Summary of recent events
MLST, DNA microarrays, and genome sequencing has allowed for a greater understanding of the metabolic capacity and epidemiology of Campylobacter jejuni. While strain-specific genes may provide an isolate a selective advantage in environments and contribute to the organism's pathogenicity, recent work indicates that C. jejuni pathogenicity is dictated by variations in the nucleotide sequence of core genes. Challenges facing C. jejuni researchers include determining: a) the degree to which genomic diversity enables this bacterium to persist in particular environments; b) if C. jejuni virulence and disease severity can be predicted based on genotype; c) the set of core and variable genes whose products contribute to virulence; and d) the genes in which nucleotide changes can affect a strain's pathogenicity.
Introduction
Campylobacter jejuni, a Gram-negative, spiral shaped bacterium [1], is one of the leading bacterial causes of food-borne human gastroenteritis. C. jejuni is currently estimated to cause 5 to 14% of diarrhea worldwide, which translates into 400−500 million cases per year [2]. Most cases of C. jejuni mediated gastroenteritis (campylobacteriosis) are characterized by nausea, abdominal cramps, diarrhea and fatigue. Although campylobacteriosis is most often self-limiting, certain strains of C. jejuni have been implicated as an antecedent to the development of Guillain-Barré Syndrome (GBS), an acute autoimmune mediated polyneuropathy characterized by ascending paralysis [3,4].
While outbreaks of campylobacteriosis occur, predominantly through consumption of contaminated milk and untreated water [5], most Campylobacter infections are sporadic in nature and linked to the improper handling and consumption of poultry. The linkage between human infection and the handling of raw poultry is not unexpected, as C. jejuni is a common commensal organism of chickens. In fact, C. jejuni colonize the intestinal tract of a variety of animals, including common livestock (cattle, sheep, pigs), domestic animals (dogs, cats), poultry, and wildlife (rabbits, pheasant, quail) [6-9]. A number of methods [e.g., Penner serotyping, Lior serotyping, fla-short variable region (SVR) sequencing, pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST)] are useful for the discrimination of C. jejuni isolates in epidemiological investigations. These methods have enabled investigators to identify the strain responsible for an outbreak [10,11]. The use of MLST, in particular, has provided researchers with the benefit of a defined molecular fingerprint to compare strains. The recent explosion of genome sequences and comparative genomic data has increased our understanding of the epidemiology and metabolic capacity of this organism.
The number of Campylobacter species and strain-specific genome sequences is increasing
The importance of Campylobacter in human gastrointestinal illness has resulted in at least eighteen isolates from eight different Campylobacter species having been or being sequenced (Table 1). The genomes of Campylobacter organisms are characterized by a low mol% (G+C) content (between 29.5% and 44.5%), small size (ranging from 1.5 Mb for Campylobacter lari RM2100 to 2.1 Mb for Campylobacter concisus 13826), and relatively few open reading frames (between 1425 and 1931 ORFs). The availability of sequenced genomes has aided in the development of methodologies to address questions concerning the relationship between genomic content and C. jejuni biology.
Table 1.
Features of Sequenced Campylobacter Genomes
| Species/Strain | Size (Mb) | %GC | ORFs | Origin | Diseasea | GenBankb | Ref. |
|---|---|---|---|---|---|---|---|
| Campylobacter jejuni subsp jejuni NCTC 11168 | 1.64 | 30.5 | 1643 | Clinical | Food Poisoning | AL111168 | [36] |
| Campylobacter jejuni subsp jejuni RM1221 | 1.78 | 30.3 | 1835 | Chicken | CP000025 | [22] | |
| Campylobacter jejuni subsp jejuni 81−176 | 1.6 | 30.6 | 1653 | Clinical | Food Poisoning | CP000538 | [24] |
| Campylobacter jejuni subsp jejuni 81116 | 1.63 | 30.5 | 1626 | Clinical | Food Poisoning | CP000814 | [43] |
| Campylobacter jejuni subsp jejuni CG8421 | 1.6 | 30.4 | 1512 | Clinical | Food Poisoning | ABGQ00000000 | |
| Campylobacter jejuni subsp jejuni HB93−13 | 1.7 | 30.6 | 1710 | Clinical | GBSc | AANQ00000000 | |
| Campylobacter jejuni subsp jejuni CG8486 | 1.65 | 30.4 | 1425 | Clinical | Food Poisoning | AASY00000000 | [44] |
| Campylobacter jejuni subsp jejuni CF93−6 | 1.67 | 30.5 | 1757 | Clinical | MFSd | AANJ00000000 | |
| Campylobacter jejuni subsp jejuni 84−25 | 1.67 | 30.4 | 1748 | Clinical | Meningitis | AANT00000000 | |
| Campylobacter jejuni subsp jejuni 260.94 | 1.65 | 30.5 | 1716 | Clinical | GBS | AANK00000000 | |
