Abstract
Mutating the rare A32-U38 nucleotide pair at the top of the anticodon loop of E. coli tRNAGGCAla to a more common U32-A38 pair results in a tRNA that performs almost normally on cognate codons but is unusually efficient in reading near-cognate codons. Pre-steady state kinetic measurements on E. coli ribosomes show that unlike the wild-type tRNAGGCAla, the misreading mutant tRNAGGCAla shows rapid GTP hydrolysis and no detectable proofreading on near-cognate codons. Similarly, tRNAGGCAla mutated to contain C32-G38, a pair which is found in some bacterial tRNAGGCAla sequences, was able to decode only the cognate codons, while tRNAGGCAla containing a more common C32-A38 pair was able to decode all cognate and near-cognate codons tested. We propose that many of the phylogenetically conserved sequence elements present in each tRNA have evolved to suppress translation of near-cognate codons.
INTRODUCTION
Numerous biochemical experiments suggest that the 45 elongator aminoacyl-tRNAs (aa-tRNAs) in E. coli act as equivalent substrates of the translational machinery. More than twenty different E. coli aa-tRNAs were found to bind Elongation Factor-Tu (EF-Tu) with similar affinities1, and eight show nearly identical rates of dissociation from the A site and the P site of encoded E. coli ribosomes2. Recent experiments have shown that ten different E. coli aa-tRNAs have nearly identical ternary complex binding affinities to the ribosomal entry site as well as similar rates of GTP hydrolysis and peptide bond formation during decoding3. In contrast to their uniform functional properties, aa-tRNAs are chemically quite different from one another. Phylogenetic analysis of tRNA sequences from 145 bacteria with fully sequenced genomes indicates that each tRNA isoacceptor has a unique set of consensus residues distributed throughout the molecule4. In addition, each tRNA species contains different types and numbers of post-transcriptional modifications in the anticodon loop and the EF-Tu tertiary core5. Several experiments have shown that when these consensus residues are mutated or when one or more of the modifications are removed, the uniform functional properties of the aa-tRNA are lost. For example, removing all the post-transcriptional modifications from aa-tRNAs weakens the binding to the ribosomal A or P sites of several aa-tRNAs2. Base pair changes in the T-stem of individual tRNAs can either weaken or strengthen their binding affinity to EF-Tu6. Removal of selected modifications or changes in the sequence within the body of a suppressor tRNA can also either increase or decrease its ability to decode a termination codon in vivo7–9. These experiments suggest that the overall chemical composition of every aa-tRNA has been “tuned” by evolution such that each aa-tRNA functions equivalently in the decoding process.
While the emerging data support the view that tRNA sequences are idiosyncratically tuned for uniform behavior during decoding, it does not explain the underlying reason why this has occurred. It is unclear what the evolutionary disadvantage would be if the different aa-tRNAs showed a range of affinities for the ribosome or proceeded through the decoding pathway at different rates. One possibility is that the uniform behavior is related to the need for aa-tRNAs to undergo accurate decoding. Each aa-tRNA must read its cognate codons, but it must not efficiently read the structurally similar near-cognate codons which contain a single nucleotide mismatch. Numerous experiments have shown that the introduction of certain tRNA mutations or the removal of an individual post-transcriptional modification can lead to misreading10–14 or translational frameshifting15–18 in vivo. However, a mechanistic understanding of this phenomenon is limited to the G24A mutation of E. coli tRNATrp, which substantially promotes misreading of several near-cognate codons19. Here, we evaluated the mutations in the anticodon loop of E. coli tRNAGGCAla known to stabilize binding to ribosomes for their ability to read near-cognate codons using kinetic and thermodynamic assays that measure different steps in the decoding process.
RESULTS
Mutating A32-U38 has little effect on cognate decoding
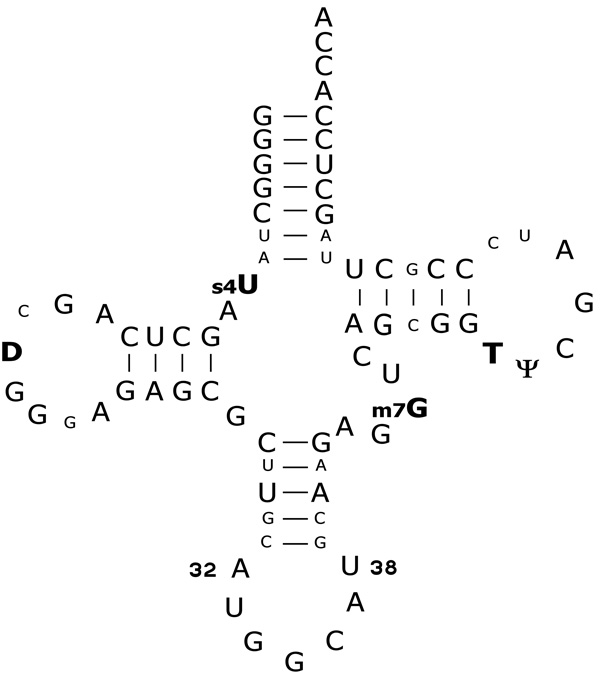
tRNAGGCAla is the minor alanine isoacceptor in E. coli which selectively reads its complementary GCC and wobble GCU codons20. Since the major isoacceptor tRNAUGCAla is capable of reading all four alanine codons, tRNAGGCAla is not essential for growth, though its deletion causes a slow growth phenotype in minimal media21. One of the distinctive structural features of tRNAGGCAla is the A–U pair at positions 32 and 38 at the top of the anticodon loop (Fig. 1). This combination of residues is quite rare in bacterial tRNAs, only being present in tRNAGGCAla and tRNAGGGPro5. Recent experiments measuring the binding of E. coli tRNAGGCAla to the A site of ribosomes encoded with a complementary GCC codon showed that the identity of the nucleotide pair at positions 32 and 38 modulates the tRNA binding affinity22. While the wild-type tRNAGGCAla demonstrated an A site binding affinity similar to other deacylated elongator tRNAs2, replacement of the A32-U38 pair by U-A, U-U, or A-A pairs caused the binding affinity of tRNAGGCAla to be four to ten-fold tighter22. Introduction of the C32-G38 pair which is present in tRNAGGCAla sequences of some other bacteria had no effect on A site binding. This suggested that the rare A32-U38 pair and its phylogenetic alternative C32-G38 have evolved to weaken the tRNAGGCAla binding affinity for ribosomes to ensure that its affinity is similar to that of other tRNAs.
