Abstract
CYP3A4 metabolizes many drugs on the market. Although transcriptional regulation of CYP3A4 is known to be tightly controlled by some nuclear receptors (NR) including vitamin D receptor (VDR/NR1I1), posttranscriptional regulation of CYP3A4 remains elusive. In this study, we show that noncoding microRNAs (miRNAs) may control posttranscriptional and transcriptional regulation of CYP3A4 by directly targeting the 3′-untranslated region (3′UTR) of CYP3A4 and indirectly targeting the 3′UTR of VDR, respectively. Luciferase reporter assays showed that CYP3A4 3′UTR-luciferase activity was significantly decreased in human embryonic kidney 293 cells transfected with plasmid that expressed microRNA-27b (miR-27b) or mouse microRNA-298 (mmu-miR-298), whereas the activity was unchanged in cells transfected with plasmid that expressed microRNA-122a or microRNA-328. Disruption of the corresponding miRNA response element (MRE) within CYP3A4 3′UTR led to a 2- to 3-fold increase in luciferase activity. Immunoblot analyses indicated that CYP3A4 protein was down-regulated over 30% by miR-27b and mmu-miR-298 in LS-180 and PANC1 cells. The decrease in CYP3A4 protein expression was associated with significantly decreased CYP3A4 mRNA levels, as determined by quantitative real-time PCR (qPCR) analyses. Likewise, interactions of miR-27b or mmu-miR-298 with VDR 3′UTR were supported by luciferase reporter assays. The mmu-miR-298 MRE site is well conserved within the 3′UTR of mouse, rat, and human VDR. Down-regulation of VDR by the two miRNAs was supported by immunoblot and qPCR analyses. Furthermore, overexpression of miR-27b or mmu-miR-298 in PANC1 cells led to a lower sensitivity to cyclophosphamide. Together, these findings suggest that CYP3A4 gene expression may be regulated by miRNAs at both the transcriptional and posttranscriptional level.
CYP3A4 is the most abundant hepatic and intestinal cytochrome P450 enzyme in humans, contributing to the metabolism of various drugs such as benzodiazepines, HIV antivirals, macrolide antibiotics, and statins (Gonzalez and Yu, 2006). Different levels of CYP3A4 transcription, which are governed by a number of nuclear receptors such as pregnane X receptor (PXR/NR1I2) (Lehmann et al., 1998; Xie et al., 2000), vitamin D receptor (VDR/NR1I1) (Schmiedlin-Ren et al., 2001; Thummel et al., 2001; Matsubara et al., 2008; Wang et al., 2008), and retinoid X receptor alpha (RXRα; NR2B1) (Wang et al., 2006, 2008), may cause substantial interindividual variability in the metabolism of these drugs and result in distinct drug effects. In contrast to the advances in understanding nuclear receptor-governed transcriptional regulation of CYP3A4, there is the lack of study on the potential regulation of CYP3A4 at its 3′-untranslated region (3′UTR) (Yu, 2007).
MicroRNAs (miRNAs) are a family of small, noncoding RNAs that govern posttranscriptional expression of target genes (Ambros, 2004; Bartel, 2004; He and Hannon, 2004). These miRNAs exhibit unique expression patterns in specific tissues and/or cells, at certain developmental stages, or in response to various stressors. They usually act through base pairing to a partially complementary segment within the 3′UTR transcript of a target gene, which causes translation inhibition and/or mRNA cleavage and leads to a reduced expression of the target gene. With the understanding of miRNA function, there is increased interest in delineating the role of miRNAs in posttranscriptional regulation of drug-metabolizing enzymes, drug transporters, and nuclear receptors (Tsuchiya et al., 2006; Yu, 2007; Kovalchuk et al., 2008; Liao et al., 2008; Takagi et al., 2008; To et al., 2008; Zhu et al., 2008; Ji et al., 2009; Pan et al., 2009), which may not only provide insight into miRNA biological function but also advance the understanding of the integrated response of cells to xenobiotic drugs. Of note, Takagi et al. (2008) has shown that CYP3A4 can be indirectly regulated by miRNA via microRNA-148a (miR-148a)-controlled regulation of PXR. However, questions remain as to whether miRNAs act directly on CYP3A4 3′UTR, and whether miRNAs affect CYP3A4 expression by targeting other nuclear receptors.
In the present study, we show that the broadly conserved micro-RNA-27b (miR-27b) targets the 3′UTR of both CYP3A4 and VDR, leading to the negative regulation of CYP3A4 in LS-180 and PNAC1 cells. In addition, there is a conserved miRNA response element (MRE) in mouse, rat, and human VDR that is targeted by mouse miR-298 (mmu-miR-298). Their interactions affect the expression of VDR and CYP3A4. Furthermore, we present data suggesting that down-regulation of CYP3A4 via miRNA-mediated pathways in PANC1 cells is translated into significantly increased sensitivity to cyclophosphamide. Theses findings may provide increased understanding of CYP3A4 regulation, and they may offer novel clues for the role of miRNAs in drug metabolism and disposition.