| Campylobacter jejuni subsp doylei 269.97 | 1.8 | 30.6 | 1731 | Clinical | Bacteremia | CP000768 | |
| Campylobacter coli RM2228 | 1.68 | 31.4 | 1765 | Chicken | [22] | ||
| Campylobacter concisus 13826 | 2.1 | 39.4 | 1929 | Clinical | Food Poisoning | CP000792 | |
| Campylobacter curvus 525.92 | 2.0 | 44.5 | 1931 | Clinical | Periodontitis | CP000767 | |
| Campylobacter fetus subsp. fetus 82−40 | 1.8 | 33.3 | 1719 | Clinical | Septicemia | CP000487 | |
| Campylobacter hominis ATCC BAA-381 | 1.7 | 31.7 | 1682 | Clinical | Non-pathogenic | CP000776 | |
| Campylobacter lari RM2100 | 1.5 | 29.6 | 1554 | Clinical | Food Poisoning | [22] | |
| Campylobacter upsaliensis RM3195 | 1.66 | 34.5 | 1782 | Clinical | GBS | [22] |
Disease associated with origin of isolate
GenBank Accession Number
Guillain-Barre Syndrome
Miller-Fisher Syndrome
MLST provides greater insight into genetic diversity and population structure
MLST allows researchers to differentiate strains based on alleles at seven “unlinked” housekeeping loci [12]. Each unique allele is assigned a number based on its order of discovery and the combination of allelic numbers is the basis for its sequence type (ST). The ST is indicative of the isolate's genotype. This method is advantageous because it yields data that are accurate, reproducible, unaffected by changes in gene order, and readily comparable among laboratories. In addition to its usefulness in investigating C. jejuni outbreaks, MLST has demonstrated that C. jejuni are genetically diverse. Dingle et al. [12] found 194 C. jejuni isolates to be genetically diverse with a weakly clonal population structure. The genetic diversity was evident in that of the 155 STs observed, 104 STs could be grouped into 11 major genetic lineages or clonal complexes whereas the remaining 51 STs were unique. Manning et al. [13] performed MLST on 266 C. jejuni veterinary and human isolates and found that the populations overlapped among the 19 clonal complexes identified. Dingle et al. [14] applied the MLST scheme used for C. jejuni to Campylobacter coli and found the two species to share approximately 86% nucleotide sequence identity at the housekeeping loci and found some evidence for horizontal gene transfer. The investigators found additional evidence for horizontal gene transfer by sequencing the short variable region (SVR) of the flaA flagellin gene [14]. Evidence for horizontal gene transfer was also reported by Meinersmann et al. [15] using flaA SVR sequence anlaysis. These studies demonstrate that recombination between C. jejuni and C. coli occurs, and supports a hypothesis that these two species are continuing to evolve [16]. Taken together, these studies demonstrate the genetic diversity of Campylobacter strains. While it may never be possible to predict an isolate's pathogenicity or an individual's clinical symptoms or disease severity based on the ST alone, the use of MLST has already contributed to a greater understanding of C. jejuni population structures and their relationships with a variety of hosts.
Identification of hypervariable plasticity regions
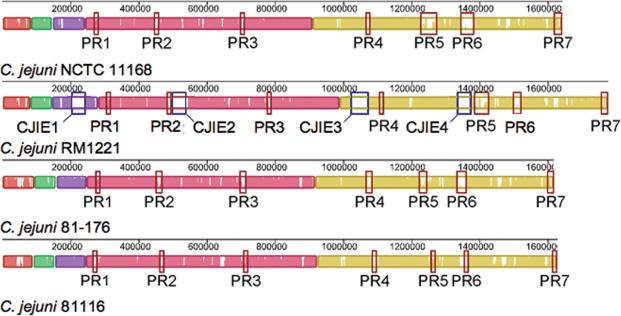
The availability of genome sequences has made it possible to construct whole genome DNA microarrays for comparative genomic hybridization (CGH) analysis. Analysis of 11 C. jejuni clinical isolates by CGH revealed extensive genetic diversity and enabled the researchers to identify the genetic core of this organism [17]. Approximately 84% of the 1654 genes analyzed were common to all strains tested and encoded proteins involved in housekeeping functions, including metabolic, biosynthetic, cellular and regulatory processes. Strain-specific gene differences were involved in the biosynthesis and modification of cell surface structures including flagella, lipooligosaccharide (LOS) and capsular polysaccharide. Pearson et al. [18] examined the genomic diversity of 18 C. jejuni strains from different sources and found that the variable genes were often present in clusters, suggesting they were acquired or lost in groups during evolution [18]. In particular, seven hypervariable plasticity regions (PR) (Figure 1) were identified (PR1 – PR7). PR1 contained genes important in the utilization of alternative electron acceptors during respiration, possibly conferring a selective advantage in restricted oxygen environments. PR2, PR3 and PR7 contained genes encoding outer membrane and periplasmic proteins, which may be linked to phenotypic variation and adaptation to different ecological niches. PR4, PR5 and PR6 contained genes involved the biosynthesis and modification of flagella, LOS, and capsule. Further studies are required to elucidate the contribution of genes within the hypervariable regions to a strain's phenotype.
