Abstract
Tyrosine hydroxylase (TH) plays a critical role in maintaining the appropriate concentrations of catecholamine neurotransmitters in brain and periphery, particularly during long-term stress, long-term drug treatment, or neurodegenerative diseases. Its expression is controlled by both transcriptional and post-transcriptional mechanisms. In a previous report, we showed that treatment of rat midbrain slice explant cultures or mouse MN9D cells with cAMP analog or forskolin leads to induction of TH protein without concomitant induction of TH mRNA. We further showed that cAMP activates mechanisms that regulate TH mRNA translation via cis-acting sequences within its 3′-untranslated region (UTR). In the present report, we extend these studies to show that MN9D cytoplasmic proteins bind to the same TH mRNA 3′-UTR domain that is required for the cAMP response. RNase T1 mapping demonstrates binding of proteins to a 27-nucleotide polypyrimidine-rich sequence within this domain. A specific mutation within the polypyrimidine-rich sequence inhibits protein binding and cAMP-mediated translational activation. UV-cross-linking studies identify a ∼44-kDa protein as a major TH mRNA 3′-UTR binding factor, and cAMP induces the 40- to 42-kDa poly(C)-binding protein-2 (PCBP2) in MN9D cells. We show that PCBP2 binds to the TH mRNA 3′-UTR domain that participates in the cAMP response. Overexpression of PCBP2 induces TH protein without concomitant induction of TH mRNA. These results support a model in which cAMP induces PCBP2, leading to increased interaction with its cognate polypyrimidine binding site in the TH mRNA 3′-UTR. This increased interaction presumably plays a role in the activation of TH mRNA translation by cAMP in dopaminergic neurons.
Tyrosine hydroxylase (TH) gene expression is tightly controlled by both transcriptional and post-transcriptional mechanisms (Kumer and Vrana, 1996; Sabban and Kvetnanský, 2001; Wong and Tank, 2007). The appropriate synthesis of dopamine, norepinephrine, and epinephrine is highly dependent on TH activity; hence, these control mechanisms play an essential role in the homeostatic regulation of catecholamine levels in both brain and periphery. Transcriptional regulation of the TH gene has been highly studied. TH gene transcription is activated in adrenal medulla, locus coeruleus, and other tissues in response to many stimuli, including stress, hypoxia, and treatment with cholinergic drugs or reserpine. If this activation is sustained (1–2 h), then TH mRNA is induced. The TH gene proximal promoter comprises numerous well-studied response elements, many of which participate in this transcriptional response.
Even though less well studied, post-transcriptional mechanisms also influence TH expression in adrenal medulla and brain. A number of in vivo studies have measured discrepancies between changes in TH gene transcription rate and TH mRNA levels in response to different stressors or drug treatments, suggesting that TH mRNA stability is regulated (Chang et al., 2000; Sun et al., 2004; Osterhout et al., 2005). Furthermore, Czyzyk-Krzeska and coworkers (Czyzyk-Krzeska et al., 1994; Czyzyk-Krzeska and Beresh, 1996; Paulding and Czyzyk-Krzeska, 1999) have shown that hypoxia induces TH mRNA in PC12 cells partly by increasing TH mRNA stability. This stabilization is due to interaction of trans-acting proteins with a polypyrimidine-rich sequence within the TH mRNA 3′-UTR. These workers have provided evidence that poly(C)-binding protein-1 (PCBP1) may participate as a trans-factor in this hypoxia-mediated TH mRNA stabilization response. Studies have also shown that the TH protein degradation rate may be regulated in response to nicotinic agonist in bovine adrenal chromaffin cells (Fernández and Craviso, 1999) and in response to ethanol or glial-derived neurotrophic factor in neuroblastoma SH-SY5Y cells (He and Ron, 2008). However, these latter responses are complex and require more investigation to establish their mechanism and physiological relevance.
Post-transcriptional regulation of TH expression also occurs at the level of translational control. Many reports have measured major discrepancies between changes in TH mRNA and TH protein levels in response to stress or drug treatments in rodents (Kaneda et al., 1991; Nankova et al., 1994; Yoshimura et al., 2004; Xu et al., 2007). In a recent report, our laboratory has shown that short-term stress is associated with 2-fold or greater increases in adrenal TH mRNA and that these increases persist for at least 24 h; however, TH protein levels are not up-regulated by these short-term stressors (Xu et al., 2007). In contrast, long-term treatments with these same stressors elicit induction of both TH mRNA and TH protein. Using polysome distribution assays, our results suggest that mechanisms regulating TH mRNA translation may be rate-limiting for TH protein synthesis and that the lack of induction of adrenal TH protein after short-term stress is due to inadequate activation or inhibition of TH mRNA translation. More recently, we have shown regulation of TH mRNA translation by cAMP in rat midbrain slice explant cultures and mouse MN9D cells (Chen et al., 2008). In this latter report, we have shown that treatment with the cAMP analog, 8-CPT-cAMP, or forskolin leads to induction of TH protein without concomitant induction of TH mRNA. This induction is due to the increased rate of TH protein synthesis, suggesting that the regulation occurs at the level of TH mRNA translation. Furthermore, we have shown that the TH mRNA 3′-UTR confers cAMP-mediated inducibility to the luciferase reporter gene, when it is fused downstream of the luciferase coding sequence. The TH mRNA 3′-UTR mediates the induction of luciferase activity, but not luciferase mRNA, indicating that cAMP is activating mechanisms that regulate TH mRNA translation via cis-acting sequences within its 3′-UTR. The present studies continue to investigate this cAMP-mediated translational response in MN9D cells. We show that this response is mediated by sequences within the polypyrimidine tract in the TH mRNA 3′-UTR and provide evidence that PCBP2 may participate as a trans-factor mediating this response.
Materials and Methods
Mutant TH (3′U)-Luc Constructs.
The TH (3′U)-Luc and modified GL3 plasmids were constructed as described previously (Chen et al., 2008). The deletion mutant constructs TH (3′UΔAS)-Luc, TH (3′UΔKA)-Luc, and TH (3′UΔKS)-Luc were generated by excising the TH mRNA 3′-UTR sequences from ApaI to SphI, KpnI to ApaI, or KpnI to SphI from TH (3′U)-Luc, respectively, followed by blunt-ending and religation. The 3′-UTR mutant constructs TH (A10,11,12)-Luc and TH (A1,2,3)-Luc were generated by site-directed mutagenesis of TH (3′U)-Luc using QuikChange II Site-Directed Mutagenesis Kit purchased from Stratagene (La Jolla, CA). The mutagenic primers used for generating these constructs were as follows: A10,11,12, forward primer, 5′-ctt tcc caa agt ctc cat cca aat ctc caa cct ttc ctg gcc c-3′, and reverse primer, 5′-ggg cca gga aag gtt gga gat ttg gat gga gac ttt ggg aaa g-3′; A1,2,3, forward primer, 5′-gcc agg gcc ttt ccc aaa gaa acc atc ccc ttc tcc aac c-3′, and reverse primer, 5′-ggt tgg aga agg gga tgg ttt ctt tgg gaa agg ccc tgg c-3′. The sequences of all constructs were confirmed using the DNA sequencing facilities at the University of Rochester Medical Center Molecular Biology Core Facility (Rochester, NY).
Cell Culture and Transfection of MN9D Cells.
MN9D cells were obtained from Dr. Lisa Opanashuk in the Department of Environmental Medicine at the University of Rochester Medical Center. These cells were created by Heller and coworkers (Choi et al., 1992) and are a hybrid cell line derived from the fusion of mouse neuroblastoma N18TG2 cells with embryonic mouse mesencephalic neurons. These cells express high levels of TH mRNA and TH protein under basal culture conditions. MN9D and PC12 cells were cultured as described previously (Nagamoto-Combs et al., 1997; Chen et al., 2008). For transfections, MN9D cells were cultured in 6- or 12-well dishes and transfected with 1 to 2 μg of TH (3′U)-Luc plasmid DNA and 0.033 to 0.066 μg of Renilla reniformis luciferase expression vector RL-SV40 (Promega, Madison, WI) using Lipofectamine (Invitrogen, Carlsbad, CA) as described by the manufacturer. Twenty-four hours after the transfection, the medium was removed, and the cells were incubated for another 24 h in the presence or absence of 0.5 mM 8-CPT-cAMP in fresh medium. The cells were then washed once with ice-cold PBS, harvested, and centrifuged. The cell pellets were stored at −80°C.
Enzyme Assays, Immunoblotting, and mRNA Quantitation.
