Abstract
Previous work has shown that bioactivation of the cigarette smoke pneumotoxicant 3-methylindole (3MI) by pulmonary cytochrome P450 enzymes is directly associated with formation of DNA adducts. Here, we present evidence that normal human lung epithelial cells, exposed to low micromolar concentrations of 3MI, showed extensive DNA damage, as measured by the comet assay, with similar potency to the prototypical genotoxic agents, doxorubicin and irinotecan. The DNA damage caused by 3MI was predominantly caused by single-strand breaks. Furthermore, we show that this damage decreased with time, given a subtoxic concentration, with detectable DNA fragmentation peaking 4 h after exposure and diminishing to untreated levels within 24 h. Pretreatment with an inhibitor of poly(ADP-ribose) polymerase 1 (PARP1), NU1025, nearly doubled the DNA damage produced by 5μM 3MI, implying that PARP1, which among other activities, functions to repair single-strand breaks in DNA, repaired at least some of the 3MI-induced DNA fragmentation. A key cellular response to DNA damage, phosphorylation, and nuclear localization of p53 was seen at subtoxic levels of 3MI exposure. 3MI was highly mutagenic, with essentially the same potency as the prototype carcinogen, benzo[a]pyrene, only when a lung-expressed CYP2F3 enzyme was used to dehydrogenate 3MI to its putative DNA-alkylating intermediate. Conversely, a rat liver S9 metabolic system did not bioactivate 3MI to its mutagenic intermediate(s). Concentrations higher than 25μM caused apoptosis, which became extensive at 100μM, similar to the response seen with 10μM doxorubicin. Our findings indicate that there is a low concentration window in which 3MI can cause extensive DNA damage and mutation, without triggering apoptotic defenses, reinforcing the hypothesis that inhaled 3MI from cigarette smoke may be a potent lung-selective carcinogen.
Keywords: 3-methylindole, DNA damage, DNA repair mechanisms, chemical carcinogenesis, mutagen, p53, lung cancer, cytochrome P450
3-Methylindole (3MI) is an anaerobic fermentation product of tryptophan, formed in the large intestine of man (Fordtran et al., 1964; Yokoyama and Carlson, 1979). In addition to natural production, cigarette smoke provides a significant source of exposure to 3MI for man because 3MI is formed by pyrolysis of tryptophan in burning tobacco and has been identified in cigarette smoke at a range of concentrations between 0.4 and 1.4 μg/cigarette (Wynder and Hoffmann, 1967). A heavy smoker (3 packs/day) could be exposed to amounts of 84 μg/day (1.4 μg 3MI/cigarette × 20 cigarettes/pack × 3 packs/day). These amounts of 3MI are substantially higher than the amounts of two prototypical cigarette smoke carcinogens, benzo[a]pyrene and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. It is possible that 3MI in cigarette smoke could contribute to lung cancer from smoking, but we are not aware of any studies to determine if 3MI exposures by inhalation or any other route can produce lung tumors in experimental animals. Pathological damage from 3MI exposure in laboratory animals and ruminants is distinctively organ specific for respiratory tissues, whereas many other pneumotoxins, such as the pyrrolizidine alkaloids and the substituted furans, can cause significant hepatic damage (Yost, 1989). Pulmonary cellular damage by 3MI is highly selective to Clara cells and alveolar epithelial cells in all species (Yost, 1989), and this selectivity has helped to determine the functional significance of cytochrome P450 enzyme expression in these cells. The toxicity of 3MI is entirely dependent on cytochrome P450-mediated bioactivation by dehydrogenation within susceptible airway cells. 3MI can be used as a selective probe for dehydrogenation reactions, which are typically bioactivation events, catalyzed by certain cytochrome P450 enzymes that are selectively expressed in the lung tissues of animals and man (Carr et al., 2003; Kartha et al., 2008; Skiles and Yost, 1996; Yan et al., 2007). These attributes of 3MI-induced toxicity provide unique advantages for the determinations of pneumotoxic mechanisms and the extrapolation of these mechanisms to man.
Vaccinia-expressed human cytochrome P450 enzymes catalyzed the formation of several metabolites of 3MI, including indole-3-carbinol, 3-methyloxindole, and 3-methyleneindolenine (3MEIN) (Thornton-Manning et al., 1993). Previous work in our laboratory has demonstrated that the cytochrome P450–catalyzed dehydrogenation metabolite, 3MEIN, forms adducts with the cysteine thiols of proteins in human liver microsomes, as well as a thioether adduct with glutathione, from incubations with goat lung microsomes (Ruangyuttikarn et al., 1991). Treatment with the cytochrome P450 suicide substrate inhibitor 1-aminobenzotriazole (ABT) was sufficient to inhibit adduct formation, indicating a cytochrome P450–dependent mechanism of adduct formation. Furthermore, the cytochrome P450–catalyzed metabolite 3MEIN has been shown to form adducts with the deoxyguanine, deoxyadenine, and deoxycytidine nucleotides of DNA in incubations with both hepatic human microsomes and rat hepatocytes (Regal et al., 2001).
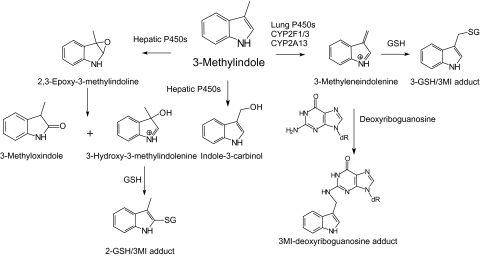
Figure 1 illustrates the metabolic pathways for formation of oxygenated and dehydrogenated intermediates and excreted metabolites, as well as the structures of the two primary glutathione adducts and the deoxyguanosine adduct, formed by trapping of 3MEIN. Also shown are the putative disparate pathways of metabolism between hepatic and respiratory P450 enzymes that theoretically explain the major divergent mechanisms between detoxication and bioactivation, respectively.
FIG. 1.
Divergent metabolic routes for hepatic and respiratory P450 enzymes. Illustrated are the predominant pathways for oxygenation of 3MI to the major excreted metabolites, 3-methyloxindole and indole-3-carbinol by hepatic P450 enzymes, and dehydrogenation of 3MI to 3MEIN by respiratory P450 enzymes. 3MEIN is the putative toxic electrophilic intermediate that is produced almost exclusively by lung P450s such as CYP2F1, CYP2F3, and CYP2A13. Also shown are the two major glutathione adducts and one of the nucleoside adducts of 3MEIN, formed from deoxyriboguanosine residues of DNA.