Figure 1.
Whole genome alignments of C. jejuni NCTC 11168, RM1221, 81−176 and 81116 performed using Mauve [42]. Each genome is laid out horizontally and homologous segments are shown as colored blocks. The average sequence identity is proportional to the height of the colored region with in each horizontal block. C. jejuni integrated elements (CJIEs) in strain RM1221 are indicated by blue rectangles. The seven plasticity regions (PR1-PR7) are indicated by red rectangles.
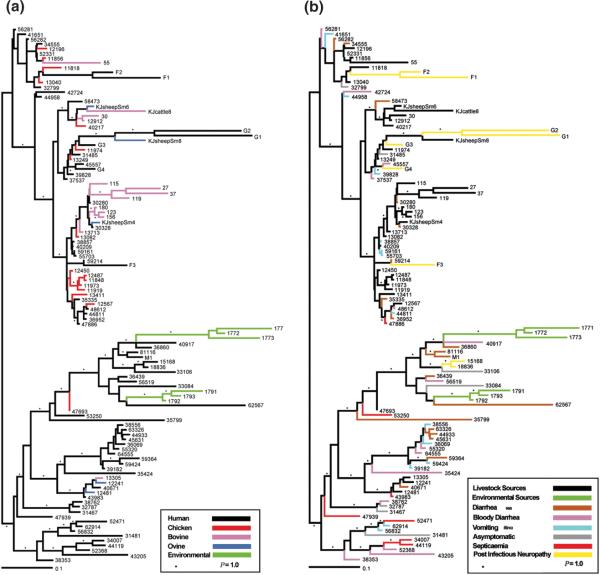
Meta-analysis of CGH data from four separate data sets revealed that many of the variable genes were absent or divergent in only one of the 97 strains [19]. In a separate study, phylogenomic analysis of 111 isolates from human, animals, and environmental sources provided insight into the population structure of C. jejuni. The investigators identified livestock and non-livestock associated clades (Figure 2A) [20]. Although analysis of C. jejuni isolates recovered from individuals with different clinical symptoms did not cluster (Figure 2B), more than half of the clinical isolates were phylogenomically related to strains from non-livestock sources. The investigators concluded that environmental sources serve as an important reservoir for C. jejuni infectious isolates. In contrast, Wilson et al. [21] concluded that livestock is the principal source of C. jejuni infection based on modeling DNA sequence evolution and rates of zoonotic transmission [21]. They suggested that the frequency of recombination in C. jejuni makes a single phylogenetic tree an inappropriate representation of the relationship between C. jejuni genomes [21]. Based on these studies, it is apparent that additional work is needed to determine the relative importance of reservoirs for C. jejuni.
Figure 2.
Phylogenomic relationship of C. jejuni strains. Strains are designated at the end of branches and are colored according to the (A) ecological niche from which the C. jejuni strain was isolated or (B) clinical symptoms or livestock/environmental source. P = 1.0 represents 100% of all phylogenies showing a given topology. Adapted with permission from the authors [20].
C. jejuni genomes are syntenic and some contain integrated elements
Presumably, all C. jejuni strains sequenced to date are pathogenic. As such, it is not possible to compare the genomic sequence of pathogenic and non-pathogenic strains. However, sequencing and comparative genomic analysis of five Campylobacter genomes (C. jejuni NCTC 11168, C. jejuni RM1221, C. coli RM2228, C. lari RM2100, and Campylobacter upsaliensis RM3195) revealed major structural differences between the strains [22]. While the genome of C. jejuni RM1221 was syntenic with the previously sequenced C. jejuni NCTC 11168, it contained four inserted genomic islands termed Campylobacter jejuni-integrated elements (CJIEs) (Figure 1). The CJIEs from C. jejuni RM1221 were not present among the other three C. jejuni sequenced strains (NCTC 11168, 81116, and 81−176). CJIE1 was found to be a Campylobacter Mu-like phage while CJIE2 and CJIE4 contained genes predicted to encode phage related endonucleases, methylases and repressors. A total of 73% of the CJIE3 predicted proteins showed sequence similarity with those encoded on the C. coli RM2228 megaplasmid, suggesting that it may be an integrated plasmid. Comparative genomic analysis of 67 C. jejuni and 12 C. coli strains from various geographical, clinical and veterinary sources revealed the CJIEs widely distributed, and more than half of the strains tested contained at least one CJIE [23]. C. coli RM2228 was also highly syntenic with C. jejuni RM1221, while C. lari and C. upsaliensis were not [22]. Noteworthy is that the regions of sequence variability aligned with the previously identified PR in C. jejuni NCTC 11168, suggesting the PR are likely physical loci within C. jejuni genomes. Additionally, CJIEs 2 and 4 appeared at loci adjacent to PR 2 and 5, respectively. The relevance of the CJIEs in the biology of C. jejuni remains to be determined.