Firefly and R. reniformis luciferase activities were assayed using the Promega Dual-Luciferase Reporter Assay System according to the manufacturer's specifications. Firefly luciferase activity was expressed as relative light units for firefly luciferase normalized to relative light units for R. reniformis luciferase for each well of cells. TH enzyme assays and immunoblotting for TH protein were performed as described previously (Chen et al., 2008). For detection of PCBP isoforms, pellets of MN9D cells were homogenized in ice-cold radioimmunoprecipitation assay buffer (25 mM Tris-HCl, pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS) with freshly added protease inhibitor cocktail (Sigma, St. Louis, MO) and incubated on ice for 30 min. The supernatant was collected by centrifugation at 10,000g for 10 min at 4°C. Supernatant proteins (30–120 μg) were loaded onto a 10% SDS-polyacrylamide gel, subjected to electrophoresis, and transferred to nitrocellulose using standard protocols. Western blots were performed under conditions of linearity with respect to protein concentration and autoradiographic signals. The antibody specific for PCBP2 was generated in rabbits by New England Peptide Corporation against the peptide sequence encoding amino acids 200 to 213 of mouse PCBP2 (GenBank accession number X78136). The antibody specific for PCBP1 was a gift from Dr. Maria Czyzyk-Krzeska (University of Cincinnati Medical Center, Cincinnati, OH) and has been described previously (Paulding and Czyzyk-Krzeska, 1999). The goat anti-hnRNP K antibody was from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Protein determinations were made by the method of Bradford (Bradford, 1976) using bovine serum albumin as standard.
TH mRNA and luciferase mRNA were measured using a quantitative RT-PCR assay using SYBR Green dye (BioRad Laboratories, Hercules, CA) at a concentration recommended by the manufacturer and an Applied Biosystems Prism 7000 real-time PCR cycler exactly as described previously (Chen et al., 2008). Glyceraldehyde 3-phosphate dehydrogenase mRNA was measured in each sample for normalization purposes, using sample cDNA transcribed from the same reverse transcription reaction as that used for measuring TH mRNA or luciferase mRNA.
Isolation of Cell Fractions Enriched in S-100 Cytoplasmic Proteins for RNA Binding Assays.
Frozen MN9D or PC12 cell pellets were thawed on ice for 10 min in hypotonic buffer (15 mM Tris, pH 7.5, 10 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, 0.7 μg/ml pepstatin and 1.0 μg/ml aprotinin) and then homogenized with 20 strokes of a Dounce homogenizer (B pestle). The nuclei were separated by centrifugation, and 0.11 volume of 10× cytoplasmic extraction buffer (0.3 M HEPES, pH 7.9, 1.4 M KCl, and 30 mM MgCl2) was added to each supernatant. The extracts were then centrifuged at 100,000g at 4°C for 60 min, and the resulting supernatants were dialyzed overnight in 2× binding buffer (2 mM HEPES, pH 7.9, 0.6 mM MgCl2, 10 mM KCl, 2% glycerol, 40 ng/μl tRNA, 0.1 mM phenylmethylsulfonyl fluoride, and 0.2 mM dithiothreitol). The extracts were aliquoted and stored frozen at −80°C.
In Vitro Transcription of TH mRNA 3′-UTR Probes.
To generate radiolabeled single-stranded RNAs expressing rat TH mRNA 3′-UTR sequences for use as probe or competitors in the RNA-protein binding assays, wild-type and mutagenized 3′-UTR sequences between the KpnI and SphI sites (see Fig. 1) were excised from the appropriate TH (3′U)-Luc constructs and ligated into the same enzyme sites within the multiple cloning region of pGEM 7Zf (Promega). These cloned constructs were subsequently cut with Sph1, and the linearized DNA templates were used to synthesize RNAs for the binding assays. Single-stranded wild-type 3′-UTR sequence was used as a probe in the RNA-protein binding assays (see below). This probe was synthesized using the MAXIScript kit from Ambion Corporation (Austin, TX) according to the manufacturer's recommended protocols, using 36 μCi of [α-32P]UTP (800 Ci/mmol) at a final concentration of 25 μM in a 20-μl reaction volume. The resulting radiolabeled RNA product was purified by electrophoresis on a 6% denaturing polyacrylamide/8 M urea gel. After electrophoresis, the RNA was eluted from the gel using a buffer containing 0.5 M ammonium acetate, 1 mM EDTA, and 0.2% SDS, precipitated with isopropanol, and washed once with 100% ethanol. The radioactivity of the pellet was counted, and the yield of purified radiolabeled RNA was calculated. The pellet was then suspended in DEPC-treated distilled water to a concentration of 2 ng/μl (80,000–120,000 cpm/μl). Competitor RNAs were similarly synthesized using the appropriate pGEM 7Zf recombinant plasmid as a template and a trace amount of [α-32P]UTP to calculate the final yield of competitor RNA. These competitors were suspended in DEPC-treated distilled water to a final concentration of 200 ng/μl.
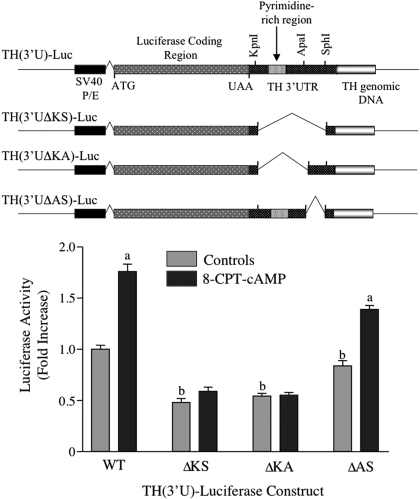
Fig. 1.
Activation of TH (3′U)-luciferase expression by cAMP requires sequences between the KpnI and ApaI sites of TH mRNA 3′-UTR. MN9D cells were transfected with one of the TH-luciferase plasmids depicted at the top of the figure, along with the R. reniformis luciferase expression vector, RL-SV40. After overnight incubation, cells were treated with vehicle or 0.5 mM 8-CPT-cAMP for 24 h. Firefly luciferase expression was normalized to R. reniformis luciferase expression in each dish of cells. Luciferase expression was expressed as the -fold increase over that observed in control cells transfected with the wild-type TH-luciferase construct. The results represent the means ± S.E. from 10 to 13 dishes. a, p < 0.01 compared with untreated control cells transfected with the same TH-luciferase vector; b, p < 0.01 compared with untreated control cells transfected with wild type TH-luciferase vector.
RNA-Protein Binding Assays.
Binding assays (final volume of 30 μl) contained 10 μg of S-100 cytoplasmic proteins, 3 μl of 10× binding buffer (10 mM HEPES, pH 7.9, 3 mM MgCl2, 50 mM KCl, and 1 mM dithiothreitol), and 4 ng of radiolabeled RNA probe expressing wild-type TH RNA 3′-UTR sequences. The assays were carried out at 30°C for 30 min. For competition experiments, proteins were incubated for 15 min with the indicated amount of competitor RNA before the addition of radiolabeled probe. After completion of the binding reaction, 1 μl of RNase T1 (50 U/μl) and 1 μl of heparin sulfate (1 μg/μl) were added sequentially to the reaction mixture for 10 min each at 30°C. Electrophoresis of RNA-protein complexes was carried out using 6% nondenaturing polyacrylamide gels. After electrophoresis, the gels were dried and exposed to film. Quantitative assessment of RNA-protein bands was performed by optical density measurements using GelEval 1.22 (Frogdance Software, Dundee, UK).
RNase T1 Mapping.
RNA-protein complexes were separated by electrophoresis as described above. The gel piece containing the specifically bound complex was excised, and the complex was eluted from the gel with elution buffer (0.5 M ammonium acetate, 1 mM EDTA, and 0.2% SDS) overnight at 37°C. The eluted RNA was ethanol-precipitated, resuspended in DEPC-treated water, subjected to phenol/chloroform extraction, and reprecipitated with ethanol. The final RNA pellet was resuspended in 20 μl of sterile buffer (10 mM Tris, pH 7.4, 1 mM EDTA), and an aliquot was further digested with 10 units of RNase T1 for 30 min at 37°C. For comparison purposes, free radiolabeled RNA probe was also digested with RNase T1. The digested RNA was boiled for 5 min in denaturing gel-loading buffer (95% formamide, 0.025% xylene cyanol, 0.025% bromphenol blue, 18 mM EDTA, and 0.025% SDS), and the digestion products were separated by electrophoresis on 6% denaturing polyacrylamide/8 M urea gels. The wet gel (wrapped in plastic wrap) was exposed to film. Radiolabeled RNA size markers (10, 22, and 47 nt) were synthesized from pGEM-7ZF linearized with ApaI (10 nt), SphI (22 nt), and EcoRI (47 nt) using T7 RNA polymerase.
UV-Cross-Linking Experiments.
RNA-protein binding assays were performed as described above. The reaction mixtures were then placed on ice in a 96-well flat-bottomed plate and exposed to UV light using a Stratlinker UV Crosslinker instrument (Stratagene) set at 1 J/cm2 in Energy Mode. The cross-linked products were then digested for 20 min at 37°C with RNase A (30 μg/ml) and RNase T1 (750 U/ml). The reaction products were separated on a 10% SDS polyacrylamide gel and detected using autoradiography.
Biotin Pull-Down Assay.
