Abstract
HIV-1 integrase (IN) is a validated target for developing antiretroviral inhibitors. Using affinity acetylation and mass spectrometric (MS) analysis, we previously identified a tetra-acetylated inhibitor (2E)-3-[3,4-bis(acetoxy)phenyl]-2-propenoate-N-[(2E)-3-[3,4-bis(acetyloxy)phenyl]-1-oxo-2-propenyl]-l-serine methyl ester; compound 1] that selectively modified Lys173 at the IN dimer interface. Here we extend our efforts to dissect the mechanism of inhibition and structural features that are important for the selective binding of compound 1. Using a subunit exchange assay, we found that the inhibitor strongly modulates dynamic interactions between IN subunits. Restricting such interactions does not directly interfere with IN binding to DNA substrates or cellular cofactor lens epithelium-derived growth factor, but it compromises the formation of the fully functional nucleoprotein complex. Studies comparing compound 1 with a structurally related IN inhibitor, the tetra-acetylated-chicoric acid derivative (2R,3R)-2,3-bis[[(2E)-3-[3,4-bis(acetyloxy)phenyl]-1-oxo-2-propen-1-yl]oxy]-butanedioic acid (compound 2), indicated striking mechanistic differences between these agents. The structures of the two inhibitors differ only in their central linker regions, with compounds 1 and 2 containing a single methyl ester group and two carboxylic acids, respectively. MS experiments highlighted the importance of these structural differences for selective binding of compound 1 to the IN dimer interface. Moreover, molecular modeling of compound 1 complexed to IN identified a potential inhibitor binding cavity and provided structural clues regarding a possible role of the central methyl ester group in establishing an extensive hydrogen bonding network with both interacting subunits. The proposed mechanism of action and binding site for the small-molecule inhibitor identified in the present study provide an attractive venue for developing allosteric inhibitors of HIV-1 IN.
Recent development of the first clinically useful strand-transfer inhibitor (STI) Raltegravir (MK-0518) has validated HIV-1 integrase (IN) as a important antiretroviral target (Summa et al., 2008). The enzyme functions as a multimer to insert the reverse-transcribed RNA genome into the host chromosome through two reaction steps. In the first step, termed 3′-processing, IN cleaves a GT dinucleotide from each end of the viral DNA. In the second step, termed strand transfer, IN catalyzes concerted integration of the processed viral DNA ends into chromosomal DNA. Studies with purified recombinant protein and model DNA substrates indicated that the individual protein monomers establish complementary contacts with DNA substrates with the subunit-subunit contacts playing a crucial role in the formation of the functional nucleoprotein complexes (Engelman et al., 1993; van Gent et al., 1993; van den Ent et al., 1999; Zhao et al., 2008). Although a dimeric protein is sufficient to process each 3′-end, a tetramer is needed to carry out the concerted integration of both viral ends (Faure et al., 2005; Guiot et al., 2006; Li et al., 2006).
After the discovery of the transcriptional coactivator lens epithelium-derived growth factor (LEDGF) as a key cellular cofactor for HIV-1 integration, LEDGF-IN interactions have become a new venue for antiviral drug design (De Rijck et al., 2006; Al-Mawsawi et al., 2008; Hou et al., 2008; Busschots et al., 2009). LEDGF directly interacts with IN through a C-terminal region termed the integrase binding domain (IBD) (Cherepanov et al., 2004; Vanegas et al., 2005) and tethers the preintegration complex to chromatin (Maertens et al., 2003; Emiliani et al., 2005; Llano et al., 2006b; Shun et al., 2007). Overexpression of the IBD, which tightly binds IN but lacks an N-terminal nuclear localization signal or the chromatin binding domain of LEDGF, severely inhibits HIV-1 replication in cell culture assays (De Rijck et al., 2006). Furthermore, HIV-1 strains resistant to the STI were fully susceptible to inhibition by the IBD (De Rijck et al., 2006). It is intriguing that the IBD is significantly more effective at suppressing HIV-1 replication in LEDGF-deficient cells (555-fold) compared with cells containing LEDGF at normal levels (∼30-fold) (Llano et al., 2006a). These observations suggest that in the absence of competing LEDGF levels, the IBD could effectively engage IN and adversely affect its function.
We recently observed that the high exchange rate among IN subunits in multimers can be reduced by IBD binding (McKee et al., 2008). The preformed IN-IBD complex is capable of binding donor DNA but fails to catalyze concerted integration (McKee et al., 2008). These findings suggested that the dynamic interplay between IN subunits is essential for the assembly of the fully functional nucleoprotein complex and that restricting the molecular movement of individual subunits within a multimer could compromise catalytic processes. Consistent with this, IBD-derived peptides have been shown to stabilize a multimeric form of IN and impair HIV-1 infection (Hayouka et al., 2007; Al-Mawsawi et al., 2008).
Although studies with the IBD have provided proof-of-concept for targeting the dynamic structure of free IN, a focus of developmental efforts is to discover small-molecule inhibitors that can modulate the subunit-subunit interactions. In this regard, small-molecule inhibitors that have been shown previously to selectively bind at the IN dimer interface are worth revisiting. For example, 3,4-dihydroxyphenyltriphenylarsonium bromide and a coumarin-based inhibitor have been reported to bind IN at sites that partly overlap the IBD binding pocket (Molteni et al., 2001; Al-Mawsawi et al., 2006). Using affinity acetylation and MS analysis, we previously mapped one contact between the small-molecule inhibitor (2E)-3-[3,4-bis(acetoxy)phenyl]-2-propenoate-N-[(2E)-3-[3,4-bis(acetyloxy)-phenyl]-1-oxo-2-propenyl]-l-serine methylester (compound 1; Fig. 1) and Lys173 located at the IN dimer interface (Shkriabai et al., 2004). However, the mechanisms of action of this and similar compounds have not been elucidated.
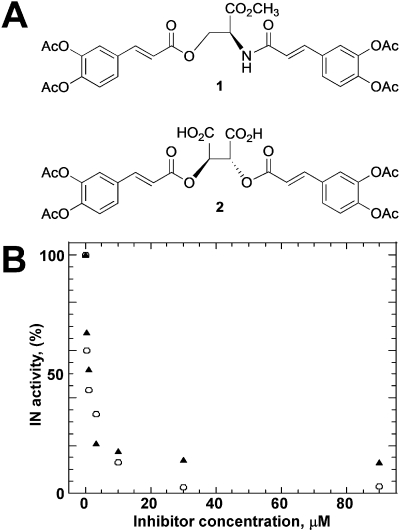
Fig. 1.