Figure 1.
Secondary structure of E. coli tRNAGGCAla. The nucleotides in bold with post-transcriptional modifications were not modified in the tRNAs used for this study. Residues in smaller font are present in E. coli tRNAGGCAla but are not conserved among all bacterial tRNAGGCAla. Positions 32 and 38 in the anticodon loop are numbered.
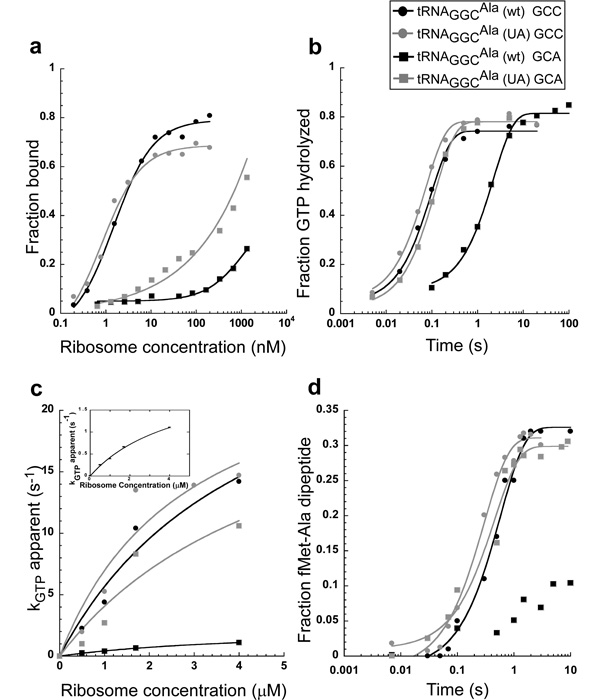
Since the binding affinity of aa-tRNAs to the ribosomal A site does not measure a step in the normal decoding process, discerning the relevance of the 32–38 pair for tRNAGGCAla function requires assays that measure decoding directly. As mutated tRNAGGCAla sequences are most easily tested using unmodified tRNA transcripts, the decoding properties of unmodified tRNAGGCAla were first compared to previous data of its fully modified counterpart. Unmodified Ala-tRNAGGCAla was assayed on E. coli ribosomes programmed with a 27 nucleotide derivative of the initiation region of T4 gp32 mRNA displaying the cognate GCC codon in the A site and an AUG in the P site. As previously described in greater detail3, three different assays were used to evaluate the ability of Ala-tRNAGGCAla to undergo decoding. First, the equilibrium dissociation constant (Kd) of a catalytically inactive ternary complex bound to the entry site of E. coli ribosomes containing tRNAfMet in the P site was determined (Fig. 2a)23. Second, the rate of GTP hydrolysis by the ternary complex was determined at several encoded ribosome concentrations in order to deduce kGTPmax, the GTPase rate at saturating ribosome concentrations (Fig. 2b, c). Finally, kpep, the observed rate of peptide bond formation between fMet-tRNAfMet and Ala-tRNAGGCAla was measured (Fig. 2d). The unmodified tRNAGGCAla and the previously assayed modified tRNAGGCAla showed similar values of Kd (2.3 nM vs. 1.7 nM), kGTPmax (31 s−1 vs. 45 s−1), and kpep (1.7 s−1 vs. 2.0 s−1) when determined under identical reaction conditions3. Thus, unlike several other tRNAs2,10,24, the post-transcriptional modifications have only a small effect on the decoding process of tRNAGGCAla in the conditions used in these in vitro experiments. This is likely to reflect the fact that native tRNAGGCAla has no modifications in the anticodon loop and only has five modifications in the tertiary core which do not directly contact the ribosome (Fig. 1)20,25,26. Although removal of the modifications in the tertiary core of tRNA can destabilize tRNA structure, these effects are minimal in the buffer containing 10 mM MgCl2. However, in buffers containing lower MgCl2 concentrations, such as the high fidelity buffers often used in translation studies, transcripts are not fully folded27–30.
Figure 2.