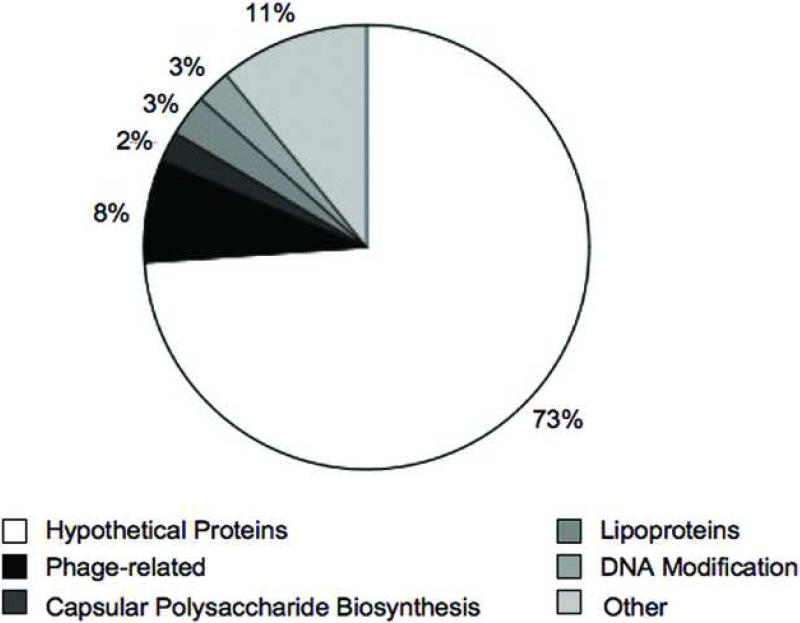
Comparative genomic analyses have helped identify dispensable genes amongst C. jejuni isolates (i.e., genes absent or highly divergent in one or more of the isolates). The genes unique among the four C. jejuni sequenced strains (NCTC 11168, RM1221, 81−176 and 81116) are listed in Supplemental Table 1. The majority of these genes mapped to the variable loci identified previously and encode hypothetical proteins (Figure 3). Some of the unique genes were predicted to be involved in capsular polysaccharide biosynthesis, DNA modification, lipoprotein synthesis, or were phage-related. Additionally, the DNA sequences flanking the variable loci were conserved, suggesting that large regions of the C. jejuni chromosome are relatively stable. Sequencing of C. jejuni 81−176 revealed aerobic and anaerobic respiratory pathways that may confer advantages in low oxygen environments (i.e., gastrointestinal tract) [24]. Additionally, an ortholog of γ-glutamyltranspeptidase was identified, which is important for Helicobacter pylori colonization [25,26]. A mutation of this gene, resulting in loss of function, significantly reduced C. jejuni colonization of mice [24]. Together, these findings indicate that strain variable genes can provide an isolate an advantage in selective environments.
Figure 3.
Classification of genes unique to one of the four C. jejuni strains (NCTC 11168, RM1221, 81−176 and 81116). Unique genes are listed in Supplemental Table 1 and were classified based upon genome annotations
Nucleotide variations alter pathogenic behavior
The availability of C. jejuni genome sequences has provided the genetic basis for investigating the metabolism, gene regulation, and physiology of these organisms. These data indicate that many of the previously identified putative virulence determinants, including cytolethal distending toxin (CDT), various adhesins (e.g., CadF, JlpA, and PEB1), and the flagellar structural proteins, are conserved amongst strains [17]. Despite conservation of these genes, it is likely that the variations in the pathogenicity of C. jejuni strains are influenced by nucleotide differences in virulence genes. In this regard, none of the techniques discussed above are capable of determining whether point mutations, nucleotide insertions, nucleotide deletions, and gene rearrangements are present in one strain versus another. Below we provide specific examples of where nucleotide variations may affect the pathogenicity of C. jejuni strains.
