposure to ambient air was limited.21 Secondly, PDMS served as a barrier between the background SAM and the aqueous chemical environments used during pattern processing. This barrier function likely prevented alkanethiolate solution exchange,17 and possibly lateral alkanethiol diffusion,15 which could have otherwise rapidly contaminated the 100% HS—EG3 background with HS—EG6—COOH. Furthermore, if HS—EG6—COOH diffusion into the surrounding PDMS during the localized replacement process did result in HS—EG6—COOH contaminants in the background SAM via alkanethiolate exchange, the hydrophilicity of the cell adhesion peptides used in this work would restrict their diffusion in the hydrophobic PDMS network during the conjugation reaction, and would preclude peptide conjugation in the background.24 Taken together, these results demonstrate that the use of PDMS microdevices, in combination with aqueous SAM chemistries, serve to protect the underlying bio-inert background from oxidation and solution exchange while facilitating localized SAM replacement. In this manner, these processing parameters allow for the creation of SAM substrates with spatially distinct regions for cell attachment.
Modulating human mesenchymal stem cell adhesion
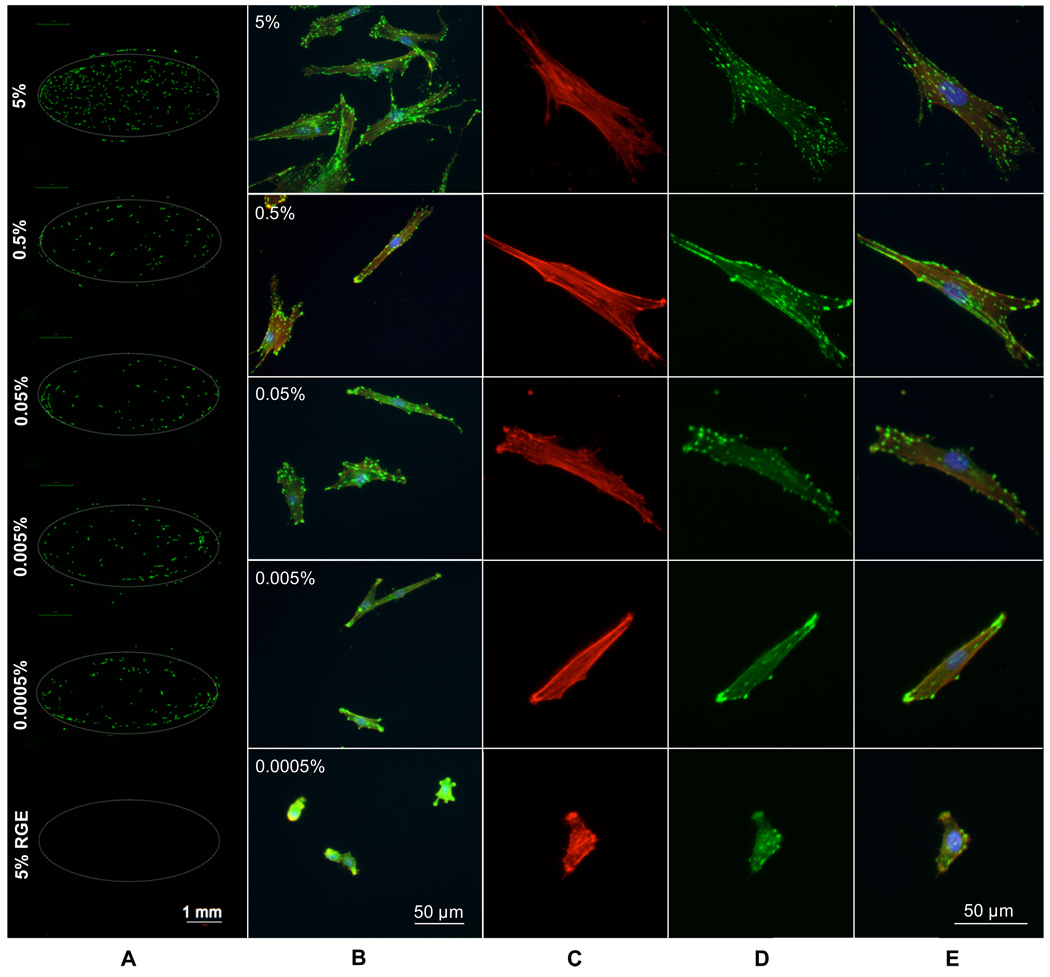
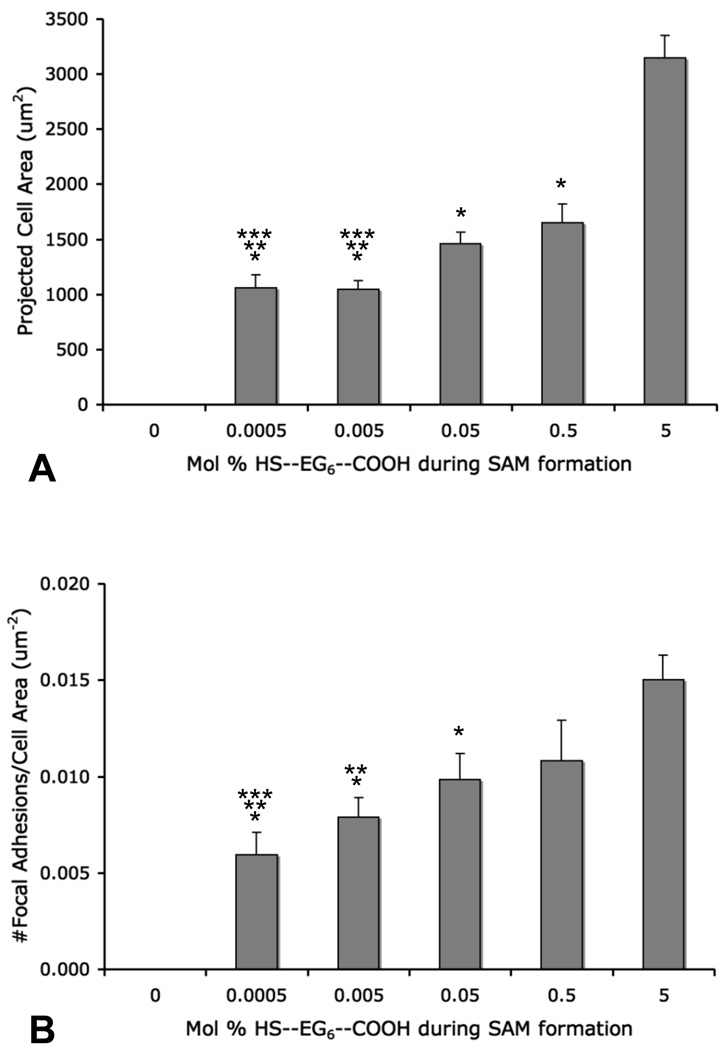
Modulation of hMSC adhesion was achieved by patterning a range of cell adhesion peptide densities on a single substrate via localized SAM replacement. hMSCs were cultured on a patterned array in which the mol percentage of HS—EG6—COOH during aqueous SAM re-formation was varied between 0.0005% and 5%. On a single chip, each density condition was run in quadruplicate with the cell adhesion peptide RGDSP conjugated to three features of a single density and the mutant peptide RGESP conjugated to the remaining feature. Cells were seeded on the entire substrate, and therefore hMSCs were exposed to a wide range of cell adhesion peptide densities as well as multiple amino acid sequences on a single substrate. Oval geometries were created for these cell adhesion studies to demonstrate the geometric adaptability of the localized replacement approach, and to increase the number of adherent cells per condition and facilitate quantification of cell adhesion. hMSCs attached to all densities of RGDSP, but did not attach to any RGESP-presenting array features (Fig. 7A). Quantitation of cell number with respect to peptide density yielded no significant trend. To examine focal adhesion complex formation, immunocytochemistry was used to stain for vinculin, a component of focal adhesion complexes, and actin filaments via labeled phalloidin binding. In these