Comparison of tRNAGGCAla (wt) to tRNAGGCAla (UA) on the GCC cognate and GCA near-cognate codons. (a) Equilibrium dissociation curves of catalytically inactive ternary complexes binding to the ribosomal entry site. (b) Timecourse of GTP hydrolysis at 1.7 µM ribosomes. (c) Ribosome saturation curve of GTP hydrolysis. (d) Timecourse of dipeptide formation between fMet-tRNAfMet and Ala-tRNAGGCAla. Dipeptide formation for tRNAGGCAla (wt) on the GCA codon could not be fit to a simple exponential, so no line was drawn.
The ability of the unmodified wild-type tRNA, tRNAGGCAla (wt), to decode its cognate codons was compared to a tight-binding double mutant where the wild-type A32-U38 pair was changed to a U32-A38 pair, tRNAGGCAla (UA)22. Both of these tRNAs were effective in decoding the GCC and GCU codons, but the ternary complex containing tRNAGGCAla (UA) bound to the perfectly complementary GCC codon approximately two-fold tighter and the wobble GCU codon about four-fold tighter than tRNAGGCAla (wt) (Table 1). However, tRNAGGCAla (UA) exhibited kGTPmax and kpep values that were indistinguishable from tRNAGGCAla (wt) on both cognate codons (Table 1). Thus, it appears that while mutating the A32-U38 pair to U32-A38 in tRNAGGCAla slightly increases the affinity of the ternary complex for the ribosome, it does not affect the subsequent kinetic steps of decoding under the conditions used here.
Table 1.
Thermodynamic and kinetic parameters for different tRNAGGCAla on cognate and near-cognate codons
| Kd (nM)a | kGTPmax (s−1)b | kpep (s−1)a | ||||||
|---|---|---|---|---|---|---|---|---|
| Codon | A-U | U-A | A-U | U-A | A-U | U-A | C-G | C-A |
| GGC | 2.3 ± 0.40 | 1.0 ± 0.23 | 31 ± 13 | 27 ± 11 | 1.7 ± 0.21 | 1.8 ± 0.69 | 1.5 ± 0.70 | 1.4 ± 0.27 |
| GCU | 5.9 ± 0.93 | 1.3 ± 0.24 | 23 ± 3.3 | 25 ± 3.1 | 1.4 ± 0.17 | 1.4 ± 0.16 | 1.5 ± 0.36 | 1.8 ± 0.15 |
| GCG | ~1000c | 175 ± 30 | ||||||
| GCA | ~1000c | 210 ± 52 | 2.4 ± 0.23 | 26 ± 18 | <0.05d | 2.1 ± 0.34 | <0.05 d | 3.4 ± 0.64 |
| GUC | <0.05 d | 1.4 ± 0.36 | <0.05 d | 1.6 ± 0.35 | ||||
| ACC | <0.05 d | 0.49 ± 0.16 | <0.05 d | 2.8 ± 0.79 | ||||
Error values are the standard error of the mean value.
Values are the average of at least three independent experiments.
Values were determined based on curves fit to at least four apparent kGTP values determined at different ribosome concentrations.
Estimated value since precise Kd determination exceeded the limits of accurate measurement.
Estimated limit (see Methods).
The A32-U38 pair in tRNAGGCAla prevents misreading
The binding affinities of the wild-type and mutant tRNAGGCAla ternary complexes to the ribosomal entry site were assayed using the two near-cognate alanine codons GCA and GCG which introduce an A–G or G-G mismatch into the third position of the codon-anticodon helix. As would be expected based on previous studies comparing wild-type tRNA binding to near-cognate codons19,31, both ternary complexes bound the mismatched codons much less well than the cognate codons (Fig. 2a, Table 1). However, ternary complex containing tRNAGGCAla (UA) bound the near-cognate codons at least five-fold tighter than complexes containing tRNAGGCAla (wt). Hence, the stabilizing effect of mutating the A32-U38 base pair to U-A is similar or even slightly greater on the near-cognate codons than was observed with the cognate codons. Since the rate of ternary complex association to ribosomes has previously been shown to be the same for both cognate and near-cognate codons32, it is likely that the stabilizing effect of the mutation is due to a slower dissociation rate of the ternary complex off the ribosome. This would result in tRNAGGCAla (UA) being less accurate in the initial selection step of decoding33. This suggests that one reason the A32-U38 pair in tRNAGGCAla has evolved to be so well conserved is to destabilize ternary complex binding to ribosomes and thereby improve the accuracy of the initial selection steps of decoding.
To assess whether the 32–38 pair also influences translation accuracy in the subsequent steps of decoding, it was first necessary to determine how well the tRNAGGCAla (wt) transcript can decode a near-cognate codon. As shown in Figures 2b and 2c, it was possible to obtain values of kGTP at several ribosome concentrations and estimate a kGTPmax using the near-cognate GCA codon, despite the fact that the ternary complex binds less well to ribosomes containing mismatched codons. The value of kGTPmax was 2.4 s−1, which is 13-fold slower than the value obtained for the cognate GCC codon. As shown in Figure 2d, the formation of dipeptide bond on the near cognate GCA codon occurs much more slowly than kpep=1.7 s−1 obtained with the cognate codon, but it was only possible to estimate kpep to be less than 0.05 s−1 (see Methods). The slower values of kGTPmax and kpep have been explained by an induced fit mechanism in which the mismatched codon-anticodon interaction causes incorrect adaptation of the tRNA to the ribosome and thereby prevents the ribosomal conformational changes needed to promote rapid catalysis33,35,36.