A, structures of compounds 1 and 2. B, the inhibition profiles of recombinant wild-type IN with compounds 1 (▲) and 2 (○). The mean values for at least three experiments are presented. The deviation for each measurement was ±10%.
In the present study, we extend our previous research with compound 1. Using a subunit exchange assay, we have observed that the inhibitor modulates dynamic interactions between IN subunits in a dose-dependent fashion. Furthermore, we have found that the central linker region of 1 is essential for selective binding of the inhibitor to the IN dimer interface. Moreover, molecular modeling experiments have enabled us to propose contact amino acids and define a potential small-molecule binding cavity within HIV-1 IN, which may represent a new therapeutic target.
Materials and Methods
Preparation of Wild-Type and Mutant Recombinant HIV-1 IN.
Full-length wild-type and soluble (F185K/C280S) integrase proteins were expressed in Escherichia coli. The point mutations were introduced in the wild-type IN sequence using a polymerase chain reaction mutagenesis kit (QIAGEN, Valencia, CA). Wild-type and mutant IN proteins were purified according to procedures described previously (McKee et al., 2008).
Assay for HIV-1 IN Inhibitors.
Compounds 1 and 2 were prepared as reported previously (Lin et al., 1999). MALDI-ToF analysis confirmed successful synthesis and a high degree of purity of the full-length products (Supplemental Fig. 1). The preparations of compound 1 and 2 were dissolved in dimethyl sulfoxide to make 10 mM stock solutions and were stored at −20°C until use. The compounds were highly stable throughout the course of our studies.
Measurement of in vitro IC50 values was carried out as described previously (Hazuda et al., 1994). In brief, a short double-stranded DNA (a hybrid of the 21- and 19-oligomer synthetic oligonucleotides) representing the preprocessed HIV-1 U5 LTR end was immobilized on a CovaLink microtiter plate (NUNC/Thermo Fisher Scientific, Rochester, NY) via carbodiimide-mediated condensation. After incubation with IN, a biotinylated target oligonucleotide was added, and the strand transfer reaction was allowed to proceed. The plates were washed thoroughly and blocked before incubation with alkaline phosphatase-conjugated streptavidin (Pierce, Rockford, IL). After additional washing, para-nitrophenylphosphate was added, and the development of p-nitrophenolate was determined spectrophotometrically at 405 nm.
Mass Spectrometric Analyses.
The acetylation reactions for short control peptides and recombinant IN have been described previously (Shkriabai et al., 2004). In brief, 5 μM peptides were incubated with 500 μM compound 1 or 2 for 30 min at 37°C, and the reaction products were analyzed with MALDI-ToF using an Axima CFR instrument (Shimadzu Biotech, Manchester, UK) and the α-cyano-4-hydroxy-cinnamic acid matrix. To identify the sites in HIV-1 IN modified by 1 and 2, the protein was first incubated with increasing concentrations of inhibitors and then digested by trypsin, and the proteolytic fragments were analyzed with Shimadzu MALDI-ToF and Waters quadrupole/time of flight-II instruments (Manchester, UK) as described previously (Shkriabai et al., 2004).
DNA-IN Interactions.
Disulfide-mediated IN-DNA cross-linking has been described previously (Zhao et al., 2008). The assays used IN(E152C) protein and a 21-oligomer specific DNA with a cross-linkable analog at the G2 position (Zhao et al., 2008 provides detailed preparation of the protein and DNA). The cross-linking reactions were performed by incubating 1 μM IN with equimolar DNA duplex in 50 mM HEPES, pH 7.0, 100 mM NaCl, 5 mM MgCl2, and 10% glycerol at 37°C for 20 min. The reactions were quenched by the addition of 20 mM methyl methanethiosulfonate and subjected to SDS-PAGE analysis. Free protein and protein-DNA complexes were visualized by Western blots using IN antiserum.
Assay for IN Subunit Exchange.
A subunit exchange assay was performed as described previously (McKee et al., 2008). In brief, histidine-tagged IN (50 nM) was preincubated in pull-down (PD) buffer [50 mM HEPES, pH 7.1, 300 mM NaCl, 100 mM imidazole, 2 mM MgCl2, 2 mM β-mercaptoethanol, and 0.1% (v/v) Nonidet P-40] for 30 min at room temperature with and without increasing amounts of ligands (1 or 2). An equimolar amount of tag-free IN (50 nM) was then added, and subunit exchange was allowed for 60 min at room temperature. Samples were briefly centrifuged for 2 min at 1000g to remove nonspecific aggregates. Supernatants were incubated with nickel-agarose beads (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) for 30 min in the presence of bovine serum albumin (0.1 mg/ml). After the affinity pull-down of protein-protein complexes, the resin was washed three times with the PD buffer and boiled in SDS-PAGE loading buffer. Histidine-tagged and tag-free INs were then separated by SDS-PAGE and visualized by Western blots using a mouse monoclonal IN antibody (from the AIDS Research and Reference Reagent Program, National Institutes of Health, Bethesda, MD).
IN Cross-Linking Assay.
The IN cross-linking assay was performed as described previously (Li et al., 2006). In brief, IN (100 nM) was preincubated in buffer [50 mM HEPES, pH 7.1, 300 mM NaCl, 2 mM MgCl2, 1 mM dithiothreitol, and 0.1% (v/v) Nonidet P-40] for 30 min at room temperature with or without compound 1. The bifunctional reagent BS3 (Pierce) was then added at 100 μM final concentration, and the mixture was incubated for 15 min at room temperature. Reactions were quenched by the addition of Tris/NaOAc and boiled in the SDS-PAGE loading buffer. INs were separated by SDS-PAGE and visualized by Western blot using a mouse monoclonal IN antibody [HIV-1 IN Monoclonal Antibody (2C11) from Dr. Dag E. Helland obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health].
Protein Solubility Test.
IN (100 nM) was preincubated in PD buffer [50 mM HEPES, pH 7.1, 300 mM NaCl, 100 mM imidazole, 2 mM MgCl2, 2 mM β-mercaptoethanol, 0.1% (v/v) Nonidet P-40] for 60 min at room temperature with or without 250 μM ligand (compound 1 or 2). Samples were then centrifuged for 10 min at 5000g. Pellets and supernatants were analyzed by SDS-PAGE and detected by Western blot as described above.