images, focal adhesion complexes, or “focal adhesions”, appear as intense punctate green staining of vinculin. High magnification fluorescence photomicrographs of stained hMSCs exhibited changes in hMSC spreading and focal adhesion density with changes in local RGDSP density (Fig. 7B–E). hMSCs adhering to RGDSP conjugated to surfaces formed from 5 mol percent HS—EG6—COOH exhibited finely distributed, punctate focal adhesion staining throughout the cell (top panel, Fig 7B, 7D). At intermediate mol percentages, such as the 0.05% condition (middle panel, Fig. 7B, 7D), hMSCs exhibited a more coarse distribution of focal adhesion complexes throughout the cell. At lower mol percentages, such as 0.005% (bottom middle panel, Fig. 7B, 7D), focal adhesion complexes were generally localized to the periphery of the cell, and at the lowest mol percentage of HS—EG6—COOH and presumably lowest density of RGDSP, 0.0005% (bottom panel, Fig. 7B, 7D), cells exhibited very few focal adhesions, and in many cases, delocalized vinculin staining. To present these results in a quantitative manner, projected cell area (Figure 8A) as well as focal adhesion density (Figure 8B) were measured. Cells attaching to RGDSP surfaces with decreasing mol % of HS—EG6—COOH showed a decreasing trend in projected cell area (at a significance of p < 0.05, 0.5 and 0.05% were lower than 5% conditions, while 0.005 and 0.0005% were lower than 5-0.05% conditions) as well as a decreasing trend in focal adhesion complex density (at a significance of p < 0.05, 0.05% was lower than 5% conditions, 0.005% was lower than 5 and 0.5% conditions, and 0.0005% was lower than 5- 0.05% conditions).
Figure 7.
Regulating hMSC adhesion to patterned RGD-presenting SAMs. (A) Calcein AM staining and (B) focal adhesion (FA) staining of hMSCs attached to patterned features containing decreasing amounts of the cell adhesion peptide RGDSP (FA stain: FITC-anti-vinculin, TRITC-phalloidin, DAPI). Higher magnification images of individual hMSCs (C) actin staining, (D) vinculin staining and (E) overlay including DAPI
Figure 8.
Quantification of (A) projected cell area and (B) normalized quantification of FA density. (Error bars represent standard error of the mean, (*) denotes significant difference compared to 5% (p < 0.05), (**) denotes significant difference compared to 0.5% (p < 0.05), and (***) denotes significant difference compared to 0.05% (p < 0.05).)