Alkyl DNA adduct formation is of interest due to its implications in the induction of mutagenesis, apoptosis, and carcinogenicity (Cavalieri and Rogan, 2004; De Bont and van Larebeke, 2004). Although 3MI induced DNA fragmentation and apoptosis in immortalized human bronchial epithelial cell lines (Nichols et al., 2003), the ability of 3MI to induce DNA damage in primary human lung cells has not been determined.
With the following study, we investigate the potential of 3MI to cause DNA damage in untransformed human bronchial epithelial cells and determine the cellular response to these adducts. We show that at sublethal levels of 3MI, extensive though repairable, single-strand DNA damage occurs and tumor suppressors are upregulated. These results imply that repeated 3MI exposure through cigarette smoke inhalation may lead to a cycle of DNA damage and repair that may lead to mutation and promotion of neoplasia.
MATERIALS AND METHODS
Cell culture.
Normal human bronchial epithelial (NHBE) cells, isolated in the presence of retinoic acid, were obtained from Lonza (Walkersville, MD) and cultured in bronchial epithelial growth medium with retinoic acid in a humidified incubator with 95% air and 5% CO2. For subculturing, cells were trypsin dissociated and plated into fibronectin-coated/collagen-coated culture plates. Cells were treated at passage three after achieving ∼80% confluence. Chemicals were dissolved in dimethyl sulfoxide (DMSO) and diluted in growth media prior to treating the cells. Control cells were treated with DMSO alone, and the maximum concentration any treatment group was exposed to was 0.5% DMSO.
Analysis of the extent of DNA damage.
DNA strand breaks were measured using alkaline single–cell gel electrophoresis (comet assay) according to the manufacturer's recommended protocol (Trevigen Inc., Gaithersburg, MD). Briefly, primary NHBE cells were cultured and treated with DMSO (vehicle), 3MI, ABT, doxorubicin, and NU1025. Both dose-response and time course assays were performed. Cells were scraped and centrifuged to remove media. Cell pellets were then resuspended in 1× PBS, combined with low-melting point agarose, and plated on CometSlides. Slides were incubated at 4°C in the dark for 30 min, followed by immersion in prechilled lysis buffer (2.5 M NaCl, 100mM EDTA, 10mM Trizma base, and 1% Triton X-100) for 1 h, and subsequently incubated in alkaline (pH > 13) electrophoresis buffer (10 N NaOH and 200mM EDTA) for 30 min at room temperature. Following the postlysis incubation, electrophoresis power was applied at 300 mV for 30 min. Upon completion of electrophoresis, slides were immersed in 100% ethanol for 5 min and air dried. Slides were then flooded with 0.5 μg/ml ethidium bromide (EtBr), visualized using a fluorescent microscope equipped with an EtBr filter, and imaged. Images were scored using CASP freeware version 1.2.2, and DNA damage was computed as the average olive tail moment (OTM) for 50 separate comets. OTM is defined as the fraction of tail DNA multiplied by the means of the head and tail DNA distributions.
Analysis of DNA damage subtypes.
To differentiate DNA damage subtypes, the comet assay was conducted at three different pHs: neutral (pH 7), pH 12.1, and alkaline (pH > 13). OTM analyzed at pH > 13 includes alkaline labile sites, single-strand breaks, and double-strand breaks. OTM at pH 12.1 represents single-strand breaks and double-strand breaks, while OTM at pH 7 represents only double-strand breaks. Comparison of the levels of DNA damage at each different pH was used to determine the most prominent subtype of DNA damage occurring upon exposure to 3MI. This assay was performed in much the same manner as the alkaline comet assay, with the following modifications to the postlysis incubation and electrophoresis steps: For neutral comets, 30-min postlysis incubation and 10-min electrophoresis were conducted in 1× TBE (90mM Tris, 90mM boric acid, 2mM EDTA) buffer. For pH 12.1 comets, 30-min postlysis incubation and 30-min electrophoresis were conducted in alkaline buffer, adjusted to the appropriate pH with 10M HCl. Five micromolar doxorubicin was used as a positive control for double-strand breaks, 5μM irinotecan for single-strand breaks, and 200μM N-nitroso-N-ethylurea for alkaline labile sites.
p53 nuclear localization.
An antibody raised in rabbits specific for p53 was obtained from Cell Signaling Technologies, Inc. (Beverly, MA). Cells were cultured in 48-well dishes and then exposed to a range of 3MI concentrations from 0.01 to 200μM for 90 min. Three wells were treated for each culture condition. Cells were aspirated, then fixed, and permeablized with ice-cold 100% methanol. Proteins were blocked overnight with PBS containing 10% sheep serum and 10% bovine serum albumin. The proteins were then exposed to the p53-specific antibody, diluted 1:100 in the blocking solution, washed three times with PBS, and then exposed to a fluorescent secondary antibody (Yo-Pro1-conjugated goat anti-rabbit; Molecular Probes, Eugene, OR). Proteins were washed and counterstained with 4′-6-diamidino-2-phenylindole to identify the nuclei. Image J (National Institutes of Health) software was used to count total nuclei and p53-positive nuclei from four separate fields, and a minimum of 500 cells were counted for each field. Data are represented as a percent of total nuclei counted.
Western blotting.