Monitoring the Effects of Compounds on IN-LEDGF Interactions.
LEDGF-IN binding assays were performed as described previously (McKee et al., 2008). In brief, histidine-tagged IN (100 nM) was preincubated in PD buffer for 30 min at room temperature with and without increasing amounts of ligands (compounds 1 or 2). Tag-free LEDGF (300 nM) was then added and incubated for 60 min at room temperature. Samples were centrifuged and pulled down by a nickel-agarose resin as described above. The bound proteins were subjected to SDS-PAGE separation, and LEDGF was visualized by Western blot using a mouse monoclonal LEDGF antibody (BD Biosciences, San Jose, CA).
LEDGF Binding to Wild-Type and Mutant IN.
LEDGF (1 μM) was incubated with 600 nM histidine-tagged IN (wild type or mutant) in binding buffer [50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 35 mM imidazole, 2 mM β-mercaptoethanol, and 0.1% (v/v) Nonidet P-40] for 60 min at room temperature. Samples were pulled down by a nickel-affinity resin for 30 min in the presence of bovine serum albumin (0.1 mg/ml). The resin was washed three times with the same buffer, and the bound proteins were subjected to SDS-PAGE separation and visualized by Coomassie blue staining.
Size-Exclusion Chromatography.
Protein solutions (10 μM) in storage buffer (50 mM HEPES, pH 7.4, 1 M NaCl, 7.5 mM CHAPS, 10 mM dithiothreitol, and 10% glycerol) were subjected to size-exclusion chromatography using a Superdex 200 10/300 GL column (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) and running buffer (50 mM HEPES, pH 7.4, 1 M NaCl, and 10% glycerol). The column was calibrated with the following proteins: conalbumin (75 kDa), carbonic anhydrase (29 kDa), ribonuclease A (13.7 kDa), and aprotinin (6.5 kDa). Proteins were detected by absorbance at 280 nm.
Molecular Docking Studies.
All simulations were performed on a Silicon Graphics O2 workstation (SGI, Mountain View, CA). The coordinates of the catalytic core domain (CCD) of IN were extracted from the crystal structure 2B4J (Cherepanov et al., 2005) obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (available at http://www.rcsb.org/pdb/home/home.do). The initial three-dimensional model of compound 1 was generated using the biopolymer module within the Insight II software package (Accelrys Inc., San Diego, CA). Molecular docking studies were performed using the Autodock program (version 3.0), which allows virtual docking of a fully flexible ligand within a rigid defined binding pocket (Goodsell and Olson, 1990). A “target grid” covering both putative binding cavities (including Lys173, His171, Asp167, Trp131, and Thr125 residues) was generated. Polar hydrogens were added, Kollman charges were assigned, and three-dimensional affinity grid maps were calculated for each atom type [C, A (aromatic C), N, O, S, and H] using the Autogrid module. For the ligand, hydrogens were added, Gasteiger charges were assigned, and the rotatable bonds were determined by the AutoTors module. The docking parameters were as follows: genetic algorithm, trials of 50 dockings, random starting position and conformation, rotation step ranges of 5°, and 1 million energy evaluations. The Autodock pose showing the lowest binding energy was extensively energy-minimized using the Discover module of Insight II software using the CFF91 force field and the steepest descent method. Both protein and ligand were left flexible, and no constraints were applied.
Results
Compounds 1 and 2 (Fig. 1A) have been synthesized previously (Lin et al., 1999) as structural analogs of chicoric acid (CA) (Supplemental Fig. 2). Compound 2 represents tetra-acetylated CA. Compound 1 also contains di-O-tetra-acetyloxycinnamoyl groups, but it differs significantly from 2 and CA in the central linker region. For example, 1 possesses a noncharged methyl ester group, whereas 2 and CA contain two polar carboxylic acids (Fig. 1). Despite these structural differences, the compounds inhibited IN with similar IC50 values (Lin et al., 1999) (Fig. 1B), and they were assumed to exhibit similar mechanisms of action. However, more recent studies have suggested that 1 may differ from authentic CA compounds in its interactions with IN. For example, using affinity acetylation coupled with MS analysis, we have demonstrated that 1 selectively acetylates Lys173 located at the protein dimer interface (Shkriabai et al., 2004) (Fig. 2). In contrast, CA has been reported to interact at the IN active site (Healy et al., 2009), which is significantly distanced from the dimer interface.
Fig. 2.
Segments of quadrupole/time of flight mass spectra demonstrating selective acetylation of Lys173 with 1. A, IN + 6.25 μM 1; B, IN + 25 μM 1; C, IN + 10 μM 2; D, IN + 200 μM 2; E, free IN. The tryptic peptide of IN containing acetylated Lys173 is indicated. C1 is unmodified tryptic peptide (AMASDFNLPPVVAK) of IN, which serves as an internal control.
To clarify these differences, we compared compounds 1 and 2 for their interactions with HIV-1 IN. We chose compound 2 as a representative CA for the following reasons. The detailed in vitro IN activity and cell culture infectivity assays indicated that 2 and hydroxylated CA exhibit the same mechanism of action (Lin et al., 1999; Pluymers et al., 2000). Furthermore, in common with 1, compound 2 contains di-O-acetyl groups that would enable us to compare interactions of the two compounds with IN using the affinity acetylation and MS analysis approach (Shkriabai et al., 2004).
Using model peptides, we have demonstrated previously that aryl di-O-acetyl compounds could effectively acetylate lysine, cysteine, and tyrosine side chains (Shkriabai et al., 2004). Therefore, to compare acetylation reactions for 1 and 2, we used a short control peptide (HDMNKVLDL) containing a single lysine residue. As expected, no acetylation was detected at low compound concentrations (5 μM) because of the lack of specific interactions. In contrast, 500 μM 1 and 2 effectively acetylated the target lysine residue (data not shown). Comparison of the modified peak intensities indicated that aryl di-O-acetyl groups in the tested compounds exhibit very similar chemical reactivities.