A patterned SAM substrate presenting surfaces formed from multiple HS—EG6—COOH mol percentages (0–5%) and multiple peptides (RGDSP and RGESP) supported a range of adherent hMSC phenotypes. The lack of significant differences in the number of attached cells with varying peptide density is similar to results from Roberts and co-workers, in which bovine capillary endothelial cell densities were similar on SAMs presenting various densities of RGD (0.001–100 mol %).2 Our observation that hMSC focal adhesion complexes localized to the cell perimeter when attaching to regions formed from lower HS—EG6—COOH mol percentages, and presumably lower RGDSP densities, is also consistent with results by Kato and Mrksich, in which focal adhesion complex distribution in swiss 3T3 fibroblasts cultured on peptide-terminated OEG alkanethiolate SAMs was also localized to the cell periphery at decreased mol percentages of linear or cyclic RGD.6 Furthermore, the correlation between focal adhesion density and HS—EG6—COOH solution mol percentage during SAM re-formation observed here is consistent with our previous work, in which hMSCs were cultured on non-patterned SAMs substrates presenting RGDSP immobilized via a “click” copper(I)-catalyzed azide-alkyne cycloaddition.43 In a slightly different method to control cell adhesion peptide density, Hoover and co-workers used dip pen nanolithography to create SAM nanoarrays of linear RGD spots and found that attached 3T3 swiss mouse fibroblasts exhibited increased focal adhesion density on arrays presenting higher densities of RGD spots.44
Patterned substrates created via localized SAM replacement may ultimately be used as a tool to investigate the relationship between hMSC adhesion and downstream cellular behavior. In particular, it is possible that the changes in focal adhesion formation, actin organization, and projected cell area observed in this work may lead to significant effects on other hMSC behaviors, such as differentiation down specific cell lineages. Engler et al. recently observed that neurogenic, myogenic, and osteogenic differentiation of hMSCs could be significantly influenced by substrate stiffness, but also noted that increased substrate stiffness correlated with increased focal adhesion expression, actin organization, and projected cell area.45 In addition, Rowlands et al. utilized combinations of ECM proteins and substrate stiffness to control hMSC shape, and demonstrated effects on osteogenic and myogenic differentiation.46 In a more explicit approach to modulate cell area, Mcbeath and co-workers used microcontact-printed islands of fibronectin to control hMSC projected cell area and found that hMSCs confined to 1024 µm2 islands of fibronectin underwent enhanced adipogenesis while hMSCs attaching to 10,000 µm2 islands underwent enhanced osteogenesis.47 Further insight into the relationship between focal adhesion complex formation and osteogenesis was recently