This assay was performed essentially as described by Lee et al. (2007). In brief, biotin-labeled RNA transcripts were synthesized from appropriate linearized plasmids, TH(KS)-7ZF, TH(KA)-7ZF, or TH(AS)-7ZF. RNA-protein binding assays were performed as described above using 500 μg of cytoplasmic protein from MN9D cells and 400 pM biotin-labeled RNA. RNA-protein reaction mixtures were placed on ice and irradiated with UV as described above. The biotin-labeled RNA-protein complexes were isolated using streptavidin-conjugated Dynabeads (Invitrogen), magnetically separated from other cell components and analyzed by Western blotting, using PCBP2 antibody.
Ribonucleoprotein Pull-Down Assay.
MN9D cells were homogenized in 100 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% Nonidet P-40, 1 mM dithiothreitol, 100 U/ml RNasin (Promega), 2 mM vanadyl ribonucleosides (Sigma), and 1 μl/ ml protease inhibitor cocktail (Sigma), and the homogenate was centrifuged at 10,000g for 10 min at 4°C. The supernatant was incubated for 2 h with either 50 μl of PCBP2 antiserum or an identical volume of preimmune serum from the same rabbit used for producing the antiserum. Immunoprecipitates were collected using protein A-conjugated Dynabeads (Invitrogen) according to the manufacturer's protocol and subjected to RT-PCR to detect mRNAs encoding TH, PCBP2, or PCBP1. The primers for the PCR reactions amplified sequences between 395 and 888 for TH mRNA (Chen et al., 2008), 696 and 943 for PCBP2 mRNA, and 428 and 933 for PCBP1 mRNA. PCR products were separated on 1% agarose gels and detected using ethidium bromide staining.
Cloning of Mouse PCBP2 from MN9D Cells and Generation of Recombinant Adenovirus Expression Viruses.
Total cellular RNA was extracted from MN9D cells and reverse-transcribed using SuperScript III RT (Invitrogen). The resulting cDNA was used as a template for amplification in PCR using forward and reverse primers designed for mPCBP2 (EcoRV-mPCBP2 and mPCBP2-Sal1) with EcoRV and Sal1 sites on the appropriate ends. The sequences of these primers were as follows: 5′-gcg ata tca tgg aca ccg gtg tga ttg-3′ and 5′-act gtc gac cta gct gct ccc cat gc-3′. The PCRs were performed as described by the manufacturer using Pfx DNA polymerase (Invitrogen). mPCBP2 cDNAs were isolated by agarose gel electrophoresis using QIAquick Gel Extraction Kit (QIAGEN, Valencia, CA). These DNA fragments and the pShuttle-IRES-hrGFP-1 vector (Stratagene) were cut with EcoRV and Sal1 and purified by phenol-chloroform extraction and ethanol precipitation. These DNAs were ligated to construct pShuttle-mPCBP2 and cloned. The resulting mPCBP2 cDNA clones were confirmed by sequence analysis.
Recombinant Adenovirus Vectors Were Generated Using the AdEasy XL Adenoviral.
Vector System (Stratagene) following the manufacturer's protocols. pShuttle-mPCBP2 vectors were linearized using Pme I. Linearized vectors were purified by agarose gel electrophoresis, treated with alkaline phosphatase for 30 min at 37°C, and then purified by phenol-chloroform extraction and ethanol precipitation. The final pellet was resuspended in sterile water at a final concentration of 0.05 μg/μl. These linearized and dephosphorylated vectors were then used to transform BJ5183-AD-1 cells that had been pretransformed with the pAdEasy-1 plasmid using standard electroporation procedures to produce recombinant adenovirus (Ad) plasmids. The recombinant Ad plasmids were amplified in DH5α Escherichia coli and then linearized with Pac I before transfection of AD293 cells using Lipofectamine 2000 (Invitrogen). The cells were incubated at 37°C for 7 to 10 days. Fresh medium was added when needed. To obtain primary viral stocks, cells were collected, washed with PBS once, and then resuspended in 0.5 ml of PBS (per 60 mM dish). Cell suspensions were subjected to four rounds of freeze/thaw, and supernatant (primary viral stock) was collected by centrifugation at 12,000g for 10 min at room temperature. To amplify recombinant adenovirus, AD293 cells were infected with primary viral stock and then harvested when cells showed evidence of cytopathic effects. The supernatant was saved either for another round of infection or was subjected to purification using CsCl gradient centrifugation. The titers of viral stock were determined by plaque assay.
Infection of MN9D Cells with Recombinant Adenovirus.
MN9D cells were split into 12-well plates the day before infection so that cells were approximately 60 to 70% confluent after overnight incubation. Recombinant adenoviruses were diluted in 0.5 ml of growth medium with a final concentration of 2 × 107 pfu/ml. The growth medium from each well was removed and replaced with 0.5 ml of adenovirus-containing medium. The plates were gently rocked and then incubated for 6 h. The medium was then removed and replaced with fresh medium. The cells were incubated under normal culture conditions, harvested at the appropriate time point, and stored at −80°C.
Statistical Analyses.
The results were analyzed by one-way analysis of variance using the computer program INSTAT. Comparisons between groups were made using the Dunnett multiple comparisons test or the Student-Neumann-Keuls test. A level of p < 0.05 was considered statistically significant.
Results
Sequences between the Kpn1 and Apa1 Sites within the TH mRNA 3′-UTR Confer cAMP Inducibility on TH-Luciferase Expression in MN9D Cells.
In a previous study, we showed that treatment of midbrain slices or MN9D cells with the cAMP analog 8CPT-cAMP or forskolin induced TH protein but not TH mRNA and that this response was due to activation of TH mRNA translation (Chen et al., 2008). To test which TH mRNA sequences mediated this translational response, luciferase reporter genes were constructed, which comprised the coding region of firefly luciferase flanked by TH mRNA 5′-UTR and 3′-UTR sequences. In our previous report (Chen et al., 2008), we showed that the effect of cAMP on the expression of this reporter gene reproduced that seen with endogenous TH expression: luciferase activity increased in response to cAMP, only when TH mRNA UTR sequences flanked the reporter gene. It is noteworthy that there was no effect of cAMP on luciferase mRNA levels. These results supported the concept that cAMP activated TH mRNA translation via sequences in its UTRs. Initial deletion analysis showed that the 35-nt 5′-UTR did not participate in this response but that sequences within the 262-nt 3′-UTR conferred cAMP-mediated translational activation of luciferase expression (Chen et al., 2008).
In the first set of experiments in the present study, we confirmed these previous findings and narrowed down the 3′-UTR sequences that mediated this response. MN9D cells were transfected with one of the TH-luciferase constructs shown in Fig. 1 and then treated with 0.5 mM 8-CPT-cAMP for 24 h. Control cells were left untreated. Luciferase activity increased ∼1.7-fold after 8-CPT-cAMP treatment of cells transfected with the wild-type construct, TH (3′U)-Luc (Fig. 1). In contrast, deletion of most of the 3′-UTR between the KpnI and SphI sites (162 nt) caused a complete inhibition of this response. Likewise, deletion of the 93 nt between the KpnI and ApaI sites resulted in complete loss of response. In addition to the loss of the cAMP response, deletion of the KpnI/SphI or KpnI/ApaI regions was associated with ∼50% decreases in basal luciferase expression. In contrast, deletion of the sequences between the ApaI and SphI sites did not significantly inhibit the cAMP response, even though basal expression was decreased slightly. These results suggest that the 93 nt region of the TH mRNA 3′-UTR between the KpnI and ApaI sites contains a cis-acting sequence that participates in the cAMP-mediated activation of TH mRNA translation in MN9D cells.
MN9D Cytoplasmic Proteins Bind to the TH mRNA 3′-UTR.