Materials and Methods
Materials.
Cyclophosphamide and 1α-hydroxycholecalciferol (VD3) were bought from MP Biochemicals (Solon, OH), and ketoconazole was obtained from Sigma-Aldrich (St. Louis, MO). Dulbecco's modified Eagle's medium (DMEM), trypsin, and antibiotics were bought from Mediatech (Manassas, VA). RPMI 1640 medium and fetal bovine serum (FBS) were purchased from Hyclone (Waltham, MA). Oligonucleotide primers were synthesized by Eurofins MWG Operon (Huntsville, AL) and Integrated DNA Technologies (Skokie, IL). All other molecular biological reagents were purchased from Invitrogen (Carlsbad, CA) or Promega (Madison, WI).
In Silico Identification of Putative miRNA Binding Sites.
The 3′UTR sequences of human CYP3A4 (GenBank sequence NM_017460) and VDR/NR1I1 (NM_000376) were searched for antisense matches to individual miRNAs using MicroInspector (http://mirna.imbb.forth.gr/microinspector/) (Rusinov et al., 2005) and TargetScan (http://www.targetscan.org/) (Lewis et al., 2005).
Cell Culture.
Human pancreas cancer PANC1 and colon carcinoma LS-180 cells (American Type Culture Collection, Manassas, VA) were cultured in DMEM and RPMI 1640 medium, respectively. The media were both supplemented with 10% FBS, 100 U/ml penicillin G sodium, and 100 μg/ml streptomycin sulfate. Human embryonic kidney (HEK) 293 cells were cultured in DMEM supplemented with 4.5 g/liter glucose, 10 mM HEPES, and 10% FBS.
Plasmids.
Construction of pS-miR-328 and the control pS-Let-7a and Lin-41-luciferease reporter plasmids were described previously (Pan et al., 2009). Likewise, miR-122a, -27b, and mmu-miR-298 precursors were cloned into pSilencer 4.1-CMV vector (Ambion, Austin, TX) using gene-specific primers carrying BamHI and HindIII restriction sites (Supplemental Table). Correct plasmids were named pS-miR-122a, pS-miR-27b, and pS-mmu-miR-298, respectively, compared with the control plasmid (pS-Neg) that consists of a scrambled sequence, and pS-GAPDH that selectively knocks down GAPDH. A CYP3A4 3′UTR segment [0–1130 base pairs (bp), from stop codon], which was amplified from human genomic DNA with primers with XbaI/FseI sites (Supplemental Table), was cloned into XbaI/FseI-digested pGL3 vector (Promega). The 3′UTR segment was inserted downstream of the Firefly luciferase gene. Likewise, VDR 3′UTR segment 1 (−59–1360 bp, from stop codon) containing miR-298 MRE, and segment 2 (1255–2263 bp) containing miR-27b MRE were cloned, respectively, downstream of the Renilla luciferase gene within psiCHECK-II (Promega) after digestion with XhoI and NotI. Mutants were created using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) and mutagenesis primers (Supplemental Table). In particular, the miR-27b MRE mutant 1 within CYP3A4 3′UTR had three nucleotide transitions (A607T, T609G, and G612C), and mutant 2 had two nucleotide transitions (A607T and T609G) within the 8-bp seed sequence; the miR-298 MRE mutant 1 had three nucleotide transitions (C234G, C235G, and G239C), and mutant 2 had two nucleotide transitions (C234G and C235G). All inserts were confirmed by direct DNA sequencing.
Luciferase Assay.
All transfection experiments were conducted with Lipofectamine 2000 (Invitrogen), following the manufacturer's instructions. In particular, 2 μl of Lipofectamine 2000 was mixed with the desired plasmids for each well in 24-well plates. HEK293 cells were cotransfected with pGL3-CYP3A4 3′UTR-luciferase reporter plasmid (0.1 μg) and miRNA precursor or pS-Neg plasmid (0.4 μg), together with pRL-TK plasmid (0.01 μg) that expresses Renilla luciferase. Luciferase activities were assayed 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega). Triplicate, quadruplicate, or hexaplicate transfections were tested. Firefly luciferase activity was normalized to Renilla luciferase activity and compared among different treatments. Likewise, HEK293 cells were cotransfected with VDR 3′UTR-luciferase reporter construct (0.1 μg), miRNA precursor or pS-Neg plasmid (0.4 μg), and selective miRNA antagomir or a scrambled control (5 nM) (Dharmacon, Chicago, IL). Renilla luciferase activity was normalized to firefly luciferase activity and compared among different treatments.
Immunoblot Analysis.