provided by Kundu et al., who demonstrated that hMSC attachment to vitronectin-coated and collagen I-coated surfaces promoted osteogenesis, and noted that hMSC attachment to vitronectin exhibited increased focal adhesion complex formation.9 Collectively, these studies highlight the importance of cell adhesion in the process of hMSC differentiation, as well as some of the underlying mechanistic details that drive such cellular processes. However, in standard cell culture it is difficult to directly correlate the presence of specific cell adhesion proteins to changes in cell phenotype, due to dynamic reorganization of the cell adhesion environment, and the wide array of epitopes that can influence stem cell adhesion. The use of well-defined SAM substrates to explore cell adhesion, combined with the rapid fabrication patterning approach described in this work, may ultimately enable the development of high-throughput experimentation capable of explicitly characterizing the roles of cell adhesion in hMSC behavior and aid in the development of mechanistic understandings of hMSC differentiation.
Conclusion
Localized SAM replacement can be used to efficiently create arrays of peptides, with control over peptide identity and density on an otherwise bio-inert SAM background. The commercial availability of the base materials, as well as the simplicity of the microfluidic approach may permit widespread use of this system to screen for the effects of cell-material interactions. Results from this study indicate that hMSCs adhere to patterned features presenting a broad range of cell adhesion peptide densities, and that peptide density strongly influences hMSC spreading and focal adhesion complex formation. The control and potential for high-throughput imparted by the patterned substrates developed in this work could ultimately be used to further dissect the role of cell-ECM interactions on regulation of human mesenchymal stem cell behavior. Therefore, this approach may ultimately be used as a platform to screen for the effects of a broad range of signals on stem cell phenotype.
Supplementary Material
Acknowledgements
The authors would like to acknowledge funding from the National Institutes of Health (5R21EB005374-02 and the Biotechnology Training Program NIGMS 5 T32 GM08349). PM-IRRAS data were obtained at the NSF-funded University of Wisconsin Materials Research Science and Engineering Center. The authors thank Tamás Gaál for assistance with fluorescence surface scanning, William King for assistance with mass spectrometry and HPLC analysis, and Keil Regehr, Jay Warrick, and David Beebe at the University of Wisconsin for assistance with microfabrication.