Comparative analysis of CDT, a multi-subunit toxin [27,28], in C. jejuni, C. coli, and Campylobacter fetus revealed that the cdt gene cluster is widely distributed in a species-specific manner [29]. CDT has been shown to induce tissue damage and fluid accumulation in the colon of infected mice [30]. However, the contribution of CDT in campylobacteriosis is not clear, as C. jejuni CDT-negative strains have been isolated from individuals with clinical signs of diarrhea [31]. Detection of the cdtABC gene cluster does not necessarily result in the expression and synthesis of a functional product [31]. AbuOun et al. [31] identified two mutations that result in a CDT-negative phenotype. One mutation was characterized by a 667 bp deletion, encompassing a portion of both the cdtA and cdtB genes, and the second mutation was a nucleotide change that resulting in nonsynonymous residue at position 95 of CdtB [31]. This example illustrates the importance of performing experiments to test for a functional product even when a gene is detected by hybridization.
While not well understood, single nucleotide variations may contribute to molecular mimicry of gangliosides concentrated in peripheral nervous tissue by the LOS of C. jejuni [32,33]. Although CGH analyses of C. jejuni strains from individuals with GBS and uncomplicated gastroenteritis have failed to identify GBS associated genes [34], direct sequence analysis has revealed information regarding phase variation in the LOS biosynthesis genes [35]. Analysis of the genome sequence of C. jejuni 81−176 identified a homopolymeric G tract in cgtA, which encodes N-acetylgalactosaminyltransferase. Variation in the number G residues resulted in differences in the LOS core structure, affecting ganglioside mimicry and the ability of this strain to invade epithelial cells in vitro [35]. This example illustrates that slipped-strand mispairing occurs during DNA replication at homopolymeric tracts consisting of consecutive guanine residues. The resulting differences in homopolymeric tract length can affect translation and result in phase variation. Given that these regions are frequently associated with genes that synthesize surface structures [22,24,36] their presence may influence a strain's pathogenicity and immunogenicity. Lastly, it is worth noting that although homopolymeric G tracts are common in the C. jejuni strains sequenced, their number varies among strains.
Comparison of the C. jejuni NCTC 11168 (11168-GS) isolate used for generating the genomic sequence and the original clinical isolate (11168-O) revealed differences in their virulence-associated phenotypes including colonization, invasion, translocation and motility [37]. Gene expression profiling of these two strains revealed dramatic differences in the expression of flagellar and motility related genes. Sequence analysis of three sigma factors (i.e., RpoD, RpoN, and FliA) in the 11168-GS and 11168-O strains identified single-nucleotide polymorphisms (SNPs) in each sigma factor gene, resulting in at least one amino acid substitution in each sigma factor. The investigators proposed that the differences in gene expression were due to changes in global gene regulation. The possibility that a nucleotide change in a sigma factor could influence gene expression on a global level was subsequently proven by the identification a defective rpoN gene in C. jejuni strain S2B and its complementation [38].
Malik-Kale et al. [38] assessed the virulence potential of two C. jejuni strains (Turkey and CS) that were indistinguishable by PFGE, MLST, and CGH. Interestingly, these two C. jejuni strains showed dramatically different virulence potential in both in vitro cell culture and piglet models. Gene expression analysis revealed dramatic differences in gene expression profile. In particular flagellar Class II genes were found to have lower expression in the C. jejuni CS strain (i.e., the less virulent isolate), suggesting expression of these genes are important to pathogenesis by C. jejuni. Additionally, DNA sequence analysis of genes that regulate flagellar synthesis revealed a point mutation in flgR may be responsible for the loss of virulence. Previous work indicated that flagellar synthesis is modulated via phase variation [39]. In particular, phase variation of FlgR is due to the loss or gain of a nucleotide in homopolymeric adenine or thymine tracts within flgR. The identified point mutation in flgR did not occur in a region previously identified as being phase variable. Based on these data, we conclude that allelic variation may also dictate a strain's pathogenic potential.