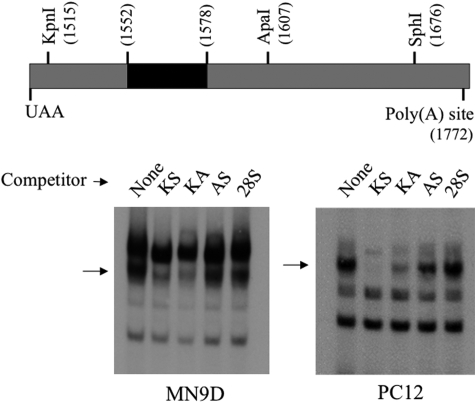
Czyzyk-Krzeska and coworkers (Czyzyk-Krzeska et al., 1994; Czyzyk-Krzeska and Beresh, 1996; Paulding and Czyzyk-Krzeska, 1999) showed that this region of TH mRNA 3′-UTR between the KpnI and ApaI sites bind to cytoplasmic proteins from PC12 cells, and that this binding participates in the regulation of TH mRNA stability in response to hypoxia. Hence, we tested whether cytoplasmic proteins from MN9D cells bind to these TH mRNA 3′-UTR sequences. We used cytoplasmic proteins from PC12 cells as a positive control for binding and to compare the binding complexes between these two cell lines. When MN9D cytoplasmic proteins were incubated with radiolabeled RNA comprising the region of TH mRNA 3′-UTR between the KpnI and SphI sites, three to four major RNA-protein complexes were observed on nondenaturing polyacrylamide gels (Fig. 2). These major bands migrated at a rate similar to that of those observed on gels derived from assays using PC12 cytoplasmic extracts (Fig. 2). When a 100-fold molar excess unlabeled competitor RNA comprising the same 3′-UTR sequence as that used for the radiolabeled probe (designated KS competitor in Fig. 2) was present in the assays, the major complex designated with an arrow in Fig. 2 was competed away, whereas the other complexes were not. In contrast, nonspecific competitor RNA comprising 160 nt of 28S rRNA did not compete away any of the RNA-protein complexes. These results suggest that the major complex designated by the arrow represented specific binding of MN9D and PC12 proteins to the TH mRNA 3′-UTR. The other bands apparently represented nonspecific binding. To further define the sequences involved in forming this specific binding complex, unlabeled RNA competitor comprising the sequences between either the KpnI and ApaI sites (KA competitor) or the ApaI and SphI sites (AS competitor) were used in the binding assays. The KA competitor effectively competed away the specific complex, whereas the AS competitor did not, using proteins from either MN9D or PC12 cells. Hence, these binding assays suggest that cytoplasmic proteins from both MN9D and PC12 cells bind to sequences within the 93-nt region of the TH mRNA 3′-UTR between the KpnI and ApaI sites.
Fig. 2.
MN9D and PC12 cytoplasmic proteins bind specifically to sequences between the KpnI and ApaI sites of TH mRNA 3′-UTR. Cytoplasmic proteins were isolated from MN9D and PC12 cells and incubated with radiolabeled RNA encoding TH mRNA 3′-UTR sequences between the KpnI and SphI sites as noted in the diagram. The black region between 1552 and 1578 designates the polypyrimidine-rich domain. Competitor RNAs were added to the incubation mixtures 15 min before the radiolabeled probe at 100-fold molar excess concentrations. The arrow designates the specifically bound RNA-protein complex.
MN9D Proteins Bind to a 27-nt Polypyrimidine-Rich Sequence within the TH mRNA 3′-UTR.
The region of TH mRNA 3′-UTR between the KpnI and ApaI sites contains a polypyrimidine-rich sequence that binds to PC12 cytoplasmic proteins (Czyzyk-Krzeska and Beresh, 1996). To test whether this same polypyrimidine-rich sequence acts as a binding site for MN9D cytoplasmic proteins, we performed an RNase T1 mapping experiment as described by Czyzyk-Krzeska and Beresh (1996). This assay takes advantage of the fact that RNase T1 cuts after G residues; because the 27-nt polypyrimidine-rich region does not possess any G residues, it should remain intact after RNase T1 digestion. The RNA-protein binding assay was performed as described above (which includes digestion with RNase T1 before electrophoresis), and the specifically bound RNA-protein complex was eluted from the gel. The proteins were removed from the eluted radiolabeled RNA by phenol extraction, and then the RNA was either immediately subjected to denaturing gel electrophoresis or digested a second time with RNase T1 before electrophoresis.
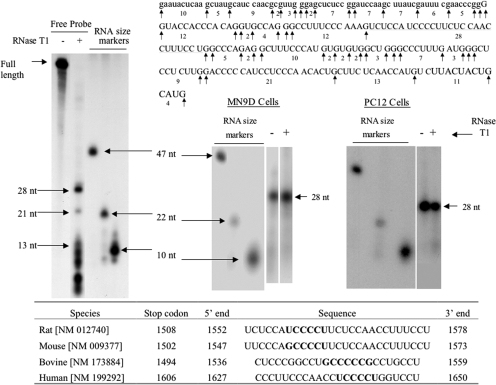
The predicted sizes of the RNase T1 digestion products of the free RNA probe are shown in Fig. 3. When the free probe was digested with RNase T1, the predicted 28- and 21-nt digestion products were observed on denaturing polyacrylamide gels (Fig. 3, left). As predicted, a large number of smaller digestion products of 13 nt or fewer were also observed but were not well resolved on these gels. The radiolabeled RNA that formed the specifically bound complex in the binding assays migrated on the denaturing gels at a rate that closely resembled that of the 28 nt digestion product (Fig. 3, right). This apparent 28-nt RNA fragment was the major TH mRNA 3′-UTR sequence bound to MN9D or PC12 cell cytoplasmic proteins. Further digestion of the eluted RNA with RNase T1 did not affect the migration rate of the RNA on the denaturing gels, suggesting that the sequence comprising this band did not contain any G residues that were blocked by protein binding during the first RNase T1 digestion. These results provide evidence that both MN9D and PC12 cell proteins bind specifically to the polypyrimidine-rich sequence comprising this 28-nt RNA fragment. The table at the bottom of the figure shows that similar polypyrimidine-rich sequences are found in the 3′-UTRs of TH mRNA in other species.
Fig. 3.
RNase T1 mapping depicts a 28-nt polypyrimidine binding site to which MN9D and PC12 cytoplasmic proteins bind to TH mRNA 3′-UTR. MN9D and PC12 cytoplasmic proteins were incubated with radiolabeled RNA probe encoding TH mRNA 3′-UTR sequences between the KpnI and SphI sites. The binding reaction was treated with RNase T1 and then subjected to electrophoresis on a nondenaturing polyacrylamide gel. The specifically bound RNA-protein complex was eluted from the gel and phenol-extracted to remove proteins. An aliquot of the eluate was subjected to electrophoresis on a denaturing polyacrylamide gel, whereas another aliquot was treated with RNase T1 before electrophoresis. The autoradiogram on the left side of the figure represents free radiolabeled RNA probe digested with RNase T1. Electrophoresis size markers are also designated on the autoradiogram. The TH mRNA (uppercase letters) and vector sequences (lowercase letters) comprising this probe along with the RNase T1 cutting sites are depicted in the figure. The two autoradiograms on the right side of the figure depict RNase T1 mapping experiments using MN9D and PC12 cytoplasmic proteins. The table at the bottom of the figure depicts polypyrimidine-rich sequences found in the 3′-UTRs of TH mRNAs from a number of species.
Mutations within the Polypyrimidine-Rich Region Inhibit Binding of MN9D Cell Proteins to the TH mRNA 3′-UTR.
Czyzyk-Krzeska and Beresh (1996) characterized in great detail the binding of PC12 cytoplasmic proteins to the polypyrimidine-rich sequence in rat TH mRNA 3′-UTR. They reported that the core sequence required for binding is UCCCCU (see underlined sequence in Fig. 3). The introduction of purines in place of pyrimidines within this core sequence totally abolished binding of PC12 cell proteins, whereas this pyrimidine-to-purine replacement outside of this core sequence, but within the polypyrimidine tract, only partially inhibited binding. We used these PC12 studies as a guide to characterize the binding of MN9D cytoplasmic proteins to the TH mRNA 3′-UTR using competitor RNAs with pyrimidine-to-purine replacements in two different parts of the pyrimidine-rich sequence.
Autoradiograms and competition displacement curves for different competitor RNAs used in the binding assays for MN9D proteins are presented in Fig. 4. As controls, wild-type KpnI-SphI 3′-UTR sequence and 28S rRNA sequence were used as specific and nonspecific competitors, respectively. As seen in the representative autoradiogram in Fig. 4, the wild-type 3′-UTR sequence competes away the specific RNA-protein complex almost completely using 100-fold molar excess competitor RNA, with a 50% displacement value of ∼15-fold excess (see graph in Fig. 4). In contrast, the nonspecific 28S rRNA sequence did not compete away the specific binding complex at any concentration tested up to 100-fold excess. Based on the PC12 cell studies, two pyrimidine-to-purine mutant RNA competitors were tested. These mutant RNAs encoded the same KpnI-SphI 3′-UTR as the wild-type sequence, but triplet pyrimidines were converted to triplet adenosines at select points in the polypyrimidine tract (see polypyrimidine tract sequence and tested mutants in Fig. 4). The A1,2,3 mutant competitor RNA competed away binding to the wild-type 3′-UTR sequence almost as effectively as the wild-type competitor (50% displacement was ∼20-fold excess). In contrast, the A10,11,12 mutant did not efficiently compete away binding to the wild-type sequence (Fig. 4). This result indicates that this latter mutation, which was within the core binding sequence in the polypyrimidine tract as defined in the studies of Czyzyk-Krzeska and Beresh (1996), almost completely inhibited MN9D cell protein binding to the TH mRNA 3′-UTR.
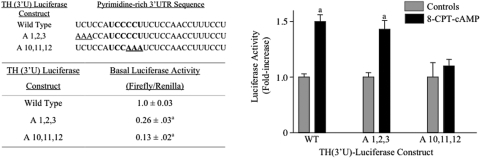
Fig. 4.