Cells were treated with 200 nM VD3 or drug vehicle, as reported elsewhere (Thummel et al., 2001). Cells were harvested at 48 h after transfection with 10 μl of Lipofectamine 2000 and miRNA-expressing or control plasmid (4 μg) in 6-well plates, and cell lysates were prepared with radioimmunoprecipitation assay lysis buffer (Rockland Immunologicals, Gilbertsville, PA) supplemented with complete protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany). Protein concentrations were determined using the BCA Protein Assay Kit (Pierce, Rockford, IL). Whole-cell proteins (50 μg) were separated on 7.5% SDS-polyacrylamide gels (PAGE) and electrophoretically transferred onto nitrocellulose membranes (Invitrogen, Grand Island, NY), which were then incubated with a selective anti-CYP3A4 monoclonal antibody (BD Biosciences, San Jose, CA) or anti-VDR polyclonal antibody C-20 (Santa Cruz Biotechnology, Santa Cruz, CA). After further incubation with a horseradish peroxidase rabbit anti-mouse IgG (BD Bioscience) or a peroxidase-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc., West Grove, PA), the proteins were visualized with an enhanced chemiluminescence detection system (Pierce). Images were acquired and densitometric analyses were conducted using Kodak Image Station (New Haven, CT), as described previously (Yu et al., 2009). GAPDH was used as a loading control.
Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction Analysis.
All experiments were conducted essentially as described previously (Pan et al., 2009), except that the reverse transcription of isolated small RNAs was carried out using a universal primer, and quantitative real-time polymerase chain reaction (qPCR) analyses of individual miRNAs were performed using a miRNA-specific primer and another universal primer (Supplemental Table). Gene-specific primers for qPCR analyses of CYP3A4 or VDR coding sequence (CDS) or 3′UTR mRNAs, GAPDH and U6 RNA controls were also included in the Supplemental Table.
Cytotoxicity.
Cytotoxicity studies were conducted as reported previously (Chen et al., 2006; Pan et al., 2009). In brief, PANC1 cells were seeded in 96-well plates in the presence of 200 nM 1α-VD3. After 48 h, cells were transfected with 0.5 μl of Lipofectamine 2000 and 0.4 μg of pS-Neg, pS-mmu-miR-298, or pS-miR-27b plasmids in fresh medium. Cyclophosphamide (0–1000 μM) was added to the media 24 h after transfection. Control treatments included ketoconazole (100 nM) or drug vehicle only [0.1% DMSO (v/v)]. Cell viability was determined by sulforhodamine B assay. Inhibition (IC50 value) of cell growth by cyclophosphamide was estimated by fitting the percentage of cell growth (vehicle control plus 0 mM of cyclophosphamide treatment as 100%) to the Hill equation (GraphPad Prism; GraphPad Software Inc., San Diego, CA). All experiments were carried out in triplicate and repeated once with separate cultures.
Statistical Analysis.
All values were expressed as the mean ± S.D. Different treatments (qPCR and luciferase data) were compared by one-way ANOVA with Dunnett's posttest, and multiple variances (cytotoxicity) were analyzed by two-way ANOVA (GraphPad Prism). Differences were considered significant if the probability was less than 0.05 (P < 0.05).
Results and Discussion
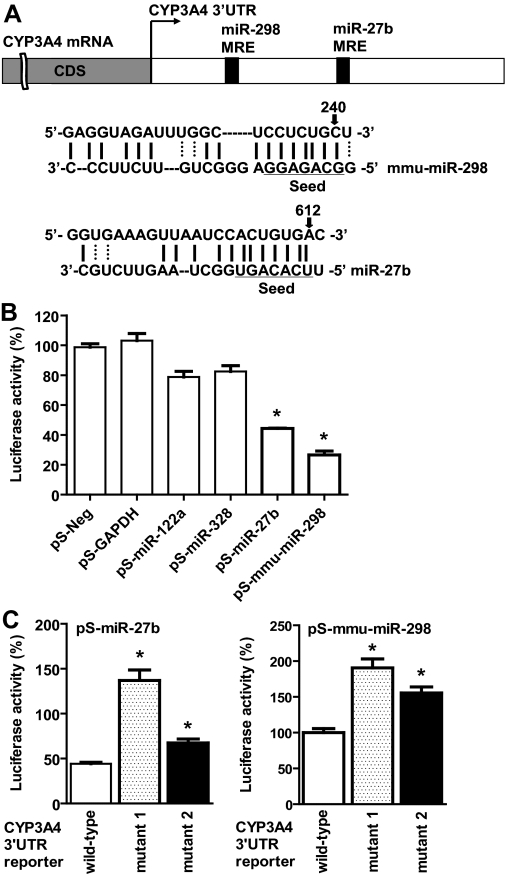
To investigate potential regulation of CYP3A4 by miRNAs, we first used MicroInspector (Rusinov et al., 2005) and TargetScan (Lewis et al., 2005) algorithms to screen antisense matches of CYP3A4 3′UTR against human and mouse miRNAs. The inclusion of mouse miRNAs was motivated by our previous observations showing that hepatic CYP3A4 was selectively silenced in the livers of male CYP3A4-transgenic (Tg-CYP3A4) mice after puberty (Granvil et al., 2003; Yu et al., 2005), which might involve miRNA mechanisms (Yu, 2007; Felmlee et al., 2008). Each algorithm identified more than 10 candidate MRE sites within the CYP3A4 3′UTR, which included the same MRE for miR-27b (Fig. 1A), a broadly conserved miRNA that has been shown to regulate CYP1B1 (Tsuchiya et al., 2006). It was hypothesized that certain miRNAs may be involved in a common pathway or biological process, e.g., the metabolism of xenobiotics. As such, miR-27b was studied. We also included mmu-miR-298 (Fig. 1A) in this study because mmu-miR-298 precursor levels, measured using real-time qPCR (Schmittgen et al., 2004), were increased in the livers of Tg-CYP3A4 mice after puberty, whereas the levels were decreased in the livers of wild-type control mice after puberty (data not shown). A role for mmu-miR-298 in the regulation of CYP3A4 might offer clues toward understanding the possible mechanisms of the silencing of CYP3A4 in Tg-CYP3A4 mouse livers (Yu, 2007). It is not surprising that a specific MRE was not identified by some algorithms but others, given the fact that each program uses different methods and databases. Indeed, this also indicates the need for experimental validation of miRNA targets.