Conclusions and future perspectives
The identification of genetic markers predictive of ecological source and virulence potential are important to detecting and preventing the dissemination of C. jejuni via food sources. As we have reviewed, MLST, DNA microarrays, and genome sequencing of C. jejuni strains have demonstrated the genetic diversity of this important food-borne pathogen. Comparative genomic studies have demonstrated C. jejuni population structure relates to ecological source (livestock versus non-livestock sources) and identified genes (e.g., Cj1321−1326 in C. jejuni NCTC 11168) that are associated with livestock sources. Additionally, DNA sequence analyses implicate phase and allelic variation as possible mechanisms for altered gene expression and protein synthesis. In spite of recent advances, significant gaps exist in our knowledge of C. jejuni biology. First, researchers have yet to uncover a correlation between genomic diversity and disease severity. Second, C. jejuni virulence and disease pathology are not yet predictable based on genotype. Third, the core genes necessary for disease and the variable (i.e., dispensable) genes whose products contribute to C. jejuni disease are not known. Fourth, based on the observation that nucleotide changes in certain genes alter a strain's pathogenicity, studies are needed to identify additional genes/proteins whose expression/function is influenced by nucleotide/residue variations. To address these questions, a small infectious disease animal model is needed to test the pathogenic potential of C. jejuni isolates. However, few animal models are currently available to assess C. jejuni virulence [40]. One possibility is the use of the interleukin-10-deficient murine model to determine the relationship between C. jejuni genetic variation and disease spectrum [41]. Continued work focusing on the relationship of genotype to phenotype is important in understanding this enigmatic organism.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Charles L. Larson (School of Molecular Biosciences, Washington State University) and Dr. Philip F. Mixter (School of Molecular Biosciences, Washington State University) for critical review of this manuscript. Comparative genomic analyses of C. jejuni is supported from funds awarded to MEK from the National Institute of Health, Department of Health and Human Services under contract number NO1-AI-30055.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miller WG, Mandrell RE. Prevalence of Campylobacter in the food and water supply: Incidence, Outbreaks, Isolation and Detection. In: Ketley JM, Konkel ME, editors. Campylobacter jejuni: Molecular and Cellular biology. Horizon Bioscience; 2005. pp. 101–164. [Google Scholar]
- 2.Friedman CR, Neimann J, Wegener HC, Tauxe RV. Epidemioogy of Camylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser MJ, editors. Campylobacter. 2nd ASM Press; 2000. pp. 121–138. [Google Scholar]
- 3.Nachamkin I, Allos BM, Ho T. Campylobacter species and Guillain-Barre syndrome. Clin Microbiol Rev. 1998;11:555–567. doi: 10.1128/cmr.11.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nachamkin I, Allos BM, Ho TW. Campylobacter jejuni infection and the association with Guillain-Barre syndrome. In: Nachamkin I, Blaser MJ, editors. Campylobacter. 2nd ASM Press; 2000. pp. 155–175. [Google Scholar]
- 5.Pebody RG, Ryan MJ, Wall PG. Outbreaks of Campylobacter infection: rare events for a common pathogen. Commun Dis Rep CDR Rev. 1997;7:R33–37. [PubMed] [Google Scholar]
- 6.Atanassova V, Ring C. Prevalence of Campylobacter spp. in poultry and poultry meat in Germany. Int J Food Microbiol. 1999;51:187–190. doi: 10.1016/s0168-1605(99)00120-8. [DOI] [PubMed] [Google Scholar]
- 7.Koene MG, Houwers DJ, Dijkstra JR, Duim B, Wagenaar JA. Strain variation within Campylobacter species in fecal samples from dogs and cats. Vet Microbiol. 2009;133:199–205. doi: 10.1016/j.vetmic.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 8.Soncini G, Valnegri VL, Vercellotti L, Colombo F, Valle D, Franzoni M, Bersanii C. Investigation of Campylobacter in reared game birds. J Food Prot. 2006;69:3021–3024. doi: 10.4315/0362-028x-69.12.3021. [DOI] [PubMed] [Google Scholar]
- 9.Stern NJ, Bannov VA, Svetoch EA, Mitsevich EV, Mitsevich IP, Volozhantsev NV, Gusev VV, Perelygin VV. Distribution and characterization of Campylobacter spp. from Russian poultry. J Food Prot. 2004;67:239–245. doi: 10.4315/0362-028x-67.2.239. [DOI] [PubMed] [Google Scholar]
- 10.Gerner-Smidt P, Hise K, Kincaid J, Hunter S, Rolando S, Hyytia-Trees E, Ribot EM, Swaminathan B. PulseNet USA: a five-year update. Foodborne Pathog Dis. 2006;3:9–19. doi: 10.1089/fpd.2006.3.9. [DOI] [PubMed] [Google Scholar]
- 11.Klena JD, Konkel ME. Methods for Epidemilogical Analysis of Campylobacter jeuni. In: Ketley JM, Konkel ME, editors. Campylobacter: Molecular and Cellular Biology. Horizon Bioscience; 2005. pp. 165–179. [Google Scholar]