Western blotting was performed as described previously (Wang et al., 1998) with some modifications. Briefly, treated cells were suspended and lysed in protein loading buffer containing 10μM dithiothreitol and a protease inhibitor cocktail using a rubber policeman, followed by heating for 5 min at 100°C and quick chilling on ice. Samples were immediately fractionated by a 10% SDS-polyacrylamide gel electrophoresis at 100 V. Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane, and the membrane was blocked with 1× PBST (1× PBS and 0.1% Tween) containing 2% nonfat milk for 1 h at room temperature. The PVDF membrane was incubated overnight with antibodies specific for the protein of interest diluted 1:1000 in 1× PBST containing 0.5% nonfat milk at 4°C. Primary antibodies used were purchased form Cell Signaling Technology and consist of Phospho-p53 (Ser20) antibody #9287 specific for the serine 20–phosphorylated forms of the protein, 53BP1 antibody #4937 for total p53, and poly(ADP-ribose) polymerase 1 (PARP1) antibody #9542, which detects both the full-length 116 kDa PARP1 and the 89-kDa cleavage fragment. After the overnight incubation, the membrane was rinsed four times with 1× PBST and was then incubated with horseradish peroxidase–conjugated secondary antibody, diluted 1:2000 in 1× PBST containing 1% nonfat milk for 1 h at room temperature. The secondary antibody was removed, and the membrane was washed four times with 1× PBST. The PVDF membrane was developed with the ECL Plus kit (GE Healthcare, Piscataway, NJ) at room temperature and exposed to Kodak film. In the case of p53, the phosphorylation-specific antibody was used first, then after development, the blot was stripped using 0.2mM sodium hydroxide for 5 min. The blot was then washed, blocked, and reprobed with the antibody, which detects total p53. Film was developed in a Konica Medical Film Processor (Model QX-130A, Konica Minolta Medical Imaging USA, Inc., Wayne, NJ), and the film was then digitalized using a BioRad Gel Doc 1000 (BioRad Labs, Hercules, CA) and QuantityOne 4.1.1 software. Digital images were cropped using PowerPoint software.
Analysis of apoptosis via measurement of annexin V binding.
The presence of phosphatidylserine on the outer membrane of cells was detected using the Annexin V-FITC Apoptosis Detection Kit I (BD Pharmigen, San Diego, CA), following the manufacturer’s instructions. This immunochemical technique is commonly used in many different types of apoptotic cells to identify this initial event of apoptosis. Briefly, cells were treated with a range of 3MI concentrations for 12 h and washed with cold PBS, followed by Annexin V–binding buffer. Cells were then incubated with Annexin V-FITC and propidium iodide in binding buffer while rocking in the dark for 10 min. Cells were rinsed twice with binding buffer, fixed for 5 min in 2% formalin, rinsed with PBS, and then quantitated using fluorescent microscopy. Masked data analysis was performed with 3 × 500 cells counted for each condition.
Cytochrome P450 2F3 expression and purification.
Expression and purification of cytochrome CYP2F3 were performed as previously published (Kartha and Yost, 2008; Kartha et al., 2008)
Microbial mutagenesis.
Bacterial mutagenesis assays were conducted using a standard reverse mutagenesis plate test with Salmonella (Ames et al., 1975), preceded by a modification step in which 100 μl of a log-phase bacterial culture of the Salmonella typhimurium strain TA98 was preincubated for 30 min at 37°C in a shaking incubator with the test compound (vehicle controls were treated with 5 μl of DMSO, and test groups were treated with increasing concentrations of either benzo[a]pyrene or 3MI dissolved in DMSO), 450 μl of 0.1M phosphate buffer (50mM Na2HPO4-7H2O, 0.04% H3PO4), 100 pmol of purified cytochrome P450 2F3 (for 3MI), or Aroclor 1254–induced rat liver S9 (Celsis, Baltimore, MD) (for benzo[a]pyrene) in the presence or absence of 18mM NADPH. Following the 30-min preincubation period, 2.5 ml of molten top agar containing 50μM histidine and 50μM biotin was added to each reaction mixture, then poured onto minimal growth agar plates, and incubated upside down for 48 h at 37°C. Test plates containing only cytochrome P450 2F3 and TA98 were used as controls. Masked colony counting was performed using a Fisher bacterial colony counter.
Statistical analysis.
All the data were reported as mean ± SD. The difference between untreated and treated cells was tested using a one-way ANOVA. The difference was considered significant with a probability of 0.05. Post hoc analysis of the differences between 3MI-treated cells and other treatment groups were calculated using Student's t-test, and differences were considered significant with a probability of p ≤ 0.05.
RESULTS
Extent of DNA Damage in Response to 3MI Exposure
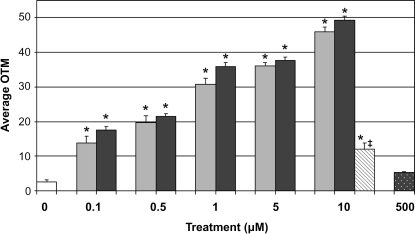
3MI-induced DNA damage in NHBEs was assessed in both a concentration- and a time-dependent manner using the alkaline comet assay. Low concentrations of 3MI, ranging from 0.1 to 10μM for 4 h, demonstrated a concentration-dependent increase in DNA damage compared to vehicle-treated control (Fig. 2). This concentration-dependent increase mirrored the damage produced by incubation with the known DNA-damaging agent doxorubicin, an inhibitor of topoisomerase II. Pretreatment for 30 min with 500μM ABT, a suicide substrate inhibitor of virtually all cytochrome P450 enzymes, significantly attenuated 3MI-mediated DNA damage. Treatment with 3MI concentrations lower than those depicted did not produce a statistically significant increase in DNA damage (data not shown).
FIG. 2.
Concentration-response of 3MI-induced DNA damage in NHBE cells. Four-hour incubation with increasing concentrations of 3MI (gray bars) and doxorubicin (black bars) produced a concentration-dependent increase in DNA damage (average OTM). Thirty-minute pretreatment with ABT (gray striped bar) inhibited the DNA damage response to 3MI treatment. Treatment with ABT alone (black dotted bar) did not significantly increase the levels of DNA damage as compared to vehicle (0.5% DMSO)-treated control (white bar). *p < 0.05 compared to vehicle-treated control, ‡p < 0.05 compared to 10μM 3MI.
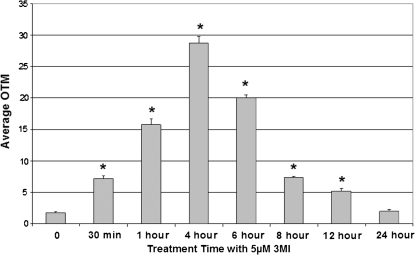
Time course analysis consisted of exposure to 5μM 3MI for 0, 0.5, 1, 4, 6, 8, 12, and 24 h. DNA damage had become significantly elevated by 30 min of exposure and peaked at 4 h (Fig. 3). By 24 h of exposure, DNA damage levels were no longer significantly elevated from time zero.