Fig. 1.
CYP3A4 3′UTR may be directly targeted by some miRNAs. A, individual MRE sites within CYP3A4 3′UTR predicted by MicroInspector or TargetScan. B, HEK293 cells transfected with pS-miR-27b or pS-mmu-miR-298 plasmids showed lower CYP3A4 3′UTR-luciferase reporter activities when compared with cells transfected with the control pS-Neg plasmid (n = 3). C, disruption of miR-27b and -298 MRE sites resulted in higher CYP3A4 3′UTR-luciferase activities (n = 6) after cotransfection with individual miRNAs and the corresponding reporter plasmids. ∗, P < 0.05, as compared with the control (one-way ANOVA with Dunnett's posttest). See Materials and Methods for more details.
To assess the putative MRE sites within CYP3A4 3′UTR, we used luciferase reporter assays. A gain-of-function approach was chosen because mmu-miR-298 is absent in HEK293 cells, and the levels of miR-27b, -122a, and -328 are low (e.g., <400 copies of miR-27b per cell), especially when compared with those in normal tissues (e.g., >3000 copies of miR-27b per cell in human liver and kidney) (Lee et al., 2008). Thus, we cloned the precursors of miRNAs of interest and constructed a CYP3A4 3′UTR-luciferase reporter plasmid. Utilization of native 3′UTR sequence containing the predicted MRE site could be more relevant compared with artificial sequences containing multiple MRE sites. As the positive control, Lin-41-luciferase activities were reduced approximately 50% when cells were transfected with pS-Let-7a plasmid (Supplemental Fig. 1). Our data showed that CYP3A4 3′UTR-luciferase activity decreased by more than 50% in cells transfected with pS-miR-27b or pS-mmu-miR-298 plasmid, when compared with cells transfected with the control pS-Neg plasmid (Fig. 1B). In contrast, transfection with pS-GAPDH, pS-miR-122a, and pS-miR-328 plasmids showed limited effects on CYP3A4 3′UTR-luciferase activity. To further investigate the miRNA-MRE interactions, the corresponding miR-27b and -298 MRE sites were selectively disrupted, namely at the segment complementary to the seed sequence. Compared with wild-type CYP3A4 3′UTR-luciferase activities, the mutants showed significantly higher luciferase activities in cells cotransfected with the corresponding miRNA-expressing plasmid (Fig. 1C). These results suggest that miR-27b and mmu-miR-298 target CYP3A4 3′UTR directly, particularly at the predicted MRE sites.
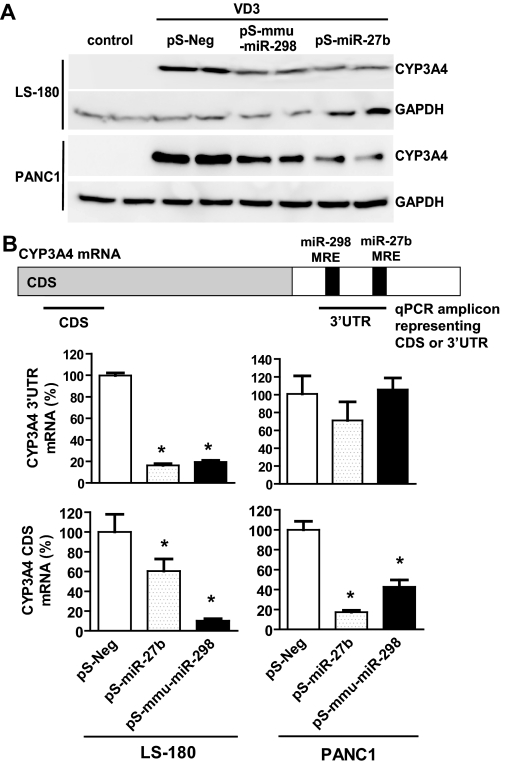
Likewise, we used a gene overexpression method to examine the effects of miR-27b and mmu-miR-298 on CYP3A4 expression. Studies on the expression of protein, CDS, and MRE-containing 3′UTR transcripts might offer some understanding of miRNA regulatory mechanisms. For instance, when the level of the target gene transcript remains unchanged and the level of protein is decreased, the effect of miRNA is probably due to translation inhibition. Differential expression of CDS and 3′UTR may further indicate whether the mechanisms involve mRNA degradation or transcriptional regulation. For example, a more dramatic reduction of the 3′UTR level compared with the CDS mRNA level may suggest involvement of mRNA degradation. Our immunoblotting analyses (Fig. 2A) indicated that, after transfection with pS-miR-27b or pS-mmu-miR-298 plasmid, both LS-180 and PANC1 cells showed over 30% decrease in CYP3A4 protein expression (Supplemental Fig. 2). Stem-loop qPCR indicated that both cell lines had over 3-fold higher mature miRNA levels, when compared with the controls (data not shown). Reduced CYP3A4 protein expression was associated with a significantly lower CYP3A4 3′UTR mRNA level in LS-180 cells (Fig. 2B). In contrast, the levels of CYP3A4 3′UTR transcript were unchanged in PANC1 cells, indicating that a different miRNA regulatory mechanism may exist in PANC1 cells. It is interesting to note that both cell lines showed significantly reduced CYP3A4 CDS mRNA expression after transfection with miRNA-expressing plasmids (Fig. 2B), suggesting that miR-27b and mmu-miR-298 may also influence the transcription of CYP3A4, e.g., by targeting nuclear receptors.