- 12.Dingle KE, Colles FM, Wareing DR, Ure R, Fox AJ, Bolton FE, Bootsma HJ, Willems RJ, Urwin R, Maiden MC. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manning G, Dowson CG, Bagnall MC, Ahmed iH, West M, Newell DG. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. Appl Environ Microbiol. 2003;69:6370–6379. doi: 10.1128/AEM.69.11.6370-6379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingle KE, Colles FM, Falush D, Maiden MC. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J Clin Microbiol. 2005;43:340–347. doi: 10.1128/JCM.43.1.340-347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meinersmann RJ, Phillips RW, Hiett KL, Fedorka-Cray P. Differentiation of Campylobacter populations as demonstrated by flagellin short variable region sequences. Appl Environ Microbiol. 2005;71:6368–6374. doi: 10.1128/AEM.71.10.6368-6374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheppard SK, McCarthy ND, Falush D, Maiden MC. Convergence of Campylobacter species: implications for bacterial evolution. Science. 2008;320:237–239. doi: 10.1126/science.1155532. [DOI] [PubMed] [Google Scholar]
- 17.Dorrell N, Mangan JA, Laing KG, Hinds J, Linton D, Al-Ghusein H, Barrell BG, Parkhill J, Stoker NG, Karlyshev AV, et al. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 2001;11:1706–1715. doi: 10.1101/gr.185801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson BM, Pin C, Wright J, I'Anson K, Humphrey T, Wells JM. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 2003;554:224–230. doi: 10.1016/s0014-5793(03)01164-5. [Using Array-CGH of 18 C. jejuni isolates from diverse sources, the authors were able to identify hypervariable plasticity regions (PRs). This is the first microarray based comparsion to include non-human isolates.] [DOI] [PubMed] [Google Scholar]
- 19.Taboada EN, Acedillo RR, Carrillo CD, Findlay WA, Medeiros DT, Mykytczuk OL, Roberts MJ, Valencia CA, Farber JM, Nash JH. Large-scale comparative genomics meta-analysis of Campylobacter jejuni isolates reveals low level of genome plasticity. J Clin Microbiol. 2004;42:4566–4576. doi: 10.1128/JCM.42.10.4566-4576.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Champion OL, Gaunt MW, Gundogdu O, Elmi A, Witney AA, Hinds J, Dorrell N, Wren BW. Comparative phylogenomics of the food-borne pathogen Campylobacter jejuni reveals genetic markers predictive of infection source. Proc Natl Acad Sci U S A. 2005;102:16043–16048. doi: 10.1073/pnas.0503252102. [Using array-CGH, the authors were able to classify 111 C. jejuni isolates from diverse clinical, animal and environmental sources into either a livestock or non-livestock clade and identify genes associated with sources of isolation. This is the first genomic comparison to identify genes associated with an isolates source.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson DJ, Gabriel E, Leatherbarrow AJ, Cheesbrough J, Gee S, Bolton E, Fox A, Fearnhead P, Hart CA, Diggle PJ. Tracing the source of campylobacteriosis. PLoS Genet. 2008;4:e1000203. doi: 10.1371/journal.pgen.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouts DE, Mongodin EF, Mandrell RE, Miller WG, Rasko DA, Ravel J, Brinkac LM, DeBoy RT, Parker CT, Daugherty SC, et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [Sequencing and comparison of C. jejuni RM1221, C. lari RM2100, C. upsalienis RM3195 and C. coli RM2228. This is the first whole genome sequence comparison to include non-jejuni Campylobacter species.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parker CT, Quinones B, Miller WG, Horn ST, Mandrell RE. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J Clin Microbiol. 2006;44:4125–4135. doi: 10.1128/JCM.01231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofreuter D, Tsai J, Watson RO, Novik V, Altman B, Benitez M, Clark C, Perbost C, Jarvie T, Du L, et al. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect Immun. 2006;74:4694–4707. doi: 10.1128/IAI.00210-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chevalier C, Thiberge JM, Ferrero RL, Labigne A. Essential role of Helicobacter pylori gamma-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol Microbiol. 1999;31:1359–1372. doi: 10.1046/j.1365-2958.1999.01271.x. [DOI] [PubMed] [Google Scholar]
- 26.McGovern KJ, Blanchard TG, Gutierrez JA, Czinn SJ, Krakowka S, Youngman P. gamma-Glutamyltransferase is a Helicobacter pylori virulence factor but is not essential for colonization. Infect Immun. 2001;69:4168–4173. doi: 10.1128/IAI.69.6.4168-4173.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muza-Moons MM, Koutsouris A, Hecht G. Disruption of cell polarity by enteropathogenic Escherichia coli enables basolateral membrane proteins to migrate apically and to potentiate physiological consequences. Infect Immun. 2003;71:7069–7078. doi: 10.1128/IAI.71.12.7069-7078.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamasaki S, Asakura M, Tsukamoto T, Faruque SM, Deb R, Ramamurthy T. Cytolethal distending toxin (CDT): genetic diversity, structure and role in diarrheal disease. Toxin Reviews. 2006;25:61. [Google Scholar]