FIG. 3.
Time course of 3MI-induced DNA damage in NHBE cells. Cells incubated with 5μM 3MI exhibited a maximal DNA damage response (average OTM) after 4 h, which was no longer significantly elevated above vehicle-treated control by 24 h. *p < 0.05 compared to vehicle-treated control.
Elucidation of the Primary Subtype of DNA Damage in Response to 3MI Exposure
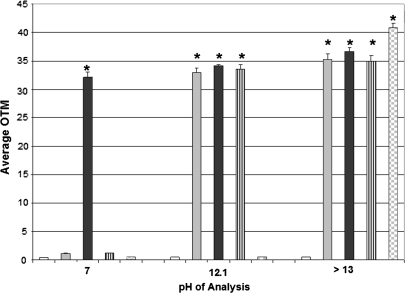
A modified comet assay was performed at three separate pHs (7, 12.1, and > 13) to differentiate the primary subtype of 3MI-induced DNA damage after 4 h. At a neutral pH of 7, only double-strand breaks in DNA were detected. As Figure 4 demonstrates, only treatment with the positive control for double-strand breaks, 5μM doxorubicin, produced an elevation in the levels of DNA damage as compared to vehicle-treated control at this pH. At pH 12.1, both single-strand breaks and double-strand breaks were detected. As shown in Figure 4, 5μM doxorubicin, 5μM 3MI, and 5μM irinotecan, the positive control for single-strand breaks, all became significantly elevated at this pH. At pH > 13, alkaline labile sites can be detected in addition to single-strand breaks and double-strand breaks. At this pH, 5μM doxorubicin, 5μM 3MI, 5μM irinotecan, and 200μM ethyl nitrosourea, the positive control for alkaline labile sites, were all significantly elevated. The largest difference in response to 3MI exposure was observed between pH 7 (double-strand breaks only) and pH 12.1 (both single- and double-strand breaks), while the difference in 3MI-induced DNA damage levels between pH 12.1 and pH > 13 was not significant. This indicated that the predominate subtype of DNA damage in response to 3MI exposure was caused by single-strand breaks.
FIG. 4.
Single-strand breaks are the predominant subtype of 3MI-induced DNA damage. Four-hour incubation with 5μM 3MI (gray bars) produced a maximal difference in DNA damage levels (average OTM) between pH 7 and 12.1, indicating that the majority of DNA damage in response to 3MI treatment results from single-strand breaks. 0.5% DMSO vehicle control (white bars), positive control for double-strand breaks 5μM doxorubicin (black bars), positive control for single-strand breaks 5μM irinotecan (black and white striped bars), and positive control for alkaline labile sites 200μM ethyl nitrosourea (gray and white checked bars) are also shown. *p < 0.05 compared to vehicle-treated control.
Inhibition of DNA Repair Increases 3MI-Induced DNA Damage
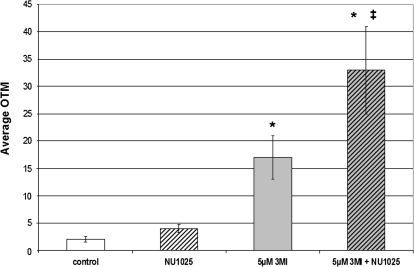
PARP1 is an enzyme that mediates single-strand break DNA repair (Ikejima et al., 1990; Soldatenkov and Smulson, 2000). The predominate subtype of 3MI-mediated DNA damage was observed to be single-strand breaks, so it was postulated that inhibition of PARP1 would increase 3MI-induced DNA damage. Pretreatment for 30 min with the PARP1 inhibitor NU1025 (Beneke et al., 2004) significantly increased the levels of 3MI-induced DNA damage as compared to both vehicle control and 5μM 3MI treatment alone (Fig. 5). Treatment with NU1025 alone did not significantly increase DNA damage.
FIG. 5.
Inhibition of single-strand break repair increases 3MI-induced DNA damage. Pretreatment for 30 min with the PARP1 inhibitor NU1025 significantly increased the levels of 3MI-induced (gray and black striped bar) DNA damage (average OTM) as compared to both the 4-h 3MI treatment alone (gray bar) and the vehicle-treated control (white bar). Treatment with the PARP1 inhibitor alone (black and white striped bar) did not significantly increase the levels of DNA damage as compared to vehicle-treated control. *p < 0.05 compared to vehicle-treated control, ‡<0.05 compared to 4-h 3MI incubation.
Submicromolar Concentrations of 3MI Induce Nuclear Localization of p53
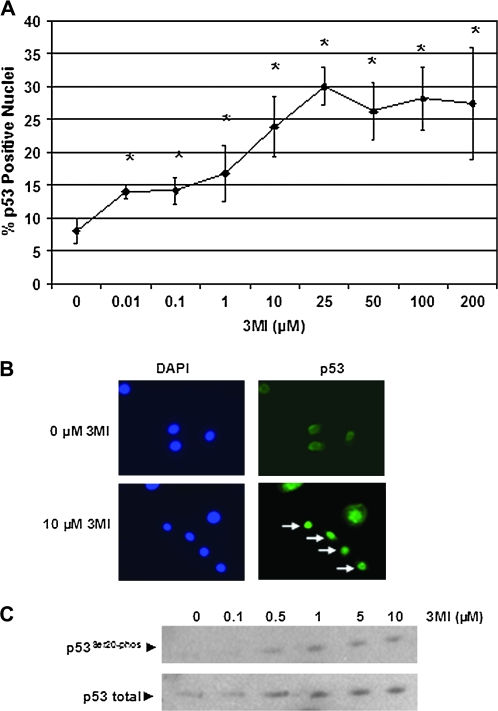
The transcription factor p53 acts as a central mediator in the cellular response to DNA damage (Kern et al., 1991). The genotoxic drug doxorubicin has previously been shown to induce nuclear localization of p53 in a lung cancer cell line (Murph et al., 2007). Since p53 nuclear localization is typically one of the first cellular responses to DNA damage (Chumakov, 2007), we assessed 3MI-mediated nuclear localization at the 1-h time point instead of the 4-h time point used to assess DNA damage in NHBE cells and identified a concentration-dependent increase in the number of cells with p53-positive nuclei (Figs. 6A and B). Likewise, we saw an increase in the phosphorylation of serine 20 of p53 (Fig. 6C), which is important in stabilizing p53 in response to DNA damage (Chehab et al., 1999). Both of these events were detected an hour after exposure to submicromolar concentrations of 3MI.