The tighter binding tRNAGGCAla (UA) shows dramatically different behavior from tRNAGGCAla (wt) in decoding the near-cognate GCA codon. As shown in Figures 2b and 2c and summarized in Table 1, tRNAGGCAla (UA) shows a value of kGTPmax of 27 s−1, which is substantially faster than the value of 2.4 s−1 observed with tRNAGGCAla (wt) and is essentially the same rate as observed on its cognate codon. Similarly, the value of kpep=2.1 s−1 on the GCA near-cognate codon is also significantly accelerated such that it is nearly equal to the value determined using the cognate GCC codon (Fig. 2d, Table 1). The fact that the fraction of dipeptide formed reaches the same level with near-cognate codons as with cognate codons indicates that the mutant tRNA is not significantly rejected off the ribosome in the presence of the near-cognate codon. In other words, tRNAGGCAla (UA) seems to evade the proofreading process by stimulating the forward reaction rates so that it efficiently makes dipeptide on the near-cognate codon.
Since the difference in initial selection rates for tRNAs on cognate versus near-cognate codons is increased in buffers containing low concentrations of MgCl2 and with polyamines33,34, it was of interest to determine if tRNAGGCAla (UA) would also be capable of decoding a near-cognate codon in such a high fidelity buffer. While the Kd of the different ternary complexes could not be determined even on the cognate codons in this buffer due to poor stability of the ribosome – ternary complex adduct in the filter retention assay, the rates of GTP hydrolysis and peptide bond formation could be measured. The apparent rate of GTP hydrolysis determined in high fidelity buffer with 2 µM ribosomes showed that, similar to the 10 mM MgCl2 buffer, tRNAGGCAla (wt) has a fast rate of hydrolysis on the cognate codon and a much slower rate on the near-cognate GCA codon, while tRNAGGCAla (UA) has a similar, fast rate of GTP hydrolysis on both codons (Supplementary Fig.1). However, unlike with the 10 mM MgCl2 buffer (Fig 2b), the extent of GTP hydrolysis achieved at long incubation times was only 20%, reflecting that a significant fraction of the EF-Tu GTP did not have an aa-tRNA stably bound. This presumably reflects the poor folding of the transcript tRNA in this buffer. Experiments measuring the rate of peptide bond formation in the high fidelity buffer showed that tRNAGGCAla (wt) rapidly formed dipeptide in the presence of the cognate codon and not the GCA near-cognate codon, while tRNAGGCAla (UA) could efficiently form dipeptide on both codons (Supplementary Fig.2). Since the data collected in the high fidelity buffer resembled the data collected in the 10 mM MgCl2 buffer in which tRNA folding was not compromised, the 10 mM MgCl2 buffer was used for the remainder of the experiments.
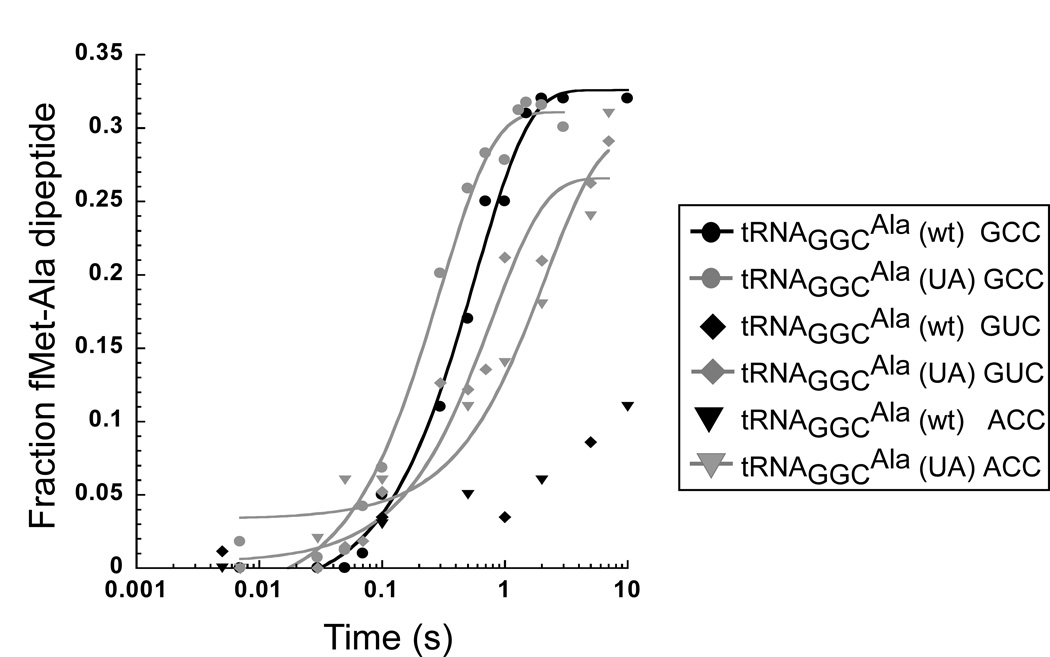
To determine if tRNAGGCAla (UA) is also capable of misreading other near-cognate codons, the kpep assay was used to monitor the rate of misincorporation at ACC (threonine) and GUC (valine) codons, which form mismatches at the first and second codon positions, respectively. Similar to the results with the mismatched GCA (alanine) codon, tRNAGGCAla (UA) was able to misread both near-cognate codons with rates, and extents of reaction similar to the cognate GCC codon while tRNAGGCAla (wt) gave very slow rates (Fig. 3, Table 1). This indicates that tRNAGGCAla (UA) has lost its ability to perform accurate decoding on any near-cognate codon.