Footnotes
Supporting Information Available: MALDI-TOF and HPLC characterization of synthesized peptides; additional views of PM-IRRAS spectra; images outlining the image processing method used to quantify human mesenchymal stem cell area and focal adhesion density. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Vroman L, Adams AL. Findings with Recording Ellipsometer Suggesting Rapid Exchange of Specific Plasma Proteins at Liquid/Solid Interfaces. Surface Science. 1969;16:438–446. [Google Scholar]
- 2.Roberts C, Chen CS, Mrksich M, Martichonok V, Ingber DE, Whitesides GM. Using mixed self-assembled monolayers presenting RGD and (EG)(3)OH groups to characterize long-term attachment of bovine capillary endothelial cells to surfaces. Journal of the American Chemical Society. 1998;120(26):6548–6555. [Google Scholar]
- 3.Houseman BT, Mrksich M. The microenvironment of immobilized Arg-Gly-Asp peptides is an important determinant of cell adhesion. Biomaterials. 2001;22(9):943–955. doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]
- 4.Kato M, Mrksich M. Rewiring cell adhesion. Journal of the American Chemical Society. 2004;126(21):6504–6505. doi: 10.1021/ja039058e. [DOI] [PubMed] [Google Scholar]
- 5.Mrksich M. Using self-assembled monolayers to model the extracellular matrix. Acta Biomaterialia. 2009;5(3):832–841. doi: 10.1016/j.actbio.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato M, Mrksich M. Using model substrates to study the dependence of focal adhesion formation on the affinity of integrin-ligand complexes. Biochemistry. 2004;43(10):2699–2707. doi: 10.1021/bi0352670. [DOI] [PubMed] [Google Scholar]
- 7.Watt FM, Hogan BLM. Out of Eden: Stem cells and their niches. Science. 2000;287(5457):1427–1430. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 8.Salasznyk RM, Klees RF, Williams WA, Boskey A, Plopper GE. Focal adhesion kinase signaling pathways regulate the osteogenic differentiation of human mesenchymal stem cells. Experimental Cell Research. 2007;313(1):22–37. doi: 10.1016/j.yexcr.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kundu AK, Putnam AJ. Vitronectin and collagen I differentially regulate osteogenesis in mesenchymal stem cells. Biochem Biophys Res Commun. 2006;347(1):347–357. doi: 10.1016/j.bbrc.2006.06.110. [DOI] [PubMed] [Google Scholar]
- 10.Klees RF, Salasznyk RM, Vandenberg S, Bennett K, Plopper GE. Laminin-5 activates extracellular matrix production and osteogenic gene focusing in human mesenchymal stem cells. Matrix Biology. 2007;26(2):106–114. doi: 10.1016/j.matbio.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klees RF, Salasznyk RM, Ward DF, Crone DE, Williams WA, Harris MP, Boskey A, Quaranta V, Plopper GE. Dissection of the osteogenic effects of laminin-332 utilizing specific LG domains: LG3 induces osteogenic differentiation, but not mineralization. Experimental Cell Research. 2008;314(4):763–773. doi: 10.1016/j.yexcr.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen XD, Dusevich V, Feng JQ, Manolagas SC, Jilka RL. Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. Journal of Bone and Mineral Research. 2007;22(12):1943–1956. doi: 10.1359/jbmr.070725. [DOI] [PubMed] [Google Scholar]
- 13.Matsubara T, Tsutsumi S, Pan H, Hiraoka H, Oda R, Nishimura M, Kawaguchi H, Nakamura K, Kato Y. A new technique to expand human mesenchymal stem cells using basement membrane extracellular matrix. Biochemical and Biophysical Research Communications. 2004;313(3):503–508. doi: 10.1016/j.bbrc.2003.11.143. [DOI] [PubMed] [Google Scholar]
- 14.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 15.Baralia GG, Pallandre A, Nysten B, Jonas AM. Nanopatterned self-assembled monolayers. Nanotechnology. 2006;17(4):1160–1165. doi: 10.1088/0957-4484/17/4/053. [DOI] [PubMed] [Google Scholar]