FIG. 6.
p53 nuclear localization and phosphorylation increase in response to submicromolar 3MI exposure. NHBE cells were exposed to a range of 3MI concentrations or vehicle for 1 h. p53 nuclear localization was determined by immunohistochemistry (A and B). White arrows (B) indicate those cells with complete p53 nuclear localization. Total p53 and p53 phosphorylated at serine 20 were determined by Western blot (C). *p < 0.05 compared to vehicle-treated control.
3MI Induces Apoptosis in NHBE Cells
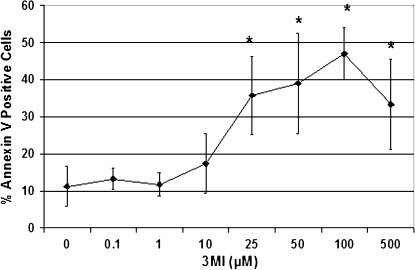
We have previously shown that 3MI can induce apoptosis at 6 and 24 h in an SV40-transformed bronchial epithelial cell line, BEAS-2B (Nichols et al., 2003). Based on these data, we chose an intermediate time point of 12 h and observed significant Annexin V binding, a hallmark of early changes in apoptotic cells, at 50μM 3MI, at least two orders of magnitude greater than concentrations at which we detect DNA damage in NHBE cells. Performing the assay using NHBE cells, we found a significant increase in Annexin V–binding cells with 25μM 3MI (Fig. 7), similar to results seen with BEAS-2B cells. The DNA repair enzyme PARP1 provides efficient protection to cells by repairing DNA single-strand breaks, and its role in protection of NHBE cells was confirmed when inhibition of PARP1 doubled 3MI-induced DNA damage (Fig. 5). After PARP1 functions in this vital protective manner, it is cleaved by caspases, and the 89-kDa fragment produced subsequent to caspase hydrolysis is often used as a marker of apoptosis. Immunochemical detection of the 89-kDa fragment was observed at high concentrations of 3MI (100 and 200μM 3MI, data not shown). As a substrate of caspases, PARP1 cleavage is another indicator that 3MI is inducing apoptosis at these higher concentrations.
FIG. 7.
Annexin V binding is detected with high concentrations of 3MI. NHBE cells were treated with a range of 3MI concentrations for 12 h. Apoptotic cells were detected with FITC-conjugated Annexin V and are expressed as percent positive cells per 3 × 500 total cells counted for each concentration. *p < 0.05 compared to vehicle-treated control.
3MI Is a Potent Mutagen in S. typhimurium Strain TA98
3MI exhibited a concentration-dependent increase in microbial mutagenesis (Table 1), with the highest number of mutants produced with 100μM 3MI (6.56 mg). This mutagenesis was shown to occur via a metabolism-dependent process because mutagenicity of 3MI was not significantly elevated above DMSO-treated control levels in the absence of NADPH, which is required for the activity of cytochrome P450 2F3. Benzo[a]pyrene exhibited a similar concentration-dependent increase in microbial mutagenesis, and the highest number of mutant colonies was found with a concentration of 100μM (12.62 mg). The mutagenicity of benzo[a]pyrene was also shown to be metabolism dependent (Table 2). However, neither compound was mutagenic with its maximal activating concentration in the absence of the appropriate activating system. Benzo[a]pyrene (100μM) was not mutagenic when it was incubated with CYP2F3, and 3MI (100μM) was not mutagenic when it was incubated with Aroclor 1254–induced rat liver S9 fractions.
TABLE 1.
Analysis of Cytochrome P450 2F3 3MI- and Benzo[a]pyrene-Mediated Mutations in Salmonella typhimurium TA98
| Treatment | Dose |
Revertants/plate |
||
| μM | Amount/plate | Average | SD | |
| DMSO | ||||
| +NADPH | 5 μl | 28.5 | 0.7 | |
| −NADPH | 5 μl | 22 | 1.4 | |
| 3MI | ||||
| +NADPH | 0.01 | 655 ng | 26 | 8.5 |
| −NADPH | 0.01 | 655 ng | 25.5 | 2.12 |
| +NADPH | 0.1 | 6.5 μg | 65 | 11.3 |
| −NADPH | 0.1 | 6.5 μg | 33 | 1 |
| +NADPH | 1 | 65.5 μg | 80.5 | 2.12 |
| −NADPH | 1 | 65.5 μg | 35 | 4.24 |
| +NADPH | 10 | 655 μg | 86.5 | 3.53 |
| −NADPH | 10 | 655 μg | 27 | 8.48 |
| +NADPH | 25 | 1.64 mg | 49 | 15.5 |
| −NADPH | 25 | 1.64 mg | 22.5 | 2.12 |
| +NADPH | 50 | 3.28 mg | 86.5 | 9.19 |
| −NADPH | 50 | 3.28 mg | 29.5 | 2.12 |
| +NADPH | 100 | 6.56 mg | 125 | 43.8 |
| −NADPH | 100 | 6.56 mg | 32.5 | 16.3 |
| +NADPH | 250 | 16.4 mg | 57.5 | 3.54 |
| −NADPH | 250 | 16.4 mg | 22.5 | 7.78 |
| +NADPH | 500 | 32.8 mg | 63.5 | 6.36 |
| −NADPH | 500 | 32.8 mg | 33 | 7.07 |
| Benzo(a)pyrene | ||||
| +NADPH | 100 | 12.62 mg | 17 | 1 |
| −NADPH | 100 | 12.62 mg | 18 | 4.14 |
TABLE 2.