Competition binding studies demonstrate that a pyrimidine-to-purine substitution in the core UCCCCU binding site does not efficiently bind MN9D proteins. MN9D cytoplasmic proteins were incubated with radiolabeled RNA probe encoding TH mRNA 3′-UTR sequences between the KpnI and SphI sites. Different amounts of wild-type or mutant RNAs encoding the same sequence as the radiolabeled probe were incubated with the proteins for 15 min before the addition of probe. A 160-nt RNA encoding 28S rRNA sequences was used as a nonspecific competitor. The sequence of the probe and the competitor RNAs are depicted in the figure.
Mutation within the Core Binding Sequence of the Polypyrimidine Tract Abolishes the cAMP-Mediated Response of TH-Luciferase Expression.
The pyrimidine-to-purine mutants characterized in the binding studies above were cloned into the TH (3′U)-luciferase backbone to produce appropriate mutant constructs to test whether these mutants inhibit the cAMP response in MN9D cells (Fig. 5). Both mutant constructs lowered basal expression of the TH-luciferase construct in MN9D cells, with the mutation within the core UCCCCU binding site producing the greatest decrease (see table in Fig. 5). This decrease is probably due to diminished stability of the TH (3′U)-luciferase mRNA molecule, based on the work of Czyzyk-Krzeska et al. (Czyzyk-Krzeska et al., 1994; Paulding and Czyzyk-Krzeska, 1999); however, this hypothesis was not directly tested in the present report. In contrast, only the A10,11,12 pyrimidine-to-purine mutation blocked the cAMP response (bar graph in Fig. 5). The A1,2,3 mutation did not inhibit the cAMP response significantly. These results suggest that the core UCCCCU sequence within the polypyrimidine tract plays a critical role in the cAMP-mediated translational response in MN9D cells.
Fig. 5.
Activation of TH (3′U)-luciferase expression by cAMP is inhibited by pyrimidine-to-purine substitution within the core UCCCCU sequence of TH mRNA 3′-UTR. MN9D cells were transfected with one of the TH-luciferase vectors along with the normalization control vector, RL-SV40. After overnight incubation, the cells were treated with vehicle or 0.5 mM 8-CPT-cAMP for 24 h. Firefly luciferase expression was normalized to R. reniformis luciferase expression in each dish of cells. In the table, luciferase activity was expressed relative to that observed in control cells transfected with the wild-type TH-luciferase construct. In the bar graph, luciferase activity was expressed as cAMP-mediated -fold increase over that observed in control cells transfected with the same TH-luciferase construct. The results represent the means ± S.E. from 6 to 12 dishes. a, p < 0.01 compared with controls.
UV-Cross-Linking Studies Implicate a ∼44-kDa Protein Binding to the TH mRNA 3′-UTR.
In initial studies to characterize the trans-acting factor(s) that mediates the cAMP translational response, we used UV-cross-linking to identify proteins binding to the KpnI–SphI region of TH mRNA 3′-UTR. One major band of ∼44 kDa and one minor band of ∼80 kDa were observed on SDS-polyacrylamide gels after UV-cross-linking of MN9D proteins to the 162-nt radiolabeled KpnI-SphI probe (Fig. 6). Analogous bands of approximately the same size were also observed in analogous binding reactions using proteins from PC12 cells, even though the 80-kDa band was much less prominent using PC12 cell proteins. Both the 44- and 80-kDa bands from the MN9D cells were completely abolished when excess competitor RNA containing the same sequences as the probe was added to the binding reactions. In contrast, nonspecific competitor RNA comprising 28S rRNA sequences (160 nt) did not compete away either band. Further competition studies demonstrated that the major ∼44-kDa complex was competed away by RNA sequences between the KpnI and ApaI sites but not by those between the ApaI and SphI sites. In contrast, the ∼80-kDa complex was not effectively competed away by either KpnI-ApaI or ApaI-SphI sequences. With respect to the major ∼44-kDa band, these competition results were similar for proteins derived from either MN9D or PC12 cells (Fig. 6), suggesting that both these sources express a protein that binds specifically to the KpnI-ApaI region of TH mRNA 3′-UTR and that this protein has a molecular mass of ∼44 kDa. The protein(s) comprising the larger, minor complex (∼80-kDa band) are found in higher concentration in MN9D cells and apparently require the entire KpnI-SphI sequence for binding.
Fig. 6.
UV-cross-linking studies depict a 40- to 44-kDa protein that binds to sequences between the KpnI and ApaI sites of TH mRNA 3′-UTR. MN9D or PC12 cytoplasmic proteins were incubated with radiolabeled RNA probe encoding TH mRNA 3′-UTR sequences between the KpnI and SphI sites. Different amounts of wild-type or deletion mutant RNAs encoding the same region as the radiolabeled probe were incubated with the proteins for 15 min before the addition of probe. A 160-nt RNA encoding 28S rRNA sequences was used as a nonspecific competitor. The reaction mixtures were exposed to UV irradiation to cross-link the RNA-protein complexes and then subjected to SDS-polyacrylamide gel electrophoresis.
PCBP2 Was Induced by cAMP in MN9D Cells and Binds to TH mRNA 3′-UTR.
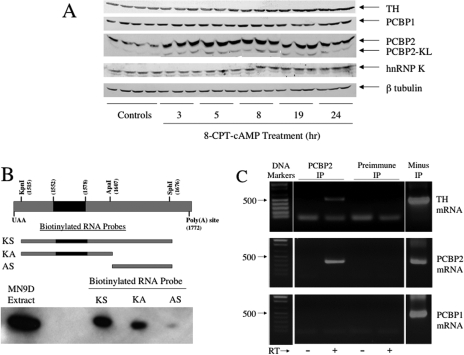
Our results suggest that the polypyrimidine-rich tract within the 3′-UTR participates in the cAMP-mediated activation of TH mRNA translation in MN9D cells and that the core UCCCCU sequence may play a pivotal role in this response. Furthermore, the region of the TH mRNA 3′-UTR encompassing the polypyrimidine tract binds to a protein with an apparent molecular size of ∼44 kDa. PCBPs are reasonable candidates for mediating this response in trans, because they bind to polypyrimidine-rich single-stranded RNA sequences and are ∼40 to 42 kDa in size (Makeyev and Liebhaber, 2002). These proteins have also been implicated in the hypoxia-inducible stabilization of TH mRNA in PC12 cells (Paulding and Czyzyk-Krzeska, 1999). To test this hypothesis, MN9D cells were treated with 8-CPT-cAMP for different periods of time, and Western analysis was used to measure changes in the levels of different PCBPs (Fig. 7).
Fig. 7.
PCBP2 is induced by cAMP in MN9D cells and binds to TH mRNA UTR sequences between the KpnI and ApaI sites. A, MN9D cells were treated with vehicle for 24 h or 0.5 mM 8-CPT-cAMP for different periods of time as designated in the figure. Different poly(C)-binding proteins were measured using Western blot analysis. B, biotin pull-down assays were used to test whether PCBP2 binds to the TH mRNA 3′-UTR. Biotin-labeled RNA transcripts were synthesized from appropriate linearized plasmids as depicted in the figure. RNA-protein binding assays were performed using 500 μg of MN9D cytoplasmic protein and 400 pM biotin-labeled RNA. RNA-protein complexes were placed on ice and irradiated with UV for 15 min at 4°C. The biotin-labeled RNA-protein complexes were isolated using streptavidin-conjugated Dynabeads and magnetic separation. The isolated proteins were analyzed by Western blot, using an antibody specific for PCBP2. C, ribonucleoprotein pull-down assays were used to test whether PCBP2 binds to TH mRNA in intact MN9D cells. Cytoplasmic extracts were incubated with PCBP2 rabbit antiserum or preimmune serum from the same rabbit. PCBP2 immunoprecipitates were isolated using protein A-conjugated Dynabeads and magnetic separation, and RT-PCR was used to amplify TH mRNA, PCBP2 mRNA, or PCBP1 mRNA present in the immunoprecipitates. PCR products were visualized using ethidium bromide staining. The PCR product derived from RT-PCR of cytoplasmic RNA from the same cell extracts but not subjected to immunoprecipitation is shown in the right lane, labeled “minus IP.”
As described previously, treatment of MN9D cells with 8-CPT-cAMP induces TH protein slowly over time, with an ∼2-fold induction observed after 24 h of treatment (see uppermost autoradiogram in Fig. 7A). Antibodies directed against three different PCBPs identified PCBP1, PCBP2, and hnRNP K antibody in MN9D cell extracts (see autoradiograms in Fig. 7A). The levels of PCBP1 and hnRNP K did not change significantly during 24 h of treatment with 8-CPT-cAMP. In contrast, PCBP2 was induced 2- to 3-fold by 8-CPT-cAMP at all time points tested between 3 and 24 h. Two PCBP2 immunoreactive bands were observed on the Western blots. The major band migrated at a rate commensurate with a molecular size of ∼42 kDa, whereas the minor band migrated as a ∼40-kDa protein. Both of these bands increased in response to cAMP to the same extent. Based on these results, we further investigated PCBP2 as a candidate for the trans-factor involved in regulating TH mRNA translation.