Figure 3.
Timecourse of peptide bond formation for tRNAGGCAla (wt) and tRNAGGCAla (UA) on the cognate GCC codon (taken from Fig. 2d) and the mismatched ACC and GUC codons. Only data that can be fit to a simple exponential is fit to a line (see Methods).
To determine whether misreading was a phenomenon specific to the U32-A38 pair, two other mutant tRNAGGCAla molecules were prepared and tested using the kpep assay with the two cognate codons and the three near-cognate codons (Table 1). tRNAGGCAla was mutated to contain two other 32–38 nucleotide pairs, one (C32-A38) which is commonly found in bacterial tRNAs other than tRNAGGCAla, and another (C32-G38) which is conserved in 22% of known bacterial tRNAGGCAla sequences but is only present in 1.2% of all bacterial tRNAs5. tRNAGGCAla (CA), representing a 32–38 pair which is present in 52% of all bacterial tRNAs5, was able to read cognate codons normally, but is also very rapid and efficient at misreading all three near-cognate codons, similarly to tRNAGGCAla (UA). In contrast, tRNAGGCAla (CG) behaves like tRNAGGCAla (wt) in effectively reading the cognate codons but shows very slow rates of kpep on near-cognate codons. These results are consistent with the fact that tRNAGGCAla in some bacteria contain the rare C32-G38 pair in place of A32-U38, but none contain the C32-A38 or U32-A38 pairs5.
DISCUSSION
The A32-U38 pair was originally identified as a sequence element that destabilized binding of tRNAs to the ribosomal A site22. It was hypothesized that the purpose of this pair in bacterial tRNAGGCAla and tRNAGGGPro was to off-set the stabilizing effect of their very GC rich codon-anticodon pairs so that they would act similarly to other tRNAs when decoding their cognate codons. Although this view may be correct, experiments presented here that measure decoding on near-cognate codons using pre-steady state kinetics make it clear that a critical role of this base pair is to prevent misreading. When this A32-U38 pair is mutated to a more common 32–38 pair, the resulting ternary complex is not only able to bind ribosomes somewhat tighter than the wild-type tRNA, but is also able to stimulate GTP hydrolysis and peptide bond formation equally well on both cognate and near-cognate codons. Since these effects would reduce the ability of tRNAGGCAla to distinguish cognate from near-cognate codons in both the initial selection and proofreading steps of decoding, it is likely that the A32-U38 was selected to maintain translational accuracy.
Once bound to the ribosome, it is astonishing how well tRNAGGCAla (UA) can function on near-cognate codons. Both the maximal rate of GTP hydrolysis and the rate and extent of peptide bond formation are indistinguishable from what is observed for tRNAGGCAla (wt) with its cognate codon. In other words, in these assays the ribosome does not detect that a mismatched codonanticodon complex has formed and allows peptide bond formation to occur normally without any proofreading. This ability of tRNAGGCAla (UA) to efficiently read near-cognate codons far exceeds the in vitro effects of error inducing antibiotics37,38. While the G24A mutation of E. coli tRNATrp also shows substantial misreading in vitro, it exhibits no difference in binding to near-cognate codons on the ribosome as a ternary complex and still shows significantly reduced levels of peptide bond formation on mismatched codons, indicating that some proofreading occurs19. tRNAGGCAla (UA) is able to rapidly undergo GTP hydrolysis and peptide bond formation on the near-cognate GCA codon even in the low magnesium high fidelity buffer in which the unmodified transcript is not as well folded as in the standard 10 mM MgCl2 buffer.
Several of the results presented here have been confirmed (H. Murakami and H. Suga, University of Tokyo, personal communication) using an in vitro translation assay with purified components to prepare oligopeptides from a defined mRNA. They found that transcripts of tRNAGGCAla containing one of the more common 32–38 pairs (C-A, U-A or U-U) were effective at incorporating alanine at a GUC (valine) codon, while tRNAGGCAla (wt) and tRNAGGCAla (CG) were not. It is interesting that when a competitor tRNA with an anticodon cognate to the GUC codon was added to the reaction, misincorporation by the tRNAGGCAla mutants was strongly suppressed, presumably because the competitor ternary complex can bind its cognate codon much tighter than the tRNAGGCAla mutants. Effective competition by correctly matched tRNAs probably also explains why expression of the misreading tRNAGGCAla (CA) in E. coli only has modest effects on bacterial growth (H. Murakami and H. Suga, University of Tokyo, personal communication).