Analysis of Aroclor 1254–Induced Rat Liver S9 Benzo[a]pyrene- and 3MI-Mediated Mutations in Salmonella typhimurium TA98
| Dose |
Revertants/plate |
|||
| Treatment | μM | Amount/plate | Average | SD |
| DMSO | ||||
| +NADPH | 5 μl | 29.5 | 2.12 | |
| −NADPH | 5 μl | 25.5 | 4.95 | |
| Benzo(a)pyrene | ||||
| +NADPH | 0.01 | 1.2 μg | 43 | 7.07 |
| −NADPH | 0.01 | 1.2 μg | 26.5 | 2.12 |
| +NADPH | 0.1 | 12.6 μg | 55 | 4.24 |
| −NADPH | 0.1 | 12.6 μg | 25.5 | 3.54 |
| +NADPH | 1 | 126.2 μg | 73.5 | 6.36 |
| −NADPH | 1 | 126.2 μg | 30 | 1.4 |
| +NADPH | 10 | 1.26 mg | 106 | 14.14 |
| −NADPH | 10 | 1.26 mg | 23 | 2.83 |
| +NADPH | 100 | 12.62 mg | 135 | 21.2 |
| −NADPH | 100 | 12.62 mg | 22 | 1.4 |
| +NADPH | 250 | 31.54 mg | 138.5 | 23.5 |
| −NADPH | 250 | 31.54 mg | 23 | 1.4 |
| 3MI | ||||
| +NADPH | 100 | 6.56 mg | 50 | 24 |
| −NADPH | 100 | 6.56 mg | 20.5 | 2.12 |
DISCUSSION
We have previously demonstrated that bioactivated 3MI can form adducts with DNA in a cell-free system and in hepatocytes (Regal et al., 2001). These adducts are consistent with nucleophilic addition of the exocyclic primary amine of the nucleosides to the methylene carbon of 3MEIN. The dGuo adduct was the most abundant, and smaller amounts of the dAdo and dCyd adducts were also efficiently produced. We have also shown that 3MI can induce apoptosis in cultured transformed lung cells (Nichols et al., 2003). With the current studies, we demonstrated for the first time that cytochrome P450–activated 3MI can cause single-strand breaks in the DNA of primary cultures of human lung epithelial cells from the bronchi, the same cells likely to be exposed to high levels of 3MI from inhaled tobacco smoke.
Other studies (Reddy et al., 2002) with a series of indole-containing chemicals, also showed that 3MI formed extensive amounts of DNA adducts. In fact, 3MI/DNA adduct amounts that were measured with the 32P-postlabeling assay in primary rat hepatocytes or with human genomic DNA (with 25μM 3MI) were almost identical to the DNA adduct levels formed by benzo[a]pyrene (50μM), a prototype genotoxin (Laws et al., 2001; Reddy et al., 2002). Surprisingly, 3MI did not produce DNA strand breaks in rat hepatocytes nor was 3MI mutagenic in a reverse mutant bacterial assay using S. typhimurium strain TA100 (Reddy et al., 2002). Incubations of 3MI with rat liver S9 fractions agreed with the results of Reddy et al. However, our results with CYP2F3 incubations demonstrated for the first time that 3MI is highly mutagenic (S. typhimurium strain TA98). Why were such disparate results obtained? We postulated that liver homogenates do not contain sufficient amounts of any cytochrome P450 enzyme capable of bioactivating 3MI to its genotoxic intermediate(s) in quantities sufficient to cause microbial reverse mutants. Rather, only incubations with lung fractions, or perhaps more appropriately, with a recombinant purified lung-expressed P450 enzyme (CYP2F3), would produce sufficient amounts of the electrophilic DNA-alkylating reactive intermediate (3MEIN) to be mutagenic. This hypothesis proved to be correct. 3MI was only mutagenic when it was metabolized by CYP2F3, not by rat liver S9, and benzo[a]pyrene was only mutagenic when it was metabolized by P450 enzymes contained in rat liver S9 fractions (Tables 1 and 2). Perhaps, the most striking result was that 3MI was shown to be as potent a mutagen as benzo[a]pyrene, the prototypical cigarette smoke mutagen and carcinogen. A comparison of rat liver Aroclor 1254–induced S9-mediated benzo[a]pyrene mutagenicity in this assay to CYP2F3-mediated 3MI mutagenicity indicated that 3MI displayed maximal mutagenicity with concentrations of ∼100μM (6.56 mg), the same approximate concentration required for maximal mutagenicity of benzo[a]pyrene.
In addition to the observed lack of 3MI-mediated mutagenesis, Reddy et al. also did not observe DNA strand breaks from 3MI treatment. Conversely, 3MI was genotoxic because it did produce extensive chromosomal aberrations in Chinese hamster ovary cells and was mutagenic to Salmonella bacteria (our results). We do not know why strand breaks were not observed in hepatocytes, even though the relative amounts of DNA adducts from 3MI incubations with pSP189 shuttle vector DNA were as high, if not higher, than the adduct levels observed when benzo[a]pyrene was incubated with the S9 bioactivation system, or with the mutagenic dihydrodiol epoxide of benzo[a]pyrene. However, the seemingly contradictory observations that 3MI did not cause DNA strand breaks, but did cause chromosomal aberrations, were actually mirrored by benzo[a]pyrene. Therefore, it seems that it is possible for a potent carcinogen to exhibit these somewhat surprising results. It is possible that large differences in DNA repair processes among disparate types of cells might explain these perplexing observations. An additional complication that might be important is that 3MI was shown by Reddy et al. to be cytotoxic to three different cell types: hepatocytes, CHO cells, and Salmonella cells. Therefore, cytotoxicity during the process of DNA repair might have altered the normal cellular response to DNA alkylation and thus changed the repair process or the ability to detect fragments of DNA.
We believe that the activating system used for the mutagenesis assay (rat hepatic S9 fraction) might not have oxidized 3MI to 3MEIN, or other DNA-reactive intermediates, in sufficient amounts to produce a positive response. Our recent studies with human liver microsomal incubations (Yan et al., 2007) demonstrated substantial differences in metabolic pathways and bioactivation events, compared to those catalyzed by CYP2F1 or CYP2F3, the predominant lung-expressed P450 enzymes that catalyze only dehydrogenation to 3MEIN (Lanza and Yost, 2001).