To test whether PCBP2 binds to TH mRNA 3′-UTR sequences, biotinylated RNA probes comprising segments of the 3′-UTR were synthesized and used in the standard binding assay with MN9D cytoplasmic proteins. The resulting complexes were UV-irradiated and bound to streptavidin beads. After extensive washing of the beads and elution of the complexes, the extracts were subjected to SDS-polyacrylamide gel electrophoresis and Western blotting for PCBP2. As seen in Fig. 7B, PCBP2 was detected in the binding complexes when KpnI-SphI or KpnI-ApaI sequences were used in the binding assays, but not when ApaI-SphI sequences were used. This result indicates that PCPB2 can bind to the same sequences within TH mRNA 3′-UTR that are required for the cAMP response, even though it remains unclear whether it is binding to the UCCCCU core sequence identified in the mutation analysis.
To test whether PCBP2 binds to TH mRNA in intact MN9D cells, a ribonucleoprotein pull-down assay was used. MN9D cell extracts were incubated with rabbit antiserum for PCBP2 or, as a control, preimmune serum isolated from the same rabbit. RT-PCR was used to identify mRNAs that were immunoprecipitated and presumably bound to PCBP2. Primers that amplified sequences 395 to 888 of TH mRNA produced a 493 bp PCR product from RNA found in immunoprecipitates using PCBP2 antiserum, but not preimmune serum (Fig. 7C, top). A second set of primers amplified TH mRNA sequences between 1534 and 1725 and yielded identical results (data not shown). As positive and negative controls, PCBP2 and PCBP1 mRNAs, respectively, were assayed in the immunoprecipitates. PCBP2 mRNA, but not PCBP1 mRNA, was shown to bind to PCBP2 in K562 cells (Waggoner and Liebhaber, 2003). In agreement with these findings, we observed PCBP2 mRNA in the PCBP2 immunoprecipitates (Fig. 7C, middle), but not PCBP1 mRNA (Fig. 7C, bottom). Neither PCBP2 nor PCBP1 mRNA was observed in immunoprecipitates isolated using preimmune serum. These results provide evidence that PCBP2 binds to TH mRNA in intact MN9D cells.
Overexpression of PCBP2 Isoforms Induced TH Protein, but Not TH mRNA in MN9D Cells.
Multiple forms of PCBP2 are expressed in mouse tissue as a result of alternative splicing. Horak and coworkers (Goller et al., 1994; Funke et al., 1996) isolated mouse PCBP2 alternatively spliced mRNA isoforms and detected two variable regions (±93 and ± 39 ± 3 bp) (see diagram in Fig. 8). The first region (93 bp) encodes 31 amino acids, and different isoforms are produced because of alternative splicing of a variable exon between exons 8 and 9 of the PCBP2 gene. The second variable region yields a diverse array of mRNA products as a result of alternative splicing of an exon between exons 10 and 11, as well as the use of alternative splice sites at the 5′ end of exon 11. PCBP2 mRNAs containing 42 bp (+ 39 + 3 bp), 39 bp (+ 39 − 3 bp), 3 bp (−39 + 3 bp) or 0 bp (−39 − 3 bp) in this variable region were identified in mouse tissues. More recently, a third variable region was identified at the 5′ end of exon 8 as a result of the use of alternative splice sites; this alternative splicing yields PCBP2 mRNAs ± 12 bp (four amino acids). All three of these variable regions are located between the second and third KH domains of mPCBP2 (Fig. 8). Because the production of these different alternatively spliced mRNAs are apparently independent of one another, 16 different isoforms of PCBP2 could exist. It is not known which of these isoforms is expressed in MN9D cells.
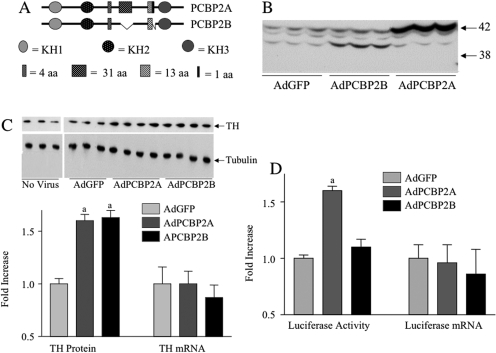
Fig. 8.
Overexpression of PCBP2 induces TH mRNA translation in MN9D cells. A, schematic diagrams of PCBP2 isoforms cloned from MN9D cells and used to produce PCPB2 adenoviral expression vectors. B, MN9D cells were infected with adenoviral constructs and 24 h after infection, proteins were isolated and subjected to Western blot analysis for PCBP2. C, MN9D cells were infected with adenoviral constructs and 24 h after infection, proteins were isolated for Western blot analysis to detect TH protein, using tubulin as a control for protein loading, or RNA was isolated and TH mRNA was measured using quantitative RT-PCR. The data in the bar graph represent the means ± S.E. from six to seven dishes. D, MN9D cells were infected with adenoviral constructs and 24 h after infection, proteins were isolated to measure luciferase activity, or RNA was isolated to measure luciferase mRNA using quantitative RT-PCR. The data represent the means ± S.E. from 11 to 15 dishes. a, p < 0.01 compared with dishes transfected with control AdGFP.
In initial attempts to address this issue, we cloned PCBP2 from MN9D cells using an RT-PCR approach and sequenced a number of resulting clones. The two most frequently isolated cDNAs were designated PCBP2A and PCBP2B (see diagram in Fig. 8). PCBP2A encoded the full-length mRNA that included all alternatively spliced exons. PCBP2B encoded an mRNA in which the 93-nt region and the 3-nt at the end of the ± 39 ± 3 region were spliced out. These two cDNAs were cloned into adenoviral expression vectors, and these were used to infect MN9D cells. Infection with the PCBP2A-expressing adenovirus produced a 4- to 5-fold increase in PCBP2 immunoreactivity that comigrated with the major 42-kDa endogenous PCBP2 band observed on the Western blots (Fig. 8B). This result suggested that the full-length, unspliced PCBP2 is the major isoform expressed in MN9D cells. Overexpression of PCBP2B yielded a 4- to 5-fold increase in PCBP2 immunoreactivity that comigrated with the minor ∼40-kDa endogenous PCBP2 band observed on the Western blots, suggesting that PCBP2B encodes this minor isoform. It should be noted in Fig. 8B that a third endogenous minor PCBP2 immunoreactive band that migrated slightly faster than the major 42-kDa band was observed. This band was observed only when the electrophoresis was run for a long period of time (as in Fig. 8), resulting in better resolution of PCBP2 immunoreactive bands. The identity of this minor band remains unclear, but based on its apparent size, it is probably the product of the PCBP2 mRNA isoform lacking 12 nt (4 amino acids).
Cell extracts from the MN9D cells infected with these different PCBP2 adenovirus expression vectors were used to test whether overexpression of these PCBP2 proteins was associated with changes in the levels of TH protein and/or TH mRNA (Fig. 8C). Infection of the cells with the control virus, AdGFP, did not affect either TH protein levels (see autoradiogram of Western blot in Fig. 8B) or TH mRNA levels measured using quantitative RT-PCR (data not shown). However, overexpression of either of the PCBP2 isoforms shown in Fig. 8B was associated with significant induction of TH protein. This induction was approximately the same as that observed when cells were treated with 8-CPT-cAMP for 24 h (1.5- to 2-fold). In contrast, TH mRNA levels were not significantly altered by overexpression of either PCPB2 isoform.
The MN9D cells used in these experiments had been stably transfected with the TH (3′U)-Luc construct as described previously (Chen et al., 2008). Treatment of these stably transfected cells with 8-CPT-cAMP for 24 h resulted in the induction of luciferase activity without increases in TH (3′U) luciferase mRNA (Chen et al., 2008). We tested whether overexpression of these PCBP2 isoforms had effects on TH (3′U)-luciferase expression (Fig. 8D). Overexpression of PCBP2A, the full-length major isoform in MN9D cells, was associated with a 1.5-fold increase in luciferase activity without any affect on TH (3′U) luciferase mRNA levels. This result was consistent with the TH expression data shown in Fig. 8C. However, overexpression of PCBP2B yielded unexpected results, in that it produced no changes in either luciferase activity or luciferase mRNA.
Discussion
Regulation of TH gene expression has been most intensely studied at the level of transcription (Kumer and Vrana, 1996; Sabban and Kvetnanský, 2001; Wong and Tank, 2007). The prevailing model posits that stimuli like stress, nicotine, or reserpine activate signaling pathways that stimulate TH gene transcription rate, leading to increased levels of TH mRNA, which are then inevitably translated into TH protein. However, numerous reports over the past 20 years have described major discrepancies between changes in the TH gene transcription rate, TH mRNA, and TH protein levels in response to these stimuli, leading to the hypothesis that regulation of post-transcriptional mechanisms may play a very important role in determining the final expression of TH protein. Yet very little is known about these post-transcriptional mechanisms.