In contrast, a striking difference was observed between compounds 1 and 2 in their interactions with HIV-1 IN (Fig. 2). Compound 1 at concentrations similar to its IC50 value effectively acetylated Lys173, whereas compound 2 failed to modify this residue even at very high concentrations (200 μM) (Fig. 2). No specific acetylation of IN was detected with low micromolar concentrations of 2, suggesting that in the protein-inhibitor complex, di-O-acetyl groups were not positioned close enough to react with the side chains of lysine, cysteine, or tyrosine residues. At elevated concentrations (200 μM) of 2, multiple lysine residues were acetylated. This could be due to nonspecific modes of modifying surface residues. To test this hypothesis, we performed control experiments using 500 μM concentration of primary amine modifying reagent sulfo-N-hydroxysuccinimide-biotin. The modification profiles obtained with 2 and sulfo-N-hydroxysuccinimide-biotin were very similar, indicating that the affected residues were indeed targeted for their surface exposure rather than for specific interactions. Thus, our experiments did not reveal the specific binding site for 2 but indicated marked differences in 1 and 2 binding to IN.
Further evidence for different mechanisms of actions for 1 and 2 emerged from IN-DNA binding studies. The results in Fig. 3 show that 2 interfered with IN-DNA complex formation in a dose-dependent manner. These results are consistent with a recently proposed molecular model (Healy et al., 2009), in which the CA binding site partly overlaps with the presumed donor DNA binding cleft. In contrast, 1 did not directly interfere with IN-DNA interactions, suggesting an alternative mechanism of action for this compound.
Fig. 3.
Effects of 1 and 2 on IN-DNA cross-linking. IN(E152C) was first incubated with increasing concentrations of 1 (A) and 2 (B) and then cross-linked to a specific DNA containing a cross-linkable analog at the G2 position (G2). The reaction products were resolved by SDS-PAGE. The bands corresponding to free IN and IN-DNA complex are indicated. A, lane 1 molecular mass markers; lane 2, IN-DNA cross-linking in the absence of 1. Lanes 3 to 10 contained increasing concentrations of 1 (1, 2, 4, 8, 16, 32, 64, and 128 μM). B, lane 1, molecular mass markers; lane 2, IN-DNA cross-linking in the absence of 2; lanes 3 to 9 contained increasing concentrations of 2 (2, 4, 8, 16, 32, 64, and 128 μM).
Further experiments were performed to elucidate a mechanism of action for 1 (Figs. 4 and 5) and define the inhibitor binding site in more detail (Figs. 6 and 7). Given that 1 selectively interacts with Lys173 located at the protein dimer interface (Fig. 2), we examined whether 1 could affect the dynamic interactions between IN subunits. For this purpose, we used the subunit exchange assay devised by our group (McKee et al., 2008) (Fig. 4A). This method monitors kinetic interactions between the two wild-type proteins: histidine-tagged IN (indicated as IN2 in the monomeric state and IN2–IN2 in the dimeric state), and tag-free IN (indicated as IN1 in the monomeric state and IN1–IN1 in the dimeric state). Because of the effective exchange between individual protein subunits, the tag-free protein within the IN1-IN2 complex can be quantitatively pulled down by a nickel-affinity resin. To reveal whether the compounds can modulate these interactions, IN2-IN2 was first preincubated with increasing concentrations of 1 or 2 and then exposed to IN1-IN1. The data in Fig. 4B (lanes 5–10) show that 1 impaired dynamic exchange between IN subunits in dose-dependent manner. In contrast, 2 tested at high concentrations had no detectable effect on IN subunit-subunit exchange (Fig. 4B, lanes 11 and 12).
Fig. 4.
Effects of 1 and 2 on IN subunit-subunit interactions. Experimental design (A) and results (B). A, the subunit exchange between IN multimers was tested by mixing the two wild-type IN proteins: IN1-IN1, a tag-free form, and IN2-IN2, containing the histidine tag at its C terminus. The full-length proteins are depicted as dimers (IN1–IN1) and (IN2–IN2). Upon subunit-subunit exchange, three IN populations can be formed: IN1–IN1, IN1–IN2, and IN2–IN2. Of these, IN2–IN2 and IN1–IN2 can be pulled down by nickel-affinity resin through binding with the histidine tag, whereas the tag-free IN1–IN1 is washed out. B, the IN1 and IN2 proteins from the bound complexes were subjected to SDS-PAGE and detected by Western blot. Lane 1, IN2 load; lane 2, IN1 load; lane 3, no ligand or IN1 (non specific pull-down control); lane 4, the subunit-subunit exchange in the absence of compounds; lanes 5 to 10, the subunit exchange reactions in the presence of increasing 1 concentrations (8, 16, 32, 64, 128, and 256 μM); lanes 11 and 12, reactions in the presence of 128 and 256 μM 2. C, order-of-addition experiments. Lanes 1 and 3, subunit exchange reactions in the absence of compounds; lane 2, IN2-IN2 was first preincubated with 1 and then exposed to IN1-IN1; lane 4, IN2-IN2 and IN1-IN1 were first mixed to carry out subunit exchange, and then 1 was added to the mixture.
Fig. 5.
Effects of 1 on IN multimerization (A), solubility (B), and LEDGF-IN binding (C). A, in parallel reactions, free IN (lane 1) and the IN-1 complex (lane 2) were subjected to cross-linking with BS3, and the reaction products were separated by SDS-PAGE. Migrations of molecular mass markers and IN bands are indicated. B, lane 1, molecular mass markers; lane 2, total (T) sample before centrifugation; lanes 3 and 4, supernatant (S) and precipitate (P) fractions formed after centrifugation of IN in the presence of 10% dimethyl sulfoxide; lanes 5 and 6, IN solubility in the presence of 256 μM 1. C, lane 1, LEDGF input; lane 2, no ligand or IN (nonspecific pull down control); lane 3, LEDGF and IN interactions in the absence of 1; lanes 4 to 8, assays with increasing 1 concentrations (8, 16, 32, 64, and 128 μM).
Fig. 6.
Molecular docking studies for 1. A and B, two potential binding pockets (colored in magenta and orange) at the CCD dimer interface. These distinct sites are located immediately adjacent to one another. A and B, two different views of the CCD dimer to better visualize individual binding pockets. Individual subunits of IN are colored green and yellow. The location of Lys173 and height (H), depth (D), and width (W) for each cavity is indicated. C, the space-filling model for the CCD dimer-1 complex showing that the inhibitor simultaneously interacts with both IN monomers (green and yellow). The central methyl ester group is deeply buried in the cavity and cannot be seen in this picture. Integrase active site residues (Asp64, Asp116, and Glu152) on each monomer are shown in red. D, key interactions with IN residues established by the central methyl ester group and the linker region. The amino acids from green or yellow subunits are colored accordingly.