The extent of DNA damage seen with 3MI in NHBE cells in the present study was similar to the extent of damage seen with the same concentrations of doxorubicin (Figs. 2 and 4), a prototype antineoplastic drug, as well as the topoisomerase-1 inhibitor irinotecan (Fig. 4), known to cause DNA single-strand breaks. The extensive DNA damage produced by such low concentrations of 3MI speaks to the potency of this alkylating agent. Most of the damage caused by 3MI was repairable, however, as demonstrated by the decrease in single-strand breaks after 4 h of exposure (Fig. 3), and the increase in damage seen when PARP1 was inhibited (Fig. 5), preventing the recruitment of base excision repair enzymes to single-strand break sites (Beneke et al., 2004; Schreiber et al., 2002). However, new DNA damage is probably not taking place after a few hours because 3MI inactivates CYP2F1. Thus, even though 3MI could still have been present in cellular incubations for longer than a few hours, bioactivation to the DNA-damaging electrophile was probably not long lasting.
Recently published results (D'Agostino et al., forthcoming) from our investigations of human lung P450 enzymes showed that the intriguing organ-selective enzyme, CYP2A13, efficiently bioactivated 3MI to its dehydrogenated intermediate, 3MEIN (Fig. 1). In addition, this enzyme was also inactivated in a mechanism-based process, similar to the inactivation of the CYP2F enzymes. In fact, 2A13 was more sensitive to 3MI-mediated inactivation than CYP2F1 or CYP2F3. Thus, it seems possible that bioactivation by multiple P450 enzymes that are expressed selectively in the lung, but are also inactivated by 3MI, could complicate the chronology of DNA-damaging events and lead to results that are difficult to explain.
DNA damage is detectable at 3MI concentrations two orders of magnitude lower than those that induce apoptosis (Figs. 2 and 7). The damage seen at sublethal concentrations of 3MI diminishes with time and increases when cells are pretreated with an inhibitor of PARP1 (Fig. 5), an enzyme thought to mediate single-strand DNA repair, among other functions (de Murcia et al., 1997; Ikejima et al., 1990). This implies that a major portion of the DNA damage caused by 3MI adduct formation can be repaired by the cells, which could permit cells with fixed mutations to replicate. Further evidence that 3MI is a potential carcinogen in human lung cells can be found in the cells’ response to exposure. Submicromolar levels of 3MI induced phosphorylation and nuclear localization of the tumor suppressor p53 (Fig. 6). This signaling molecule is involved in regulating the cell cycle and DNA repair (el-Deiry et al., 1994). It is likely that processes mediated by these signals allow the cells to repair most of the damage in their genetic material caused by 3MI adduct formation. However, it seems likely that critical mutations to oncogenes might be caused by 3MI and passed on during accelerated cell proliferation.
The DNA of cells lining the airways of smokers is repeatedly exposed to significant levels of bioactivated 3MI. Our work presented here demonstrates that this exposure may cause significant damage to normal bronchial epithelial cell DNA and that this damage elicits a biological response. Only at higher concentrations are apoptotic pathways activated, leading to the death of the damaged cell. Cycles of DNA damage and repair will inevitably lead to an increased rate of mutation due to the intrinsic tendency of DNA polymerases involved in the repair processes to insert a noncomplementary nucleotide at a low but measurable rate (Osheroff et al., 1999; Semenenko and Stewart, 2005; Semenenko et al., 2005). Detectable DNA damage occurred in vitro at concentrations as low as 100nM (Fig. 2),while a significant increase in apoptosis did not occur below 25μM (Fig. 7). Thus, there is a broad window of 3MI exposure where normal bronchial epithelial cells will have their genetic material damaged by bioactivated 3MI and yet survive and continue proliferating. We thus conclude that 3MI exposure in the smoker's lung has the potential to increase the rate of mutation within these lung cells and thus may act as a potential carcinogen. Apoptosis of 3MI-exposed lung cells at higher concentrations can be viewed as a protective mechanism and could at least partially explain why many heavy smokers never develop lung cancer.
FUNDING
National Heart, Lung, and Blood Institute (R01HL013645).
Acknowledgments
The authors wish to express appreciation for the considerable assistance of Diane L. Lanza, University of Utah, for her help with CYP2F3 enzyme activity determinations.
References
- Ames BN, McCann J, Yamasaki E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mutat. Res. 1975;31:347–364. doi: 10.1016/0165-1161(75)90046-1. [DOI] [PubMed] [Google Scholar]
- Beneke S, Diefenbach J, Burkle A. Poly(ADP-ribosyl)ation inhibitors: promising drug candidates for a wide variety of pathophysiologic conditions. Int. J. Cancer. 2004;111:813–818. doi: 10.1002/ijc.20342. [DOI] [PubMed] [Google Scholar]
- Carr BA, Ramakanth S, Dannan GA, Yost GS. Characterization of pulmonary CYP4B2, specific catalyst of methyl oxidation of 3-methylindole. Mol. Pharmacol. 2003;63:1137–1147. doi: 10.1124/mol.63.5.1137. [DOI] [PubMed] [Google Scholar]
- Cavalieri EL, Rogan EG. A unifying mechanism in the initiation of cancer and other diseases by catechol quinones. Ann. N. Y. Acad. Sci. 2004;1028:247–257. doi: 10.1196/annals.1322.029. [DOI] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 1999;96:13777–13782. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumakov PM. Versatile functions of p53 protein in multicellular organisms. Biochemistry (Mosc.) 2007;72:1399–1421. doi: 10.1134/s0006297907130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Agostino J, Zhuo X, Shadid M, Morgan DG, Zhang X, Humphreys WG, Shu YZ, Yost GS, Ding X. The pneumotoxin 3-methylindole is a substrate as well as a mechanism-based inactivator of CYP2A13, a human cytochrome P450 enzyme preferentially expressed in the respiratory tract. Drug Metab. Dispos. doi: 10.1124/dmd.109.027300. Published on July 16, 2009. Doi:10.1124/dmd.109.027300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Harper JW, O'Connor PM, Velculescu VE, Canman CE, Jackman J, Pietenpol JA, Burrell M, Hill DE, Wang Y, et al. WAF1/CIP1 is induced in p53-mediated G1 arrest and apoptosis. Cancer Res. 1994;54:1169–1174. [PubMed] [Google Scholar]