In a recent report, Chen et al. (2008) showed that a 24- to 48-h treatment of cultured rat midbrain slices with 8-CPT-cAMP produces a ∼2-fold increase in TH protein but no increase in TH mRNA. This surprising result was also observed in the dopamine neuron-like MN9D cells. Even though TH mRNA was not induced by cAMP, the rate of TH protein synthesis increased ∼2-fold, suggesting that TH mRNA translation was altered by cAMP. This conclusion was supported by polysome distribution assays. Finally, in this study we showed that 8-CPT-cAMP increased expression of luciferase activity in MN9D cells transfected with a plasmid encoding the luciferase gene fused to TH mRNA 3′-UTR sequences. It is noteworthy that the hybrid TH-luciferase mRNA levels were not altered by cAMP, indicating that this response was due to increased translation. The work in the present report builds on this earlier research, using MN9D cells as a model system.
Identification of a cis-Acting Sequence within the TH mRNA 3′-UTR that Is Essential for the cAMP-Mediated Translational Response.
We show that the sequences between the KpnI and ApaI sites within the TH mRNA 3′-UTR are essential for the translational response to cAMP. This region also interacts with cytoplasmic proteins from MN9D cells to form a specific RNA-protein complex. This KpnI-ApaI region of the 3′-UTR interacts with PC12 cell proteins and participates in the regulation of TH mRNA stability in response to hypoxia (Czyzyk-Krzeska et al., 1994). A 27-nt polypyrimidine-rich sequence within this region is essential for PC12 cell protein binding and hypoxia-mediated stabilization (Czyzyk-Krzeska and Beresh, 1996; Paulding and Czyzyk-Krzeska, 1999). Given that the binding complex observed using MN9D proteins is similar in size to that observed using PC12 proteins and that this binding is dependent on the same KpnI-ApaI region, it was reasonable to postulate that this polypyrimidine-rich sequence may also participate in the translational response to cAMP. Our results support this hypothesis. The RNase T1 mapping experiments using MN9D cell proteins show that the major specific RNA-protein complex comprises the polypyrimidine-rich domain. In addition, the results of the mutation analysis suggest that the core UCCCCU binding site within this polypyrimidine-rich sequence, which was extensively characterized by Czyzyk-Krzeska and Beresh (1996), is essential for binding of MN9D proteins to the TH mRNA 3′-UTR and for the cAMP response. Taken together, these results support the hypothesis that both PC12 and MN9D cytoplasmic proteins bind to the UCCCCU core sequence within the polypyrimidine-rich domain of TH mRNA 3′-UTR and that in MN9D cells, this binding plays a major role in cAMP-mediated translational activation.
It should be noted that both the A1,2,3 and A10,11,12 mutations are associated with significant reductions in basal TH-luciferase expression. This result is surprising, because the A1,2,3 mutation does not apparently bind MN9D proteins (based on the competition studies in Fig. 4), and it does not inhibit the cAMP response (Fig. 5). We have not explored the mechanism(s) for this reduced basal expression, but it is probably due to decreased TH mRNA stability as described by Czyzyk-Krzeska and coworkers (Czyzyk-Krzeska et al., 1994; Czyzyk-Krzeska and Beresh, 1996; Paulding and Czyzyk-Krzeska, 1999) or decreased basal TH mRNA translation. Either way, these surprising results suggest that the cis-acting 3′-UTR sequences that post-transcriptionally control basal TH mRNA expression may differ from those that control cAMP-induced expression. This differentiation between 3′-UTR sequences that regulate basal versus induced mRNA expression has also been observed for gastrin mRNA (Lee et al., 2007).
Evidence Supporting the Role of PCBP2 in the Activation of TH mRNA Translation Elicited by cAMP in MN9D Cells.
The results of the UV cross-linking studies suggest that a ∼44-kDa protein is a major component of the specifically bound RNA-protein complex. This ∼44-kDa band is observed using proteins isolated from MN9D cells or PC12 cells, suggesting that a similar protein may be regulating TH mRNA stability and/or translation in these two model systems. In PC12 cells, hypoxia induces the cytoplasmic levels of PCBP1 (Paulding and Czyzyk-Krzeska, 1999) but not PCBP2. These observations have led to the hypothesis that hypoxia stabilizes TH mRNA in PC12 cells via increased the binding of PCBP1 to 3′-UTR polypyrimidine-rich sequences. It is plausible that the RNA-protein complexes observed in Figs. 2, 3, and 6 using PC12 proteins comprises PCBP1 and/or PCBP2, because both of these proteins are expressed in PC12 cells (Paulding and Czyzyk-Krzeska, 1999). In MN9D cells, cAMP induces PCBP2 but not PCBP1. The biotin pull-down assay demonstrates that PCPB2 binds to TH mRNA 3′-UTR sequences between the KpnI and ApaI sites, which is the area responsible for the cAMP response. Furthermore, the ribonucleoprotein pull-down assay provides evidence that PCBP2 binds to TH mRNA in intact MN9D cells. Finally, overexpression of full-length, unspliced PCBP2 increases TH protein levels without altering TH mRNA levels and enhances the expression of TH-luciferase activity but not TH-luciferase mRNA. These results provide strong evidence that induction of PCBP2 plays a role in the cAMP-mediated activation of TH mRNA translation.
The simplest model to explain this activation is that PCBP2 binds to TH mRNA, promoting its association with ribosomes and resulting in enhanced translation. The precise molecular mechanism(s) mediating this enhancement remain mysterious. Translational regulation usually occurs at the level of initiation, in which the best studied examples involve translational inhibition by modification of initiation factor phosphorylation and/or inhibition of initiation factor binding to the 43S ribosomal complex (Sonenberg and Hinnebusch, 2009). Inhibition of 60S ribosome joining to the 43S complex has also been reported; indeed, PCBP1 and hnRNP K control the translation of 15-lipoxygenase mRNA by this mechanism (Ostareck et al., 2001). The molecular mechanisms responsible for translational activation are not well developed. PCBP2 is known to enhance the translation of picornavirus mRNAs via an internal ribosome entry site (Walter et al., 2002; Kempf and Barton, 2008), but how this mechanism relates to the regulation of capped mRNA translation remains unclear. Stimulus-dependent activation of capped mRNAs is known to occur and is particularly important in the nervous system, in which activation of specific protein synthesis at synapses plays a role in neuroplasticity, memory, and learning (Costa-Mattioli et al., 2009; Richter and Klann, 2009). The molecular mechanisms responsible for this stimulus-dependent translational activation remain largely unknown and may be unique to each regulated mRNA, but probably involve phosphorylation of initiation factors, removal of inhibitory factors permitting access of activating initiation factors to the capped mRNA or the 43S preinitiation complex, interactions with miRNAs, and/or translocation of the mRNA from an intracellular storage granule to active ribosomes. Which, if any, of these mechanisms are implemented by PCBP2 binding to TH mRNA 3′-UTR remains to be resolved. It is also possible that PCBP2 overexpression or induction by cAMP leads to the induction or activation of another factor that promotes TH mRNA translation in conjunction with PCBP2 binding to TH mRNA 3′-UTR. More work is needed to understand the molecular mechanisms mediating this response.
A number of PCBP2 isoforms are observed in MN9D cells on Western blots. Cloning of PCBP2 cDNAs from MN9D cells reveals multiple mRNA isoforms resulting from alternative splicing. The major isoform expressed in MN9D cells seems to be the unspliced isoform (or isoforms lacking one to three amino acids, which would be indistinguishable from the unspliced form on a Western blot). A second isoform lacking the 31 amino acid and 1 amino acid spliced regions is also observed on the Western blots, but at a much reduced level. Both of these isoforms were induced by cAMP. Other mRNA isoforms were cloned, but their protein expression seems to be undetectable using our Western procedures (data not shown). Overexpression of the (−93, +31 − 1) PCBP2 isoform (PCBP2B in Fig. 8) induces TH protein, but not TH mRNA, which is identical with that seen with the unspliced isoform (PCBP2A). These results suggest that the region of PCBP2 encoded by these alternatively spliced sequences do not play an important role in this cAMP-mediated translational response. However, these regions may play some indeterminate role in regulating mRNA function, because this splice variant (PCBP2B) does not induce either luciferase activity or luciferase mRNA (Fig. 8D). The reason for the difference in the responses of TH expression and luciferase expression to PCBP2B remains unclear. It is possible that the 3′-UTR folds differently when fused to the luciferase gene compared with that which occurs in endogenous TH mRNA and that this difference prevents binding of PCBP2B to its cognate site or access to interacting proteins that mediate translation.