Fig. 2.
MicroRNA-27b and mmu-miR-298 negatively regulate CYP3A4 expression. A, representative immunoblots showed that LS-180 and PANC1 cells transfected with pS-miR-27b or pS-mmu-miR-298 plasmid had much lower CYP3A4 protein expression, compared with cells transfected with the pS-Neg control plasmid. Whole-cell proteins were separated by SDS-PAGE, and immunoblot analyses were carried out with selective antibodies against CYP3A4 and GAPDH, respectively. GAPDH was used as the loading control. All experiments were repeated at least once, i.e., transfection with separate cultured cells and immunoblot analysis of the same samples. B, quantitative real-time PCR were carried out using gene-selective primers. The data indicated that lower CYP3A4 protein expression was associated with reduced CDS and 3′UTR mRNA levels, suggesting possible involvement of transcriptional regulation. ∗, P < 0.05, compared with the corresponding control (n = 3 in each group, which refers to the number of independent transfection samples in a representative experiment; one-way ANOVA with Dunnett's posttest). See Materials and Methods for more details.
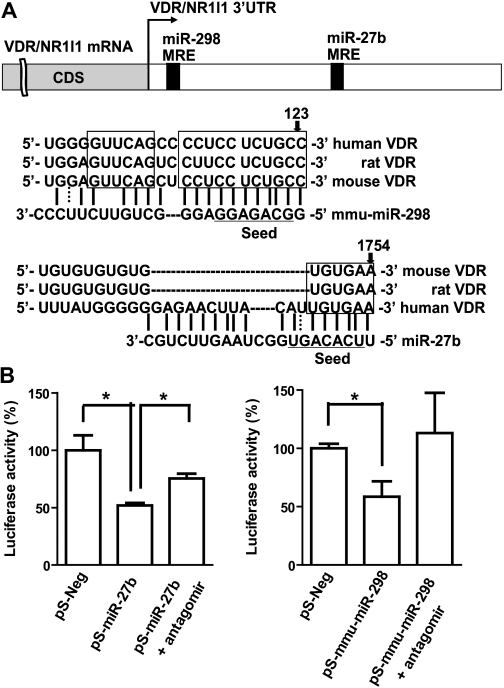
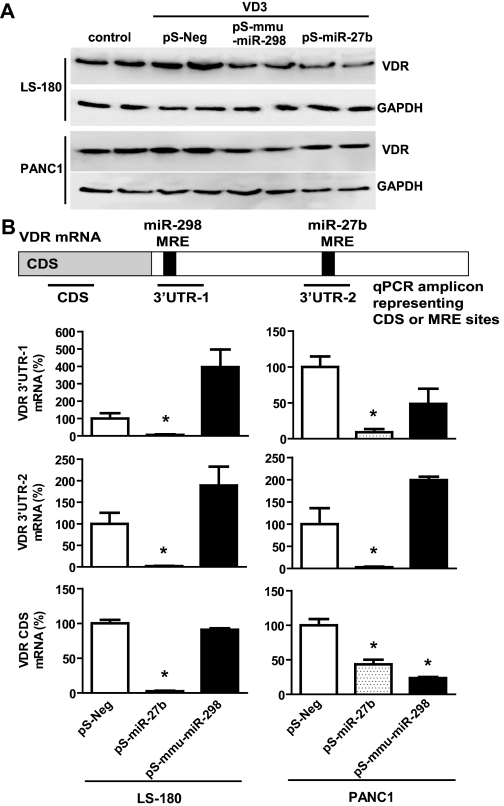
Further bioinformatic analyses suggested that the two miRNAs might regulate VDR/NR1I1 (Fig. 3A), whose role in controlling CYP3A4 transcriptional regulation is well documented (Schmiedlin-Ren et al., 2001; Thummel et al., 2001; Matsubara et al., 2008; Wang et al., 2008). It should be noted that the mmu-miR-298 MRE site within VDR 3′UTR is highly conserved in mice, rats, and humans, suggesting a potentially common regulatory mechanism by this miRNA in preclinical animal models. The predicted interactions of individual miRNAs with VDR 3′UTR were supported by the luciferase reporter assay, which showed that VDR 3′UTR-luciferase activities decreased 40 to 50% in HEK293 cells after transfection with pS-miR-27b or pS-mmu-miR-298 (Fig. 3B). In addition, the reduced luciferase activities appeared to be attenuated when cells were cotransfected with selective miRNA inhibitors (Fig. 3B). Therefore, we examined the effects of the two miRNAs on VDR protein expression in LS-180 and PANC1 cell lines (Fig. 4A). The data showed that cells transfected with pS-miR-27b or pS-mmu-miR-298 had 10 to 50% lower VDR protein expression compared with cells transfected with the pS-Neg control plasmid (Supplemental Fig. 3). To investigate whether the regulation of VDR by miRNAs involves mRNA degradation, we quantified the levels of VDR transcripts with qPCR (Fig. 4B). The expression of 3′UTR transcript containing miR-298 MRE was not altered by mmu-miR-298, suggesting that this miRNA may down-regulate VDR through translational inhibition mechanisms. In contrast, the consistent decrease in VDR 3′UTR and CDS by miR-27b in both cell lines suggests that miR-27b might also affect the transcription of VDR, which awaits further investigation.