Fig. 7.
Biochemical analysis of IN mutants. A, size-exclusion chromatography of wild-type and mutant proteins. Peaks corresponding to tetramer (Tet) IN with estimated molecular masses of ∼111 kDa and a dimeric (Dim) protein with estimate molecular masses of ∼54 kDa, are indicated. B, effects of amino acid substitutions on recombinant IN activities. Top image depicts strand transfer activities. Positions of 21-oligomer substrate (21-S) and reaction products (STP) are indicated. Bottom image displays 3′-processing activities. The positions of 21-S and specific 19-oligomer products (19-P) are shown. B, LEDGF binding to wild-type and mutant INs: lane 1, molecular mass markers; lane 2, LEDGF input; lane 3, assay without IN (nonspecific pull-down control); lane 4, assay with wild-type IN; lane 5, assay with E87A mutant; lane 6, assay with E96A mutant; lane 7, assay with Y99A mutant; lane 8, assay with K103A mutant.
Dynamic modulation of subunit-subunit exchange can be achieved either through 1 stabilizing the interacting IN monomers into a multimeric state or by preventing multimer formation through interference with subunit-subunit assembly. To differentiate between these possibilities, order-of-addition experiments were performed (Fig. 4C). Preincubation of 1 with IN2-IN2 and subsequent addition of IN1-IN1 to the reaction mixture effectively interfered with the subunit-subunit exchange (Fig. 4C, lane 2). However, 1 did not significantly affect the preassembled IN1–IN2 complex (Fig. 4C, lane 4). These findings suggest that 1 does not dissociate IN multimers. Instead, 1 binds to multimeric IN and restricts the ability of interacting subunits to exchange freely among multimers.
The effects of 1 on IN multimerization were also tested using bifunctional cross-linkers and SDS-PAGE separation (Fig. 5A). This approach has been instrumental in probing IN structures in the synaptic complex and infected cells (Cherepanov et al., 2003; Li et al., 2006). As expected (Li et al., 2006), different multimeric species were observed with the free protein. Upon the addition of 1, the intensity of the monomeric band was significantly reduced. Similar quantities of dimeric IN bands were observed in free IN and the IN-1 complex samples. Whereas a band corresponding to a tetramer was enhanced at least by 50% in the protein-inhibitor complex compared with free IN. The observed cross-linking patterns (Fig. 5A) were highly reproducible. Because in the 1-IN complex, a monomer band was reduced and multiple oligomeric species were detected, it is logical to suggest that the inhibitor promotes protein multimerization in general rather than stabilizes one particular multimeric form of IN. For example, 1 could affect conformations of interacting subunits so that the 1-IN complex is less flexible (Fig. 4) and more susceptible to multimerization (Fig. 5A) than free IN.
Next, we examined whether 1-induced protein multimerization could affect IN solubility. Figure 5B shows that under our assay conditions, free IN remained in solution and that 1 did not alter IN solubility. Given that LEDGF binds at IN dimer interface adjacent to Lys173, we checked whether 1 could affect the IN-LEDGF complex formation. The results in Fig. 5C indicate that 1 did not significantly affect protein-protein interactions indicating that the primary binding sites of 1 and LEDGF differ.
The following efforts were devoted to a more detailed characterization of the sites of 1 binding to IN. Specific acetylation of Lys173 provided an important clue regarding one of the contacts between the terminal di-O-acetyl group of the inhibitor and the protein. However, comparative SAR studies of 1 and 2 revealed an essential role of the central linker region for determining binding and mechanistic properties of these inhibitors. Indeed, 1 containing the methyl ester group within the linker interacted with the IN dimer and modulated dynamic interactions between the subunits (Figs. 2 and 4), but it did not directly affect IN-DNA interactions (Fig. 3). In contrast, 2 possessing two central carboxylic acid groups failed to bind to the IN dimer interface (Fig. 2) or modulate subunit-subunit interactions (Fig. 4). Instead, 2 interfered with the IN-DNA complex formation (Fig. 3).
To identify additional contact amino acids contributing to specific binding of 1 to the protein dimer interface, we used molecular modeling. Although full-length IN has not been amenable to high-resolution structural studies, atomic structures of individual protein domains are available. Lys173 is located in the CCD. Crystallographic studies indicated a dimeric organization of this domain (Dyda et al., 1994). The CCD contains the DDE motif coordinating the catalytic divalent metals and an IBD binding site (Fig. 6). Analysis of the protein surface surrounding Lys173 revealed two adjacent pockets depicted in orange and magenta (Fig. 6, A and B). The pocket in orange has been shown to tightly interact with the LEDGF (Cherepanov et al., 2005; Hare et al., 2009). Furthermore, the two small-molecule inhibitors (3,4-dihydroxyphenyltriphenylarsonium bromide and a coumarin-based compound) have also been demonstrated to target this site (Molteni et al., 2001; Al-Mawsawi et al., 2006). In contrast, the similarly sized cavity depicted in magenta has not been implicated previously in small-molecule binding. It is noteworthy that Lys173 is located at the boundary separating these distinct pockets (Fig. 6 A and B). Therefore, 1 could potentially target either of the two similarly sized cavities.
To delineate between interactions at the two possible binding sites (magenta and orange), docking simulations were performed using the Autodock software and a “target grid” covering both putative binding cavities on the dimeric CCD structure (see virtual docking details under Materials and Methods). Table 1 shows the results for the 10 lowest binding energy, which indicate a striking preference for 1 binding to the site represented by “magenta” cavity (Fig. 6). The lowest docking pose (−5.30 kcal/mol) was further minimized using Insight II software (see Materials and Methods).
TABLE 1.
Binding energy results for docking 1 in the CCD dimer
| Conformation | Binding Energy | Cavity Selected |
|---|---|---|
| kcal/mol | ||
| 1 | −5.30 | Magenta |
| 2 | −4.34 | Magenta |
| 3 | −3.81 | Magenta |
| 4 | −3.25 | Magenta |
| 5 | −2.69 | Magenta |
| 6 | −2.69 | Magenta/orange |
| 7 | −2.64 | Magenta |
| 8 | −2.09 | Magenta/orange |
| 9 | −1.49 | Magenta |
| 10 | −1.42 | Magenta |
The resulting molecular model for the CCD dimer-1 complex is shown in Fig. 6, C and D. The inhibitor interacts with both IN monomers. One di-O-acetyl group of 1 is located at 3.8 Å within Lys173 (subunit yellow), a distance compatible with the specific acetylation of this position observed in the MS experiments (Fig. 2). The central methyl ester group of the inhibitor is buried deeply within the cavity where it binds to IN through tight electrostatic and hydrophobic interactions (Fig. 6, C and D). IN amino acids engaging the central linker region include side chains of Glu87, Glu96, Tyr99, and Lys103. These interactions could explain the importance of the linker region for the selective binding of 1 to the IN dimer.