- 16.Gannon G, Larsson JA, Greer JC, Thompson D. Quantification of ink diffusion in microcontact printing with self-assembled monolayers. Langmuir. 2009;25(1):242–247. doi: 10.1021/la802548u. [DOI] [PubMed] [Google Scholar]
- 17.Baralia GG, Duwez AS, Nysten B, Jonas AM. Kinetics of exchange of alkanethiol monolayers self-assembled on polycrystalline gold. Langmuir. 2005;21(15):6825–6829. doi: 10.1021/la050245v. [DOI] [PubMed] [Google Scholar]
- 18.Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz SA, Chen CS. Microcontact printing: A tool to pattern. Soft Matter. 2007;3(2):168–177. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]
- 20.Schoenfisch MH, Pemberton JE. Air stability of alkanethiol self-assembled monolayers on silver and gold surfaces. Journal of the American Chemical Society. 1998;120(18):4502–4513. [Google Scholar]
- 21.Willey TM, Vance AL, van Buuren T, Bostedt C, Terminello LJ, Fadley CS. Rapid degradation of alkanethiol-based self-assembled monolayers on gold in ambient laboratory conditions. Surface Science. 2005;576(1–3):188–196. [Google Scholar]
- 22.Derda R, Li LY, Orner BP, Lewis RL, Thomson JA, Kiessling LL. Defined substrates for human embryonic stem cell growth identified from surface arrays. Acs Chemical Biology. 2007;2(5):347–355. doi: 10.1021/cb700032u. [DOI] [PubMed] [Google Scholar]
- 23.Orner BP, Derda R, Lewis RL, Thomson JA, Kiessling LL. Arrays for the combinatorial exploration of cell adhesion. J Am Chem Soc. 2004;126(35):10808–10809. doi: 10.1021/ja0474291. [DOI] [PubMed] [Google Scholar]
- 24.McDonald JC, Whitesides GM. Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc Chem Res. 2002;35(7):491–499. doi: 10.1021/ar010110q. [DOI] [PubMed] [Google Scholar]
- 25.Walker GM, Beebe DJ. A passive pumping method for microfluidic devices. Lab on a Chip. 2002;2(3):131–134. doi: 10.1039/b204381e. [DOI] [PubMed] [Google Scholar]
- 26.Berthier E, Beebe DJ. Flow rate analysis of a surface tension driven passive micropump. Lab on a Chip. 2007;7(11):1475–1478. doi: 10.1039/b707637a. [DOI] [PubMed] [Google Scholar]
- 27.Prime KL, Whitesides GM. Adsorption of Proteins onto Surfaces Containing End-Attached Oligo(Ethylene Oxide) - a Model System Using Self-Assembled Monolayers. Journal of the American Chemical Society. 1993;115(23):10714–10721. [Google Scholar]
- 28.Jo BH, Van Lerberghe LM, Motsegood KM, Beebe DJ. Three-dimensional micro-channel fabrication in polydimethylsiloxane (PDMS) elastomer. Journal of Microelectromechanical Systems. 2000;9(1):76–81. [Google Scholar]
- 29.Thibault C, Severac C, Mingotaud A-F, Vieu C, Mauzac M. Poly(dimethylsiloxane) Contamination in Microcontact Printing and Its Influence on Patterning Oligonucleotides. Langmuir. 2007;23(21):10706–10714. doi: 10.1021/la701841j. [DOI] [PubMed] [Google Scholar]
- 30.Frey BL, Corn RM, Weibel SC. New York: Wiley & Sons; 2002. [Google Scholar]
- 31.Yuan M, Zhan S, Zhou X, Liu Y, Feng L, Lin Y, Zhang Z, Hu J. A Method for Removing Self-Assembled Monolayers on Gold. Langmuir. 2008;24(16):8707–8710. doi: 10.1021/la800287e. [DOI] [PubMed] [Google Scholar]
- 32.Sotiropoulou PA, Perez SA, Salagianni M, Baxevanis CN, Papamichail M. Characterization of the optimal culture conditions for clinical scale production of human mesenchymal stem cells. Stem Cells. 2006;24(2):462–471. doi: 10.1634/stemcells.2004-0331. [DOI] [PubMed] [Google Scholar]
- 33. Nelson KE, Gamble L, Jung LS, Boeckl MS, Naeemi E, Golledge SL, Sasaki T, Castner DG, Campbell CT, Stayton PS. Surface Characterization of Mixed Self-Assembled Monolayers Designed for Streptavidin Immobilization. Langmuir. 2001;17(9):2807–2816. There may differences in adsorption kinetics between the HS—EG3 and HS—EG6—COOH that would result in preferential adsorption of one species and result in some difference between solution mol percentage and resultant surface mol percentage.