Fig. 3.
MicroRNA-27b and mmu-miR-298 may interact with VDR/NR1I1 3′UTR. A, individual MRE sites within VDR 3′UTR predicted by MicroInspector and/or TargetScan. The miR-27b MRE is well conserved in different species. B, HEK293 cells transfected with pS-miR-27b or pS-mmu-miR-298 plasmids showed lower VDR 3′UTR-luciferase reporter activities than the control. This effect appeared to be attenuated when cells were cotransfected with selective miRNA antagomir. ∗, P < 0.05 (n = 4 in each group; one-way ANOVA with Dunnett's posttest). See Materials and Methods for more details.
Fig. 4.
MicroRNA-27b and mmu-miR-298 negatively regulate VDR/NR1I1 expression. A, representative Western blot analyses indicated that LS-180 and PANC1 cells transfected with pS-miR-27b or pS-mmu-miR-298 plasmid had lower VDR protein expression. B, quantitative real-time PCR showed that lower VDR protein expression caused by miR-27b was associated with reduced CDS and 3′UTR mRNA levels. ∗, P < 0.05, compared with the corresponding control (n = 3 in each group, which refers to the number of independent transfection samples in a representative experiment; one-way ANOVA with Dunnett's posttest). Whole-cell proteins were separated by SDS-PAGE, and immunoblot analyses were carried out with selective antibody against VDR and GAPDH, respectively. See Materials and Methods for more details.
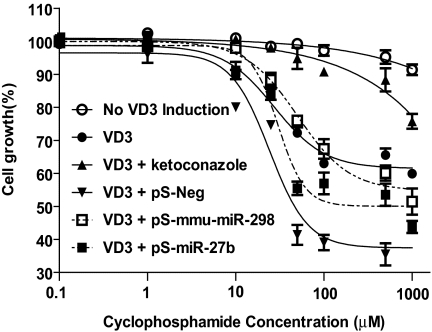
Lastly, we assessed the impact of miRNA pathways on the sensitivity of cells to cyclophosphamide, which may be activated by CYP3A4 to produce cytotoxic alkylating mustards (Chen et al., 2005; Chen et al., 2006). As expected, the induction of CYP3A4 by VD3 led to a significantly higher cyclophosphamide-induced cytotoxicity (Fig. 5), as manifested by a much lower IC50 value (27.4 μM). The increased cytotoxicity was almost completely attenuated by coadministration of the selective CYP3A4 chemical inhibitor, ketoconazole. Compared with cells transfected with the pS-Neg plasmid, cells transfected with pS-miR-27b and pS-mmu-miR-298 became less sensitive to cyclophosphamide (Fig. 5), which was indicated by significantly higher IC50 values (30.4 ± 1.1 μM for miR-27b, and 50.5 ± 1.2 μM for mmu-miR-298) in cells transfected with miRNA-expressing plasmids, when compared with that (24.1 ± 1.2 μM) in cells transfected with pS-Neg. Decreased cyclophosphamide cytotoxicity was presumably caused by down-regulation of CYP3A4 protein when cells were transfected with pS-miR-27b and pS-mmu-miR-298 plasmids (Fig. 2A). These results suggest that intervention of miRNA pathways may modify CYP3A4 expression and alter CYP3A4-catalyzed drug activation.
Fig. 5.