The 32–38 pair modulates the binding of the Ala-tRNAGGCAla ternary complex to the ribosomal entry site in a manner similar to how it modulates binding of the deacylated tRNAGGCAla to the ribosomal A site22. It is likely that in both cases the explanation of the sequence specificity lies in the structure of the anticodon loop since the 32–38 pair of tRNAPhe present in high resolution crystal structures does not appear to interact directly with the 30S ribosome in either complex36,39. As discussed previously40, the A32-U38 pair may form a stable Watson-Crick pair that in turn allows U33 and A37 to form a base pair resulting in a three nucleotide anticodon loop. The observed weaker binding of the wild-type A32-U38 pair would then be due to the energy required to break these base pairs to rearrange the loop into a more open conformation upon codon binding. An alternate, less specific explanation for the destabilizing effect of the A32-U38 pair may be that this particular pair is in some way less able to stabilize the codon-anticodon helix through stacking interactions than other non-conserved nucleotide pairs.
A different explanation is required to account for how tRNAGGCAla (UA) is able to efficiently stimulate its rapid forward rates on near-cognate codons once bound to ribosomes. Although no high resolution X-ray structure of the ternary complex bound to ribosomes is available, medium resolution cryoelectron microscopy structures suggest that the structures of tRNA and possibly EF-Tu are distorted when the ternary complex binds to a cognate codon in the entry site26,41,42. It has been proposed that when a mismatched codon is present, the altered structure of the codon-anticodon helix prohibits this distortion in the ternary complex, leading to weaker binding and rejection after GTP hydrolysis35. Presumably, tRNA mutations that promote misreading have altered distortability or dynamics that allows them to fit into the ribosome correctly despite the mismatched codon, as described in the original “waggle” theory43. For example, the misreading G24A mutation of tRNATrp lies close to a major site of distortion in the ribosome bound ternary complexes19,26,42. Although no obvious distortion of this complex is observed in the region of the 32–38 pair, the resolution of the structures are low. In addition, it is unclear whether each ternary complex will distort identically due to their differing tRNA sequences. However, the fact that tRNATrp and tRNAGGCAla utilize very different positions to avoid the same inaccurate decoding phenotype highlights the idea that each tRNA is tuned by different elements.
Although it appears that one important selective pressure on tRNA sequences seems to be to perform equivalently in translating their cognate codons, experiments presented here highlight the fact that tRNA consensus sequences can also maintain translational accuracy. While the A32-U38 (or C32-G38) consensus element in tRNAGGCAla functions in tuning both ternary complex affinity and decoding accuracy, it is uncertain whether this will always be the case. Mutating other tRNAGGCAla consensus elements do not seem to greatly affect ribosome binding22, but their ability to misread remains to be tested. It is possible that the extensive and complex tRNA sequence requirements associated with each anticodon reflect the apparent need for tRNA to show a characteristic deformability to ensure accurate decoding. Other elements, such as posttranscriptional modifications and the identity of the amino acid, are likely important for how the aa-tRNA functions on the ribosome, similar to how the nature of the amino acid is important for aa-tRNA binding to EF-Tu44. In fact, this has recently been shown to be the case for proline, which has a slower rate of dipeptide formation if esterified to tRNAPhe rather than tRNAPro 45. If this is the case, mutations of tRNA consensus elements may not always directly affect aa-tRNA function on cognate codons, but may instead affect their ability to avoid translating near-cognate codons.
METHODS
Materials
We prepared tight-coupled 70S ribosomes from E. coli MRE600 cells as described46. Final ribosome pellets were resuspended in ribosome binding buffer (RB buffer: 50 mM HEPES [pH 7.0], 30 mM KCl, 70 mM NH4Cl, 10 mM MgCl2, and 1 mM DTT) and were stored and activated as previously described2. The mRNAs used were derivatives of the initiation region of the T4 gp32 mRNA with the following sequence: 5’-GGCAAGGAGGUAAAAAUGXXXGCACGU-3’, where XXX indicates the codon complementary to the anticodon of the A site tRNA and the codon 3’ of the A site has been changed from GCA to AAA for all mRNAs with an alanine codon in the A site.
We prepared EF-Tu (H84A) as described3. E. coli tRNAGGCAla was transcribed from templates generated by primer extension of overlapping DNA oligonucleotides (IDT) and was purified via denaturing polyacrylamide gel electrophoresis. [3’-32P] labeling and aminoacylation was performed as previously described47 with typical aminoacylation yields of 70% for all tRNAs including tRNAfMet.
Ternary Complex Binding Assay
Equilibrium binding of ternary complexes to the entry site of the ribosome was determined as previously described3,47. Ternary complex was formed by first converting 0.6 µM EF-Tu (H84A) to its GTP-bound form by incubating it with 100 µM GTP, 3 mM phosphoenolpyruvate, and 12 U ml−1 pyruvate kinase in RB buffer at 37°C for 20 minutes. The GTP activated EF-Tu (H84A) was incubated with 3’32P labeled Ala-tRNA on ice for 20 minutes. A final concentration of <1 nM ternary complex was incubated for at 20°C two minutes with 0.5 – 1300 nM ribosomes, programmed with an excess of mRNA and tRNAfMet. Ribosome bound ternary complex was separated from free ternary complex by filtering over nitrocellulose (Whatman 0.45 µm) and positively charged nylon (Amersham 0.45 µm) membranes in duplicate and washing with 10-fold excess RB buffer. Further washing did not affect the amount of ternary complex retained on the nitrocellulose filter. Since filter saturation made data collection with ribosome concentrations above 1300 nM unfeasible, the Kd values for weak binding complexes were estimated assuming the extent of binding would reach the same level as the tighter cognate complexes. Data was quantified using a phosphoimager (Molecular Dynamics), and binding constants were determined by fitting the data to a single Michaelis-Menten binding isotherm using KaleidaGraph software (Synergy Software).