In summary, our results support a model in which cAMP induces PCBP2, which binds to the polypyrimidine-rich domain of the 3′-UTR of TH mRNA and promotes its translation in MN9D cells. This response seems to be cell type-specific, because it is not observed in PC12 or CATH.a cells (Chen et al., 2008). It is plausible that other mRNAs in dopaminergic neurons with polypyrimidine-rich domains in their 3′-UTRs may also be regulated by this mechanism. More work is required to determine the precise mechanisms by which PCBP2 binding mediates this translational response, by which cAMP induces PCBP2, what other cellular factors participate in this response, and what other neuronal mRNAs are regulated by this mechanism. A better understanding of these translational mechanisms may help to explain discrepancies between changes in TH mRNA and TH protein levels in response to different stimuli, such as stress, hypoxia, and long-term drug treatment (Kaneda et al., 1991; Nankova et al., 1994; Yoshimura et al., 2004; Gozal et al., 2005; Xu et al., 2007). It may also provide novel approaches to control TH expression in dopaminergic neurons in neurodegenerative diseases.
Acknowledgments
We greatly appreciate the gift of PCBP1 and PCBP2 antibodies from Dr. Maria Czyzyk-Krzeska (University of Cincinnati School of Medicine, Cincinnati, OH). We also thank Dr. Pinwei Huang for guidance in constructing the adenovirus expression vectors and performing the pull-down assays.
The work was supported by the National Institutes of Health National Institute on Drug Abuse [Grant DA05014] and the National Institutes of Health National Institute of Neurological Disorders and Stroke [Grant NS39415].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- TH
- tyrosine hydroxylase
- PCBP
- poly(C)-binding protein
- 8-CPT-cAMP
- 8-chlorophenylthio-cAMP
- PBS
- phosphate-buffered saline
- UTR
- untranslated region
- PCR
- polymerase chain reaction
- RT-PCR
- reverse transcription-polymerase chain reaction
- DEPC
- diethyl pyrocarbonate
- nt
- nucleotide
- Ad
- adenosine
- KA
- KpnI and ApaI sites
- AS
- ApaI and SphI
- bp
- base pair.
References
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254 [DOI] [PubMed] [Google Scholar]
- Chang MS, Sved AF, Zigmond MJ, Austin MC. (2000) Increased transcription of the tyrosine hydroxylase gene in individual locus coeruleus neurons following footshock stress. Neuroscience 101:131–139 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu L, Radcliffe P, Sun B, Tank AW. (2008) Activation of tyrosine hydroxylase mRNA translation by cAMP in midbrain dopaminergic neurons. Mol Pharmacol 73:1816–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HK, Won L, Roback JD, Wainer BH, Heller A. (1992) Specific modulation of dopamine expression in neuronal hybrid cells by primary cells from different brain regions. Proc Natl Acad Sci U S A 89:8943–8947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. (2009) Translational control of long-lasting synaptic plasticity and memory. Neuron 61:10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czyzyk-Krzeska MF, Beresh JE. (1996) Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J Biol Chem 271:3293–3299 [DOI] [PubMed] [Google Scholar]
- Czyzyk-Krzeska MF, Dominski Z, Kole R, Millhorn DE. (1994) Hypoxia stimulates binding of a cytoplasmic protein to a pyrimidine-rich sequence in the 3′-untranslated region of rat tyrosine hydroxylase mRNA. J Biol Chem 269:9940–9945 [PubMed] [Google Scholar]
- Fernández E, Craviso GL. (1999) Protein synthesis blockade differentially affects the degradation of constitutive and nicotinic receptor-induced tyrosine hydroxylase protein level in isolated bovine chromaffin cells. J Neurochem 73:169–178 [DOI] [PubMed] [Google Scholar]
- Funke B, Zuleger B, Benavente R, Schuster T, Goller M, Stévenin J, Horak I. (1996) The mouse poly(C)-binding protein exists in multiple isoforms and interacts with several RNA-binding proteins. Nucleic Acids Res 24:3821–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goller M, Funke B, Gehe-Becker C, Kröger B, Lottspeich F, Horak I. (1994) Murine protein which binds preferentially to oligo-C-rich single-stranded nucleic acids. Nucleic Acids Res 22:1885–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozal E, Shah ZA, Pequignot JM, Pequignot J, Sachleben LR, Czyzyk-Krzeska MF, Li RC, Guo SZ, Gozal D. (2005) Tyrosine hydroxylase expression and activity in the rat brain: differential regulation after long-term intermittent or sustained hypoxia. J Appl Physiol 99:642–649 [DOI] [PubMed] [Google Scholar]
- He DY, Ron D. (2008) Glial cell line-derived neurotrophic factor reverses ethanol-mediated increases in tyrosine hydroxylase immunoreactivity via altering the activity of heat shock protein 90. J Biol Chem 283:12811–12818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda N, Sasaoka T, Kobayashi K, Kiuchi K, Nagatsu I, Kurosawa Y, Fujita K, Yokoyama M, Nomura T, Katsuki M. (1991) Tissue-specific and high-level expression of the human tyrosine hydroxylase gene in transgenic mice. Neuron 6:583–594 [DOI] [PubMed] [Google Scholar]
- Kempf BJ, Barton DJ. (2008) Poly(rC) binding proteins and the 5′ cloverleaf of uncapped poliovirus mRNA function during de novo assembly of polysomes. J Virol 82:5835–5846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumer SC, Vrana KE. (1996) Intricate regulation of tyrosine hydroxylase activity and gene expression. J Neurochem 67:443–462 [DOI] [PubMed] [Google Scholar]
- Lee PT, Liao PC, Chang WC, Tseng JT. (2007) Epidermal growth factor increases the interaction between nucleolin and heterogeneous nuclear ribonucleoprotein K/poly(C) binding protein 1 complex to regulate the gastrin mRNA turnover. Mol Biol Cell 18:5004–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev AV, Liebhaber SA. (2002) The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA 8:265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamoto-Combs K, Piech KM, Best JA, Sun B, Tank AW. (1997) Tyrosine hydroxylase gene promoter activity is regulated by both cyclic AMP-responsive element and AP1 sites following calcium influx. Evidence for cyclic amp-responsive element binding protein-independent regulation. J Biol Chem 272:6051–6058 [DOI] [PubMed] [Google Scholar]
- Nankova B, Kvetnanský R, McMahon A, Viskupic E, Hiremagalur B, Frankle G, Fukuhara K, Kopin IJ, Sabban EL. (1994) Induction of tyrosine hydroxylase gene expression by a nonneuronal nonpituitary-mediated mechanism in immobilization stress. Proc Natl Acad Sci U S A 91:5937–5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW. (2001) Lipoxygenase mRNA silencing in erythroid differentiation: the 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell 104:281–290 [DOI] [PubMed] [Google Scholar]
- Osterhout CA, Sterling CR, Chikaraishi DM, Tank AW. (2005) Induction of tyrosine hydroxylase in the locus coeruleus of transgenic mice in response to stress or nicotine treatment: lack of activation of tyrosine hydroxylase promoter activity. J Neurochem 94:731–741 [DOI] [PubMed] [Google Scholar]
- Paulding WR, Czyzyk-Krzeska MF. (1999) Regulation of tyrosine hydroxylase mRNA stability by protein-binding, pyrimidine-rich sequence in the 3′-untranslated region. J Biol Chem 274:2532–2538 [DOI] [PubMed] [Google Scholar]
- Richter JD, Klann E. (2009) Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev 23:1–11 [DOI] [PubMed] [Google Scholar]
- Sabban EL, Kvetnanský R. (2001) Stress-triggered activation of gene expression in catecholaminergic systems: dynamics of transcriptional events. Trends Neurosci 24:91–98 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136:731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Chen X, Xu L, Sterling C, Tank AW. (2004) Chronic nicotine treatment leads to induction of tyrosine hydroxylase in locus coeruleus neurons: the role of transcriptional activation. Mol Pharmacol 66:1011–1021 [DOI] [PubMed] [Google Scholar]
- Waggoner SA, Liebhaber SA. (2003) Identification of mRNAs associated with alphaCP2-containing RNP complexes. Mol Cell Biol 23:7055–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter BL, Parsley TB, Ehrenfeld E, Semler BL. (2002) Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J Virol 76:12008–12022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong DL, Tank AW. (2007) Stress-induced catecholaminergic function: transcriptional and post-transcriptional control. Stress 10:121–130 [DOI] [PubMed] [Google Scholar]
- Xu L, Chen X, Sun B, Sterling C, Tank AW. (2007) Evidence for regulation of tyrosine hydroxylase mRNA translation by stress in rat adrenal medulla. Brain Res 1158:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura R, Xu L, Sun B, Tank AW. (2004) Nicotinic and muscarinic acetylcholine receptors are essential for the long-term response of tyrosine hydroxylase gene expression to chronic nicotine treatment in rat adrenal medulla. Mol Brain Res 126:188–197 [DOI] [PubMed] [Google Scholar]