Intervention of the miRNA pathway can alter the sensitivity of PANC1 cells to cyclophosphamide, which is activated to cytotoxic agents by CYP3A4. Note that drug sensitivities are significantly (P < 0.001; two-way ANOVA) different between the absence and presence of VD3, between VD3 only and VD3 combined with ketoconazole, and between pS-miR-27b/mmu-miR-298 and pS-Neg treatments (n = 3 at each concentration), respectively.
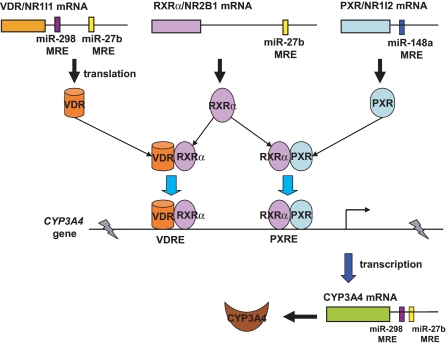
Of particular note, miR-148a has been shown to control posttranscriptional regulation of PXR and, consequently, affect the expression of CYP3A4, which may provide insight into the large interindividual variability in CYP3A4 expression observed in humans (Takagi et al., 2008). Another recent study suggests that miR-27a and miR-27b may target RXRα (Ji et al., 2009), which is a necessary component for PXR and VDR to form functional heterodimer (PXR/RXRα and VDR/RXRα, respectively) for the regulation of CYP3A4 transcriptional expression (Wang et al., 2006, 2008). In light of these observations and our current findings, miRNAs may control CYP3A4 transcriptional and posttranscriptional expression by targeting nuclear receptors, namely PXR, VDR, and RXRα, and targeting the 3′UTR of CYP3A4, respectively (Fig. 6). Nevertheless, our results obtained from gain-of-function experiments are subject to further validation by loss-of-function studies because unexpected effects might be induced by an increased level of miRNAs (Yu, 2007), and the role of miRNAs in regulation of CYP3A4 should be challenged using more complex model systems including animal models.
Fig. 6.
Involvement of miRNAs in the regulation of CYP3A4 expression. On one hand, miRNAs may directly act on CYP3A4 3′UTR (this study) and govern posttranscriptional regulation of CYP3A4. On the other hand, miRNAs can target nuclear receptors, namely PXR/NR1I2 (Takagi et al., 2008), RXRα/NR2B1 (Ji et al., 2009), and VDR/NR1I1 (this study), and control transcriptional regulation of CYP3A4. VDRE, VDR-responsive elements; PXRE, PXR-responsive elements.
In summary, our results show that miR-27b targets the 3′UTR of VDR and CYP3A4, and it negatively regulates VDR and CYP3A4 protein expression. The data suggest that regulation of VDR by miR-298 could be a common process in many animal models, which may affect CYP3A expression. Furthermore, the results indicate that intervention of miRNA pathways can be translated into an altered sensitivity of cells to xenobiotics. These findings may provide increased understanding of the complex regulation of CYP3A4 expression, as well as determine the role of miRNAs in drug metabolism and disposition.
Supplementary Material
Acknowledgments.
We are grateful to Drs. Daniel A. Brazeau and James Hong for valuable discussion and technical support. We also thank Dr. Daniel A. Brazeau for proofreading this article.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
This work was supported in part by the National Institutes of Health National Institute on Drug Abuse [Grant R01-DA021172]; and the Interdisciplinary Research Development Fund, University at Buffalo, SUNY.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
- PXR
- pregnane X receptor
- VDR
- vitamin D receptor
- RXRα
- retinoid X receptor alpha
- 3′UTR
- 3′-untranslated region
- miRNA
- microRNA
- miR-148a
- microRNA-148a
- miR-27b
- microRNA-27b
- mu-miR-298
- mouse microRNA-298
- MRE
- miRNA response element
- VD3
- 1α-hydroxycholecalciferol
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- HEK
- human embryonic kidney
- GAPDH
- glyceraldehyde 3-phosphate dehydrogenase
- bp
- base pair
- PAGE
- polyacrylamide gel
- qPCR
- quantitative real-time polymerase chain reaction
- CDS
- coding sequence
- ANOVA
- analysis of variance
- Tg
- transgenic.