- 29.Asakura M, Samosornsuk W, Taguchi M, Kobayashi K, Misawa N, Kusumoto M, Nishimura K, Matsuhisa A, Yamasaki S. Comparative analysis of cytolethal distending toxin (cdt) genes among Campylobacter jejuni, C. coli and C. fetus strains. Microb Pathog. 2007;42:174–183. doi: 10.1016/j.micpath.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Okuda J, Fukumoto M, Takeda Y, Nishibuchi M. Examination of diarrheagenicity of cytolethal distending toxin: suckling mouse response to the products of the cdtABC genes of Shigella dysenteriae. Infect Immun. 1997;65:428–433. doi: 10.1128/iai.65.2.428-433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abuoun M, Manning G, Cawthraw SA, Ridley A, Ahmed IH, Wassenaar TM, Newell DG. Cytolethal distending toxin (CDT)-negative Campylobacter jejuni strains and anti-CDT neutralizing antibodies are induced during human infection but not during colonization in chickens. Infect Immun. 2005;73:3053–3062. doi: 10.1128/IAI.73.5.3053-3062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ang CW, Laman JD, Willison HJ, Wagner ER, Endtz HP, De Klerk MA, Tio-Gillen AP, Van den Braak N, Jacobs BC, Van Doorn PA. Structure of Campylobacter jejuni lipopolysaccharides determines antiganglioside specificity and clinical features of Guillain-Barre and Miller Fisher patients. Infect Immun. 2002;70:1202–1208. doi: 10.1128/IAI.70.3.1202-1208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuki N. Infectious origins of, and molecular mimicry in, Guillain-Barre and Fisher syndromes. Lancet Infect Dis. 2001;1:29–37. doi: 10.1016/S1473-3099(01)00019-6. [DOI] [PubMed] [Google Scholar]
- 34.Leonard EE, 2nd, Tompkins LS, Falkow S, Nachamkin I. Comparison of Campylobacter jejuni isolates implicated in Guillain-Barre syndrome and strains that cause enteritis by a DNA microarray. Infect Immun. 2004;72:1199–1203. doi: 10.1128/IAI.72.2.1199-1203.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerry P, Szymanski CM, Prendergast MM, Hickey TE, Ewing CP, Pattarini DL, Moran AP. Phase variation of Campylobacter jejuni 81−176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect Immun. 2002;70:787–793. doi: 10.1128/iai.70.2.787-793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 37.Gaynor EC, Cawthraw S, Manning G, MacKichan JK, Falkow S, Newell DG. The genome-sequenced variant of Campylobacter jejuni NCTC 11168 and the original clonal clinical isolate differ markedly in colonization, gene expression, and virulence-associated phenotypes. J Bacteriol. 2004;186:503–517. doi: 10.1128/JB.186.2.503-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik-Kale P, Raphael BH, Parker CT, Joens LA, Klena JD, Quinones B, Keech AM, Konkel ME. Characterization of genetically matched isolates of Campylobacter jejuni reveals that mutations in genes involved in flagellar biosynthesis alter the organism's virulence potential. Appl Environ Microbiol. 2007;73:3123–3136. doi: 10.1128/AEM.01399-06. [Using two genotypically indistinguisible C. jejuni isolates with dramatically different virulence potentials as determined by in vitro and in vivo models, the authors were able to identify a single nucleotide substitution responisble for differences in virulence potential.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendrixson DR. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol Microbiol. 2006;61:1646–1659. doi: 10.1111/j.1365-2958.2006.05336.x. [DOI] [PubMed] [Google Scholar]
- 40.Konkel ME, Monteville MR, Klena JD, Joens LA. In vitro and in vivo models used to study Campylobacter jejuni virulence properties. In: Torrence ME, Isaacson RE, editors. Microbial Food Safety in Animal Agriculture. Iowa State Press; 2003. pp. 195–210. [Google Scholar]
- 41.Mansfield LS, Bell JA, Wilson DL, Murphy AJ, Elsheikha HM, Rathinam VA, Fierro BR, Linz JE, Young VB. C57BL/6 and congenic interleukin-10-deficient mice can serve as models of Campylobacter jejuni colonization and enteritis. Infect Immun. 2007;75:1099–1115. doi: 10.1128/IAI.00833-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson BM, Gaskin DJ, Segers RP, Wells JM, Nuijten PJ, van Vliet AH. The complete genome sequence of Campylobacter jejuni strain 81116 (NCTC11828). J Bacteriol. 2007;189:8402–8403. doi: 10.1128/JB.01404-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poly F, Read T, Tribble DR, Baqar S, Lorenzo M, Guerry P. Genome sequence of a clinical isolate of Campylobacter jejuni from Thailand. Infect Immun. 2007;75:3425–3433. doi: 10.1128/IAI.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.