Kinetic Experiments
The rate of GTP hydrolysis was determined as previously described3,19. Briefly, 300 nM ternary complex was formed with EF-Tu, γ̣-32P GTP, and Ala-tRNA on ice. Excess γ̣-32P GTP was removed by filtration through two P30 spin columns (Bio-Rad) equilibrated with RB buffer. Equal volumes of ternary complex and programmed ribosomes were mixed for set times in a KinTek quench flow apparatus and quenched with 40(v/v) formic acid to determine apparent GTP hydrolysis rates at each ribosome concentration ranging from 0.5 – 4 γM. Hydrolyzed free 32Pi was separated from γ̣-32P GTP by thin layer chromatography (TLC) using PEI cellulose plates run in 0.5 M KH2PO4. The apparent rates of hydrolysis at each ribosome concentration were determined by fitting the fraction of GTP hydrolyzed over time to a single exponential curvefit. The apparent rates were then plotted over the range of ribosome concentrations tested to extrapolate the maximal rate of GTP hydrolysis at a saturating ribosome concentration.
The rate of peptide bond formation was determined as previously described3,47. Equal volumes of 50 nM ternary complex containing EF-Tu, GTP, and 3’32P labeled Ala-tRNA was mixed with 500 nM ribosomes programmed with excess mRNA and fMet-tRNAfMet in the P site using a Kintek quench flow apparatus. Reactions were quenched in 5 mM NaOAc (pH 4.5), 100 mM EDTA. Samples were analyzed by S1 nuclease digestion followed by separating cleaved 32P AMP, 32P Ala-AMP, and 32P fMet-Ala-AMP on PEI cellulose TLC plates in glacial acetic acid/1 M NH4Cl/H2O (5:10:85). The fraction of fMet-Ala dipeptide formed was calculated compared to the total signal for deacyl, aminoacyl, and dipeptidyl tRNA present. The data for the fraction of dipeptide formed over time was fit to a single exponential curvefit to determine the rate of peptide bond formation.
In experiments that measured the time course of peptide bond formation of tRNAGGCAla (wt) or tRNAGGCAla (CG) on the mismatched codons GCA, GUC or ACC, very little dipeptide formed in the first second, but then increasing amounts of product appeared up to 10 seconds (Fig 2d, Fig 3). At longer incubation times, the amount of product slowly continued to increase until as much as 25% dipeptide was formed after 5 minutes (data not shown). This may indicate that in addition to a very slow rate of peptide bond formation on mismatched codons, tRNAGGCAla shows a rate of rejection that is unusually slow compared to tRNAPhe, tRNATrp, and tRNAUGCAla 19,31,33. However, since the kinetic curve could not be fit by a simple exponential, it is also possible that the very slow rate of dipeptide formation in these experiments is the result of multiple binding events or even EF-Tu-independent binding. As a result, we have only estimated a limit for kpep at <0.05 s−1 in these cases.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Hiroaki Suga and Hiroshi Murakami for discussions as well as graciously sharing their unpublished data. M.O. is supported by the “Homing” grant of the Foundation for Polish Science. This work was funded by National Institutes of Health Grant GM037552 (to O.C.U.)
REFERENCES
- 1.Louie A, Ribeiro NS, Reid BR, Jurnak F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 1984;259:5010–5016. [PubMed] [Google Scholar]
- 2.Fahlman RP, Dale T, Uhlenbeck OC. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 3.Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Mol. Cell. 2008;31:114–123. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saks ME, Conery JS. Anticodon-dependent conservation of bacterial tRNA gene sequences. RNA. 2007;13:651–660. doi: 10.1261/rna.345907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanderson LE, Uhlenbeck OC. The 51–63 base pair of tRNA confers specificity for binding by EF-Tu. RNA. 2007;13:835–840. doi: 10.1261/rna.485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yarus M, Cline S, Raftery L, Wier P, Bradley D. The translational efficiency of tRNA is a property of the anticodon arm. J. Biol. Chem. 1986;261:10496–10505. [PubMed] [Google Scholar]
- 8.McClain WH, Schneider J, Bhattacharya S, Gabriel K. The importance of tRNA backbone-mediated interactions with synthetase for aminoacylation. Proc. Natl. Acad. Sci. USA. 1998;95:460–465. doi: 10.1073/pnas.95.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saks ME, et al. An engineered Tetrahymena tRNAGln for in vivo incorporation of unnatural amino acids into proteins by nonsense suppression. J. Biol. Chem. 1996;271:23169–23175. doi: 10.1074/jbc.271.38.23169. [DOI] [PubMed] [Google Scholar]
- 10.Krüger MK, Pedersen S, Hagervall TG, Sørensen MA. The modification of the wobble base of tRNAGlu modulates the translation rate of glutamic acid codons in vivo. J. Mol. Biol. 1998;284:621–631. doi: 10.1006/jmbi.1998.2196. [DOI] [PubMed] [Google Scholar]
- 11.Nasvall SJ, Chen P, Bjork GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–1673. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claesson C, et al. Glycine codon discrimination and the nucleotide in position 32 of the anticodon loop. J. Mol. Biol. 1995;247:191–196. doi: 10.1006/jmbi.1994.0132. [DOI] [PubMed] [Google Scholar]