References
- Ambros V. (2004) The functions of animal microRNAs. Nature 431:350–355 [DOI] [PubMed] [Google Scholar]
- Bartel DP. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- Chen CS, Jounaidi Y, Waxman DJ. (2005) Enantioselective metabolism and cytotoxicity of R-ifosfamide and S-ifosfamide by tumor cell-expressed cytochromes P450. Drug Metab Dispos 33:1261–1267 [DOI] [PubMed] [Google Scholar]
- Chen J, Yang XX, Huang M, Hu ZP, He M, Duan W, Chan E, Sheu FS, Chen X, Zhou SF. (2006) Small interfering RNA-mediated silencing of cytochrome P450 3A4 gene. Drug Metab Dispos 34:1650–1657 [DOI] [PubMed] [Google Scholar]
- Felmlee MA, Lon HK, Gonzalez FJ, Yu AM. (2008) Cytochrome P450 expression and regulation in CYP3A4/CYP2D6 double transgenic humanized mice. Drug Metab Dispos 36:435–441 [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Yu AM. (2006) Cytochrome P450 and xenobiotic receptor humanized mice. Annu Rev Pharmacol Toxicol 46:41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granvil CP, Yu AM, Elizondo G, Akiyama TE, Cheung C, Feigenbaum L, Krausz KW, Gonzalez FJ. (2003) Expression of the human CYP3A4 gene in the small intestine of transgenic mice: in vitro metabolism and pharmacokinetics of midazolam. Drug Metab Dispos 31:548–558 [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531 [DOI] [PubMed] [Google Scholar]
- Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. (2009) Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett 583:759–766 [DOI] [PubMed] [Google Scholar]
- Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, Pogribny IP. (2008) Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther 7:2152–2159 [DOI] [PubMed] [Google Scholar]
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. (2008) Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. Rna 14:35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102:1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20 [DOI] [PubMed] [Google Scholar]
- Liao R, Sun J, Zhang L, Lou G, Chen M, Zhou D, Chen Z, Zhang S. (2008) MicroRNAs play a role in the development of human hematopoietic stem cells. J Cell Biochem 104:805–817 [DOI] [PubMed] [Google Scholar]
- Matsubara T, Yoshinari K, Aoyama K, Sugawara M, Sekiya Y, Nagata K, Yamazoe Y. (2008) Role of vitamin D receptor in the lithocholic acid-mediated CYP3A induction in vitro and in vivo. Drug Metab Dispos 36:2058–2063 [DOI] [PubMed] [Google Scholar]
- Pan YZ, Morris ME, Yu AM. (2009) MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol 75:1374–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinov V, Baev V, Minkov IN, Tabler M. (2005) MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic Acids Res 33:W696–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedlin-Ren P, Thummel KE, Fisher JM, Paine MF, Watkins PB. (2001) Induction of CYP3A4 by 1 alpha,25-dihydroxyvitamin D3 is human cell line-specific and is unlikely to involve pregnane X receptor. Drug Metab Dispos 29:1446–1453 [PubMed] [Google Scholar]
- Schmittgen TD, Jiang J, Liu Q, Yang L. (2004) A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32:e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi S, Nakajima M, Mohri T, Yokoi T. (2008) Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J Biol Chem 283:9674–9680 [DOI] [PubMed] [Google Scholar]
- Thummel KE, Brimer C, Yasuda K, Thottassery J, Senn T, Lin Y, Ishizuka H, Kharasch E, Schuetz J, Schuetz E. (2001) Transcriptional control of intestinal cytochrome P-4503A by 1alpha,25-dihydroxy vitamin D3. Mol Pharmacol 60:1399–1406 [DOI] [PubMed] [Google Scholar]
- To KK, Zhan Z, Litman T, Bates SE. (2008) Regulation of ABCG2 expression at the 3′ untranslated region of its mRNA through modulation of transcript stability and protein translation by a putative microRNA in the S1 colon cancer cell line. Mol Cell Biol 28:5147–5161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya Y, Nakajima M, Takagi S, Taniya T, Yokoi T. (2006) MicroRNA regulates the expression of human cytochrome P450 1B1. Cancer Res 66:9090–9098 [DOI] [PubMed] [Google Scholar]
- Wang K, Chen S, Xie W, Wan YJ. (2008) Retinoids induce cytochrome P450 3A4 through RXR/VDR-mediated pathway. Biochem Pharmacol 75:2204–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Mendy AJ, Dai G, Luo HR, He L, Wan YJ. (2006) Retinoids activate the RXR/SXR-mediated pathway and induce the endogenous CYP3A4 activity in Huh7 human hepatoma cells. Toxicol Sci 92:51–60 [DOI] [PubMed] [Google Scholar]
- Xie W, Barwick JL, Downes M, Blumberg B, Simon CM, Nelson MC, Neuschwander-Tetri BA, Brunt EM, Guzelian PS, Evans RM. (2000) Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nature 406:435–439 [DOI] [PubMed] [Google Scholar]
- Yu AM. (2007) Small interfering RNA in drug metabolism and transport. Curr Drug Metab 8:700–708 [DOI] [PubMed] [Google Scholar]
- Yu AM, Fukamachi K, Krausz KW, Cheung C, Gonzalez FJ. (2005) Potential role for human cytochrome P450 3A4 in estradiol homeostasis. Endocrinology 146:2911–2919 [DOI] [PubMed] [Google Scholar]
- Yu AM, Qu J, Felmlee MA, Cao J, Jiang XL. (2009) Quantitation of human cytochrome P450 2D6 protein with immunoblot and mass spectrometry analysis. Drug Metab Dispos 37:170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wu H, Liu X, Evans BR, Medina DJ, Liu CG, Yang JM. (2008) Role of MicroRNA miR-27a and miR-451 in the regulation of MDR1/P-glycoprotein expression in human cancer cells. Biochem Pharmacol 76:582–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.