To examine significance of these residues for IN function and 1 binding, we prepared purified recombinant proteins containing the following point substitutions: E87A, E96A, Y99A, and K103A in the wild-type IN sequence. Size-exclusion chromatography results in Fig. 7A show that these mutations affected IN multimerization. For example, a peak corresponding to a tetrameric protein was reduced in all mutant proteins compared with wild-type IN. We also examined acetylation of the mutant proteins with 1. In sharp contrast with wild-type IN, Lys173 was not acetylated in any of the four mutant proteins, even in the presence of elevated concentrations (200 μM) of 1 (data not shown). These experiments, however, did not fully delineate whether Glu87, Glu96, Tyr99, and Lys103 directly or indirectly contribute to 1 binding to IN as their substitutions affected IN multimerization. Nevertheless, our findings highlight the importance of a correctly assembled IN multimer for selective binding of 1. Figure 7B shows that all of the mutant proteins were catalytically inactive. Furthermore, E87A failed to bind LEDGF, and the E96A, Y99A, and K103A substitutions reduced IN affinity for the cellular cofactor (Fig. 7C). Taken together, the mutagenesis studies highlighted the architectural importance of this newly identified small-molecule binding cavity for IN function and provide proof-of-concept for targeting this site with allosteric inhibitors.
Discussion
Here we show for the first time that a small-molecule compound inhibits HIV-1 IN catalytic function through allosteric modulation of dynamic interactions between the individual protein subunits (Fig. 4). The highly dynamic nature of free IN subunits is essential for the productive assembly of the fully functional IN-vDNA complex (McKee et al., 2008). Binding of 1 to free IN stabilizes the interacting subunits (Fig. 4) and promotes protein multimerization (Fig. 5A). The preformed protein-protein complex is capable of binding DNA substrates (Fig. 3) because 1 does not seem to directly mask the substrate binding channel in the retroviral enzyme (Figs. 2 and 6). Instead, 1 could compromise IN catalytic activities by the following allosteric mechanisms. The restricted flexibility of the preformed 1-IN complex may impede correct positioning of the catalytic site on the cognate DNA substrate. Alternatively, 1 could limit a conformational flexibility of IN, which is needed for the formation of the fully functional nucleoprotein complex (Zhao et al., 2008).
Proposed mechanisms of action for 1 are reminiscent of modulation of IN structure and function by LEDGF and its IBD. For example, in vitro experiments have indicated that the cellular cofactor effectively impairs IN subunit-subunit exchange and promotes IN tetramerization (McKee et al., 2008). The preassembled protein-protein complexes are capable of binding donor DNA but are defective for the concerted integration (Pandey et al., 2007; Raghavendra and Engelman, 2007; McKee et al., 2008). These findings have suggested that IN tetramers formed in IN-LEDGF and IN-vDNA complexes are not identical. In contrast, the full-length cofactor stimulates the catalytic reactions when IN-vDNA complexes are preformed and then LEDGF is provided (Pandey et al., 2007; Raghavendra and Engelman, 2007; Hare et al., 2009). These in vitro observations, highlighting the importance of the order of vDNA and LEDGF binding to IN, corroborate with the sequence of events during early stages of HIV-1 replication. IN first encounters vDNA in the cytoplasm, in which the exchange of subunits in multimers may be essential for the effective assembly of the fully functional nucleoprotein complex termed the preintegration complex. LEDGF then engages the preassembled IN-vDNA complex in the nucleus and tethers PICs to the chromatin without disturbing the structural arrangements of IN with its DNA substrates.
This chronology of events would suggest that IN before its binding to vDNA rather than PICs could be a plausible target for allosteric inhibitors. Indeed, PICs have a very stable structure and are likely to be resistant to conformational challenges with small allosteric compounds. In contrast, unliganded IN at earlier stages of viral replication, before synthesis of vDNA by reverse transcriptase is complete, could be vulnerable to the attack by allosteric inhibitors. This notion is supported by the published data (De Rijck et al., 2006; Llano et al., 2006a) indicating that the overexpression of the enhanced green fluorescent protein-IBD, which lacks nuclear localization signal and interacts with unliganded IN in the cytoplasm, effectively inhibits HIV-1 integration. The IBD-derived short peptides have also been shown to consistently stabilize IN multimers in vitro and impair HIV-1 integration in the infected cells (Hayouka et al., 2007; Al-Mawsawi et al., 2008).
Despite certain mechanistic similarities between the IBD- and 1-dependent inactivation of IN activities, binding modes of and oligomerization pathways induced by these ligands could differ. For example, the IBD establishes extensive contacts with the CCD of one dimer and the N-terminal domain of another dimer to stabilize a tetramer of IN (McKee et al., 2008). In contrast, the binding site of 1 is likely to be restricted to a much smaller area. Our results (Figs. 2 and 6) suggest that 1 binds at the IN dimer interface at an allosteric pocket, which differs from the IBD binding sites. The resulting inhibitor-IN complex may lack sufficient flexibility to carry out catalytic reactions. Future structural studies of the inhibitor-protein complex are necessary to gain important details for how 1 affects the multimeric structure of IN.