- Fordtran JS, Scroggie WB, Polter DE. Colonic absorption of tryptophan metabolites in man. J. Lab. Clin. Med. 1964;64:125–132. [PubMed] [Google Scholar]
- Ikejima M, Noguchi S, Yamashita R, Ogura T, Sugimura T, Gill DM, Miwa M. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J. Biol. Chem. 1990;265:21907–21913. [PubMed] [Google Scholar]
- Kartha JS, Skordos KW, Sun H, Hall C, Easterwood LM, Reilly CA, Johnson EF, Yost GS. Single mutations change CYP2F3 from a dehydrogenase of 3-methylindole to an oxygenase. Biochemistry. 2008;47:9756–9770. doi: 10.1021/bi8005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartha JS, Yost GS. Mechanism-based inactivation of lung-selective cytochrome P450 CYP2F enzymes. Drug Metab. Dispos. 2008;36:155–162. doi: 10.1124/dmd.107.017897. [DOI] [PubMed] [Google Scholar]
- Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991;252:1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- Lanza DL, Yost GS. Selective dehydrogenation/oxygenation of 3-methylindole by cytochrome p450 enzymes. Drug Metab. Dispos. 2001;29:950–953. [PubMed] [Google Scholar]
- Laws GM, Skopek TR, Reddy MV, Storer RD, Glaab WE. Detection of DNA adducts using a quantitative long PCR technique and the fluorogenic 5’ nuclease assay (TaqMan) Mut. Res. 2001;484:3–18. doi: 10.1016/s0027-5107(01)00263-9. [DOI] [PubMed] [Google Scholar]
- Murph MM, Hurst-Kennedy J, Newton V, Brindley DN, Radhakrishna H. Lysophosphatidic acid decreases the nuclear localization and cellular abundance of the p53 tumor suppressor in A549 lung carcinoma cells. Mol. Cancer Res. 2007;5:1201–1211. doi: 10.1158/1541-7786.MCR-06-0338. [DOI] [PubMed] [Google Scholar]
- Nichols WK, Mehta R, Skordos K, Mace K, Pfeifer AM, Carr BA, Minko T, Burchiel SW, Yost GS. 3-methylindole-induced toxicity to human bronchial epithelial cell lines. Toxicol. Sci. 2003;71:229–236. doi: 10.1093/toxsci/71.2.229. [DOI] [PubMed] [Google Scholar]
- Osheroff WP, Jung HK, Beard WA, Wilson SH, Kunkel TA. The fidelity of DNA polymerase beta during distributive and processive DNA synthesis. J. Biol. Chem. 1999;274:3642–3650. doi: 10.1074/jbc.274.6.3642. [DOI] [PubMed] [Google Scholar]
- Reddy MV, Storer RD, Laws GM, Armstrong MJ, Barnum JE, Gara JP, McKnight CG, Skopek TR, Sina JF, DeLuca JG, et al. Genotoxicity of naturally occurring indole compounds: correlation between covalent DNA binding and other genotoxicity tests. Environ. Mol. Mutagen. 2002;40:1–17. doi: 10.1002/em.10088. [DOI] [PubMed] [Google Scholar]
- Regal KA, Laws GM, Yuan C, Yost GS, Skiles GL. Detection and characterization of DNA adducts of 3-methylindole. Chem. Res. Toxicol. 2001;14:1014–1024. doi: 10.1021/tx0100237. [DOI] [PubMed] [Google Scholar]
- Ruangyuttikarn W, Appleton ML, Yost GS. Metabolism of 3-methylindole in human tissues. Drug Metab. Dispos. 1991;19:977–984. [PubMed] [Google Scholar]
- Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, Menissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 2002;277:23028–23036. doi: 10.1074/jbc.M202390200. [DOI] [PubMed] [Google Scholar]
- Semenenko VA, Stewart RD. Monte Carlo simulation of base and nucleotide excision repair of clustered DNA damage sites. II. Comparisons of model predictions to measured data. Radiat. Res. 2005;164:194–201. doi: 10.1667/rr3414. [DOI] [PubMed] [Google Scholar]
- Semenenko VA, Stewart RD, Ackerman EJ. Monte Carlo simulation of base and nucleotide excision repair of clustered DNA damage sites. I. Model properties and predicted trends. Radiat. Res. 2005;164:180–193. doi: 10.1667/rr3402. [DOI] [PubMed] [Google Scholar]
- Skiles GL, Yost GS. Mechanistic studies on the cytochrome P450-catalyzed dehydrogenation of 3-methylindole. Chem. Res. Toxicol. 1996;9:291–297. doi: 10.1021/tx9501187. [DOI] [PubMed] [Google Scholar]
- Soldatenkov VA, Smulson M. Poly(ADP-ribose) polymerase in DNA damage-response pathway: implications for radiation oncology. Int. J. Cancer. 2000;90:59–67. doi: 10.1002/(sici)1097-0215(20000420)90:2<59::aid-ijc1>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Thornton-Manning JR, Nichols WK, Manning BW, Skiles GL, Yost GS. Metabolism and bioactivation of 3-methylindole by Clara cells, alveolar macrophages, and subcellular fractions from rabbit lungs. Toxicol. Appl. Pharmacol. 1993;122:182–190. doi: 10.1006/taap.1993.1186. [DOI] [PubMed] [Google Scholar]
- Wang H, Lanza DL, Yost GS. Cloning and expression of CYP2F3, a cytochrome P450 that bioactivates the selective pneumotoxins 3-methylindole and naphthalene. Arch. Biochem. Biophys. 1998;349:329–340. doi: 10.1006/abbi.1997.0479. [DOI] [PubMed] [Google Scholar]
- Wynder EL, Hoffmann D. Tobacco and Tobacco Smoke: Studies in Experimental Carcinogenesis. New York, NY: Academic Press; 1967. [Google Scholar]
- Yan Z, Easterwood LM, Maher N, Torres R, Huebert N, Yost GS. Metabolism and bioactivation of 3-methylindole by human liver microsomes. Chem. Res. Toxicol. 2007;20:140–148. doi: 10.1021/tx060239e. [DOI] [PubMed] [Google Scholar]
- Yokoyama MT, Carlson JR. Microbial metabolites of tryptophan in the intestinal tract with special reference to skatole. Am. J. Clin. Nutr. 1979;32:173–178. doi: 10.1093/ajcn/32.1.173. [DOI] [PubMed] [Google Scholar]
- Yost GS. Mechanisms of 3-methylindole pneumotoxicity. Chem. Res. Toxicol. 1989;2:273–279. doi: 10.1021/tx00011a001. [DOI] [PubMed] [Google Scholar]