- 34.Harder P, Grunze M, Dahint R, Whitesides G, Laibinis P. Molecular Conformation in Oligo (ethylene glycol)-Terminated Self-Assembled Monolayers on Gold and Silver Surfaces Determines Their Ability To Resist Protein Adsorption. Journal of Physical Chemistry B. 1998;102:426–436. [Google Scholar]
- 35.Lahiri J, Isaacs L, Grzybowski B, Carbeck JD, Whitesides GM. Biospecific Binding of Carbonic Anhydrase to Mixed SAMs Presenting Benzenesulfonamide Ligands: A Model System for Studying Lateral Steric Effects. Langmuir. 1999;15(21):7186–7198. [Google Scholar]
- 36.Lahiri J, Ostuni E, Whitesides GM. Patterning Ligands on Reactive SAMs by Microcontact Printing. Langmuir. 1999;15(6):2055–2060. [Google Scholar]
- 37.Frey BL, Corn RM. Covalent attachment and derivatization of poly(L-lysine) monolayers on gold surfaces as characterized by polarization-modulation FT-IR spectroscopy. Analytical Chemistry. 1996;68(18):3187–3193. [Google Scholar]
- 38.An important finding during aqueous SAM formation was that detectable incorporation of EG6-COOH and subsequent peptide incorporation required the use of pH conditions below the pKa of the carboxylic acid moiety at ~ pH 3. Therefore, all aqueous SAM formation processes were performed at pH 2.
- 39.Li L, Chen S, Zheng J, Ratner BD, Jiang S. Protein Adsorption on Oligo(ethylene glycol)-Terminated Alkanethiolate Self-Assembled Monolayers: The Molecular Basis for Nonfouling Behavior. The Journal of Physical Chemistry B. 2005;109(7):2934–2941. doi: 10.1021/jp0473321. [DOI] [PubMed] [Google Scholar]
- 40.Clare TL, Clare BH, Nichols BM, Abbott NL, Hamers RJ. Functional Monolayers for Improved Resistance to Protein Adsorption: Oligo(ethylene glycol)-Modified Silicon and Diamond Surfaces. Langmuir. 2005;21(14):6344–6355. doi: 10.1021/la050362q. [DOI] [PubMed] [Google Scholar]
- 41.Strong L, Whitesides G. Structures of self-assembled monolayer films of organosulfur compounds adsorbed on gold single crystals: electron diffraction studies. Langmuir. 1988;4(3):546–558. [Google Scholar]
- 42.Deans RJ, Moseley AB. Mesenchymal stem cells: Biology and potential clinical uses. Experimental Hematology. 2000;28(8):875–884. doi: 10.1016/s0301-472x(00)00482-3. [DOI] [PubMed] [Google Scholar]
- 43.Hudalla GA, Murphy WL. Using "Click" Chemistry to Prepare SAM Substrates to Study Stem Cell Adhesion. Langmuir. 2009;25(10):5737–5746. doi: 10.1021/la804077t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoover DK, Chan EWL, Yousaf MN. Asymmetric Peptide Nanoarray Surfaces for Studies of Single Cell Polarization. Journal of the American Chemical Society. 2008;130(11):3280–3281. doi: 10.1021/ja711016m. [DOI] [PubMed] [Google Scholar]
- 45.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 46.Rowlands AS, George PA, Cooper-White JJ. Directing osteogenic and myogenic differentiation of MSCs: interplay of stiffness and adhesive ligand presentation. Am J Physiol Cell Physiol. 2008;295(4):C1037–C1044. doi: 10.1152/ajpcell.67.2008. [DOI] [PubMed] [Google Scholar]
- 47.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.