Our MS (Fig. 2) and molecular modeling experiments (Fig. 6) have provided initial clues about the amino acids interacting with 1. Unfortunately, the mutagenesis studies did not allow us to elucidate whether the proposed residues directly engage the inhibitor because the single amino acid substitutions adversely affected both the inhibitor binding and protein multimerization (Fig. 7). Nevertheless, mutagenesis experiments have highlighted the architectural importance of residues within the proposed inhibitor binding site and suggest that a correctly assembled IN multimer is a selective target for 1. There are the following key advantages for targeting the proposed allosteric pocket with small-molecule compounds. The pocket is significantly distanced from presumed STI binding sites. For example, IN mutations conferring resistance to Raltegravir are localized in the vicinity of the catalytic DDE motif (Cooper et al., 2008). Therefore, agents selectively binding at the protein dimer interface are likely to be active against IN phenotypes resistant to the STI. Furthermore, the inhibitors targeting the allosteric cavity (Fig. 6) would not have to compete with IN–IN or IN–LEDGF interactions. Structural studies indicate extensive monomer-monomer interfaces within dimeric IN (Chen et al., 2000; Wang et al., 2001). The LEDGF contacts in IN include two separate areas in the CCD and the N-terminal domain (Cherepanov et al., 2005; McKee et al., 2008; Hare et al., 2009). Therefore, finding small compounds that interfere with intra- or interprotein-protein interactions could be challenging. In contrast, 1 is not required to overcome large energy barriers created by numerous interacting amino acids. Instead, the inhibitor exploits the preformed dimer interface, which is fully distinct from the LEDGF binding site.
The other two small-molecule inhibitors (3,4-dihydroxyphenyltriphenylarsonium bromide and a coumarin-based compound) of IN have also been shown to interact with the CCD dimer interface (Molteni et al., 2001; Al-Mawsawi et al., 2006). However, their mechanisms of inhibition have not been elucidated. It is possible that these agents also function by dynamic modulation of IN multimeric structure. Our recently reported subunit-subunit exchange assay (Fig. 4) (McKee et al., 2008) provides a means to examine SAR for these and related compounds.
Our assays enabled us to delineate mechanisms of action for 1 and authentic CA class inhibitors (see Figs. 2–4 and supplemental Fig. 2). Unlike 1, 2 failed to interact with the IN dimer interface and instead impaired IN-DNA complex formation. The hydroxylated and tetra-acetylated CAs have been examined previously in the cell culture assays (King and Robinson, 1998; Pluymers et al., 2000). Robinson and coworkers (King and Robinson, 1998) isolated a CA resistant strain containing a mutation in the IN gene corresponding to the G140S IN variant. However, subsequent studies with hydroxylated and tetra-acetylated CAs mapped resistant mutations to the gp120 gene, suggesting that these compounds may target multiple sites in infected cells (Pluymers et al., 2000).
1 has not been tested in the cell culture assays. However, the presence of di-O-acetyl groups in this inhibitor is likely to adversely affect its specificity. For example, these groups are substrates for cellular acetyltransferases, and the resulting catechols could deter 1 from its retroviral target. Comparative SAR and molecular modeling studies (Figs. 2, 4, and 6) suggest that the central linker establishes extensive hydrogen bonding network with IN subunits, whereas contributions of di-O-acetyl groups to the 1-IN complex formation are relatively limited. Therefore, the tetra-acetate groups, in theory, could be replaced by alternative structures without compromising the binding specificity. In fact, continued optimization of the 1 structure could lead to the identification of more potent inhibitors with desired specificity in the infected cells.
Taken together, the mechanism of action and binding site for the small-molecule inhibitor described herein provides an attractive venue for targeting IN. Furthermore, our findings facilitate wider ongoing efforts in the field to develop new types of clinically useful allosteric inhibitors of IN that could effectively complement current antiretroviral therapies.
Supplementary Material
Acknowledgments
HIV-1 IN Monoclonal Antibody (2C11) was obtained from Dr. Dag E. Helland through the AIDS Research and Reference Reagent Program, Division of AIDS, the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported by National Institutes of Health National Institute of Allergy and Infectious Diseases [Grants AI062520, AI077341]; and by the Intramural Research Programs of the National Institutes of Health Center for Cancer Research, the National Institutes of Health National Cancer Institute, and the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases.
Experiments by S.H. were conducted at the Proteomics and Mass Spectrometry Facility of National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- STI
- strand-transfer inhibitor
- IN
- integrase
- CCD
- catalytic core domain
- LEDGF
- lens epithelium-derived growth factor
- IBD
- integrase binding domain
- MS
- mass spectrometry
- MALDI-ToF
- matrix-assisted laser desorption ionization/time of flight
- CA
- chicoric acid
- PAGE
- polyacrylamide gel electrophoresis
- PD
- pull down
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate
- SAR
- structure-activity relationship.
References
- Al-Mawsawi LQ, Christ F, Dayam R, Debyser Z, Neamati N. (2008) Inhibitory profile of a LEDGF/p75 peptide against HIV-1 integrase: insight into integrase-DNA complex formation and catalysis. FEBS Lett 582:1425–1430 [DOI] [PubMed] [Google Scholar]
- Al-Mawsawi LQ, Fikkert V, Dayam R, Witvrouw M, Burke TR, Jr, Borchers CH, Neamati N. (2006) Discovery of a small-molecule HIV-1 integrase inhibitor-binding site. Proc Natl Acad Sci U S A 103:10080–10085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busschots K, De Rijck J, Christ F, Debyser Z. (2009) In search of small molecules blocking interactions between HIV proteins and intracellular cofactors. Mol Biosyst 5:21–31 [DOI] [PubMed] [Google Scholar]
- Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, Leavitt AD, Stroud RM. (2000) Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci U S A 97:8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. (2005) Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. Proc Natl Acad Sci U S A 102:17308–17313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov P, Devroe E, Silver PA, Engelman A. (2004) Identification of an evolutionarily conserved domain in human lens epithelium-derived growth factor/transcriptional co-activator p75 (LEDGF/p75) that binds HIV-1 integrase. J Biol Chem 279:48883–48892 [DOI] [PubMed] [Google Scholar]
- Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, Engelborghs Y, De Clercq E, Debyser Z. (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem 278:372–381 [DOI] [PubMed] [Google Scholar]
- Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Kumar PN, Eron JE, et al. (2008) Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N Engl J Med 359:355–365 [DOI] [PubMed] [Google Scholar]
- De Rijck J, Vandekerckhove L, Gijsbers R, Hombrouck A, Hendrix J, Vercammen J, Engelborghs Y, Christ F, Debyser Z. (2006) Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J Virol 80:11498–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981–1986 [DOI] [PubMed] [Google Scholar]
- Emiliani S, Mousnier A, Busschots K, Maroun M, Van Maele B, Tempé D, Vandekerckhove L, Moisant F, Ben-Slama L, Witvrouw M, et al. (2005) Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J Biol Chem 280:25517–25523 [DOI] [PubMed] [Google Scholar]
- Engelman A, Bushman FD, Craigie R. (1993) Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J 12:3269–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, Tarrago-Litvak L, Litvak S, Parissi V. (2005) HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res 33:977–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell DS, Olson AJ. (1990) Automated docking of substrates to proteins by simulated annealing. Proteins 8:195–202 [DOI] [PubMed] [Google Scholar]
- Guiot E, Carayon K, Delelis O, Simon F, Tauc P, Zubin E, Gottikh M, Mouscadet JF, Brochon JC, Deprez E. (2006) Relationship between the oligomeric status of HIV-1 integrase on DNA and enzymatic activity. J Biol Chem 281:22707–22719 [DOI] [PubMed] [Google Scholar]
- Hare S, Shun MC, Gupta SS, Valkov E, Engelman A, Cherepanov P. (2009) A novel co-crystal structure affords the design of gain-of-function lentiviral integrase mutants in the presence of modified PSIP1/LEDGF/p75. PLoS Pathog 5:e1000259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayouka Z, Rosenbluh J, Levin A, Loya S, Lebendiker M, Veprintsev D, Kotler M, Hizi A, Loyter A, Friedler A. (2007) Inhibiting HIV-1 integrase by shifting its oligomerization equilibrium. Proc Natl Acad Sci U S A 104:8316–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuda DJ, Hastings JC, Wolfe AL, Emini EA. (1994) A novel assay for the DNA strand-transfer reaction of HIV-1 integrase. Nucleic Acids Res 22:1121–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy EF, Sanders J, King PJ, Robinson WE., Jr (2009) A docking study of l-chicoric acid with HIV-1 integrase. J Mol Graph Model 27:584–589 [DOI] [PubMed] [Google Scholar]
- Hou Y, McGuinness DE, Prongay AJ, Feld B, Ingravallo P, Ogert RA, Lunn CA, Howe JA. (2008) Screening for antiviral inhibitors of the HIV integrase-LEDGF/p75 interaction using the AlphaScreen luminescent proximity assay. J Biomol Screen 13:406–414 [DOI] [PubMed] [Google Scholar]
- King PJ, Robinson WE., Jr (1998) Resistance to the anti-human immunodeficiency virus type 1 compound l-chicoric acid results from a single mutation at amino acid 140 of integrase. J Virol 72:8420–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Mizuuchi M, Burke TR, Jr, Craigie R. (2006) Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J 25:1295–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Neamati N, Zhao H, Kiryu Y, Turpin JA, Aberham C, Strebel K, Kohn K, Witvrouw M, Pannecouque C, et al. (1999) Chicoric acid analogues as HIV-1 integrase inhibitors. J Med Chem 42:1401–1414 [DOI] [PubMed] [Google Scholar]
- Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. (2006a) An essential role for LEDGF/p75 in HIV integration. Science 314:461–464 [DOI] [PubMed] [Google Scholar]
- Llano M, Vanegas M, Hutchins N, Thompson D, Delgado S, Poeschla EM. (2006b) Identification and characterization of the chromatin-binding domains of the HIV-1 integrase interactor LEDGF/p75. J Mol Biol 360:760–773 [DOI] [PubMed] [Google Scholar]
- Maertens G, Cherepanov P, Pluymers W, Busschots K, De Clercq E, Debyser Z, Engelborghs Y. (2003) LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J Biol Chem 278:33528–33539 [DOI] [PubMed] [Google Scholar]
- McKee CJ, Kessl JJ, Shkriabai N, Dar MJ, Engelman A, Kvaratskhelia M. (2008) Dynamic modulation of HIV-1 integrase structure and function by cellular lens epithelium-derived growth factor (LEDGF) protein. J Biol Chem 283:31802–31812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molteni V, Greenwald J, Rhodes D, Hwang Y, Kwiatkowski W, Bushman FD, Siegel JS, Choe S. (2001) Identification of a small-molecule binding site at the dimer interface of the HIV integrase catalytic domain. Acta Crystallogr D Biol Crystallogr 57:536–544 [DOI] [PubMed] [Google Scholar]
- Pandey KK, Sinha S, Grandgenett DP. (2007) Transcriptional coactivator LEDGF/p75 modulates human immunodeficiency virus type 1 integrase-mediated concerted integration. J Virol 81:3969–3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluymers W, Neamati N, Pannecouque C, Fikkert V, Marchand C, Burke TR, Jr, Pommier Y, Schols D, De Clercq E, Debyser Z, et al. (2000) Viral entry as the primary target for the anti-HIV activity of chicoric acid and its tetra-acetyl esters. Mol Pharmacol 58:641–648 [DOI] [PubMed] [Google Scholar]
- Raghavendra NK, Engelman A. (2007) LEDGF/p75 interferes with the formation of synaptic nucleoprotein complexes that catalyze full-site HIV-1 DNA integration in vitro: implications for the mechanism of viral cDNA integration. Virology 360:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkriabai N, Patil SS, Hess S, Budihas SR, Craigie R, Burke TR, Jr, Le Grice SF, Kvaratskhelia M. (2004) Identification of an inhibitor-binding site to HIV-1 integrase with affinity acetylation and mass spectrometry. Proc Natl Acad Sci U S A 101:6894–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. (2007) LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev 21:1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez Paz O, Hazuda DJ, et al. (2008) Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem 51:5843–5855 [DOI] [PubMed] [Google Scholar]
- van den Ent FM, Vos A, Plasterk RH. (1999) Dissecting the role of the N-terminal domain of human immunodeficiency virus integrase by trans-complementation analysis. J Virol 73:3176–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, Vink C, Groeneger AA, Plasterk RH. (1993) Complementation between HIV integrase proteins mutated in different domains. EMBO J 12:3261–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanegas M, Llano M, Delgado S, Thompson D, Peretz M, Poeschla E. (2005) Identification of the LEDGF/p75 HIV-1 integrase-interaction domain and NLS reveals NLS-independent chromatin tethering. J Cell Sci 118:1733–1743 [DOI] [PubMed] [Google Scholar]
- Wang JY, Ling H, Yang W, Craigie R. (2001) Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J 20:7333–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, McKee CJ, Kessl JJ, Santos WL, Daigle JE, Engelman A, Verdine G, Kvaratskhelia M. (2008) Subunit-specific protein footprinting reveals significant structural rearrangements and a role for N-terminal Lys-14 of HIV-1 Integrase during viral DNA binding. J Biol Chem 283:5632–5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.