- 13.Tsai F, Curran JF. tRNA(2Gln) mutants that translate the CGA arginine codon as glutamine in Escherichia coli. RNA. 1998;4:514–522. doi: 10.1017/s1355838298981274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diaz I, Ehrenberg M. ms2i6A deficiency enhances proofreading in translation. J Mol Biol. 1991;222:1161–1171. doi: 10.1016/0022-2836(91)90599-2. [DOI] [PubMed] [Google Scholar]
- 15.Bjork GR, et al. Transfer RNA modification: influence on translational frameshifting and metabolism. FEBS Lett. 1999;452:47–51. doi: 10.1016/s0014-5793(99)00528-1. [DOI] [PubMed] [Google Scholar]
- 16.Qian Q, Bjork GR. Structural alterations far from the anticodon of the tRNAProGGG of Salmonella typhimurium induce +1 frameshifting at the peptidyl-site. J. Mol. Biol. 1997;273:978–992. doi: 10.1006/jmbi.1997.1363. [DOI] [PubMed] [Google Scholar]
- 17.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herr AJ, Atkins JF, Gesteland RF. Mutations which alter the elbow region of tRNA2Gly reduce T4 gene 60 translational bypassing efficiency. EMBO J. 1999;18:2886–2896. doi: 10.1093/emboj/18.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cochella L, Green R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mims BH, Prather NE, Murgola EJ. Isolation and nucleotide sequence analysis of tRNAAlaGGC from Escherichia coli K-12. J. Bacteriol. 1985;162:837–839. doi: 10.1128/jb.162.2.837-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gabriel K, Schneider J, McClain WH. Functional evidence for indirect recognition of G.U in tRNA(Ala) by alanyl-tRNA synthetase. Science. 1996;271:195–197. doi: 10.1126/science.271.5246.195. [DOI] [PubMed] [Google Scholar]
- 22.Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 2005;12:788–793. doi: 10.1038/nsmb978. [DOI] [PubMed] [Google Scholar]
- 23.Daviter T, Wieden HJ, Rodnina MV. Essential role of histidine 84 in elongation factor Tu for the chemical step of GTP hydrolysis on the ribosome. J. Mol. Biol. 2003;332:689–699. doi: 10.1016/s0022-2836(03)00947-1. [DOI] [PubMed] [Google Scholar]
- 24.Hagervall TG, Ericson JU, Esberg KB, Li JN, Bjork GR. Role of tRNA modification in translational fidelity. Biochim. Biophys. Acta. 1990;1050:263–266. doi: 10.1016/0167-4781(90)90178-5. [DOI] [PubMed] [Google Scholar]
- 25.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 26.Valle M, et al. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nobles KN, Yarian CS, Liu G, Guenther RH, Agris PF. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res. 2002;30:4751–4760. doi: 10.1093/nar/gkf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maglott EJ, Deo SS, Przykorska A, Glick GD. Conformational transitions of an unmodified tRNA: implications for RNA folding. Biochemistry. 1998;37:16349–16359. doi: 10.1021/bi981722u. [DOI] [PubMed] [Google Scholar]
- 30.Serebrov V, Vassilenko K, Kholod N, Gross HJ, Kisselev L. Mg2+ binding and structural stability of mature and in vitro synthesized unmodified Escherichia coli tRNAPhe. Nucleic Acids Res. 1998;26:2723–2728. doi: 10.1093/nar/26.11.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kothe U, Rodnina MV. Codon reading by tRNAAla with modified uridine in the wobble position. Mol. Cell. 2007;25:167–174. doi: 10.1016/j.molcel.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Gromadski KB, Daviter T, Rodnina MV. A uniform response to mismatches in codonanticodon complexes ensures ribosomal fidelity. Mol. Cell. 2006;21:369–377. doi: 10.1016/j.molcel.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 33.Pape T, Wintermeyer W, Rodnina M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gromadski KB, Rodnina MV. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 35.Sanbonmatsu KY. Alignment/misalignment hypothesis for tRNA selection by the ribosome. Biochimie. 2006;88:1075–1089. doi: 10.1016/j.biochi.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 37.Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pape T, Wintermeyer W, Rodnina MV. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- 39.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 40.Olejniczak M, Uhlenbeck OC. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie. 2006;88:943–950. doi: 10.1016/j.biochi.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Stark H, et al. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codonrecognition complex. Nat. Struct. Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- 42.Li W, et al. Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM. EMBO J. 2008;27:3322–3331. doi: 10.1038/emboj.2008.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Söll D, RajBhandary U. tRNA : structure, biosynthesis, and function. xiii. Washington, D.C: ASM Press; 1995. p. 572. [Google Scholar]
- 44.LaRiviere FJ, Wolfson AD, Uhlenbeck OC. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- 45.Pavlov MY, et al. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. USA. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ledoux S, Uhlenbeck OC. [3'-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods. 2008;44:74–80. doi: 10.1016/j.ymeth.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.