Abstract
It has been proposed that the architecture of protein domains has evolved by the combinatorial assembly and/or exchange of smaller polypeptide segments. To investigate this proposal, we fused DNA encoding the N-terminal half of a β-barrel domain (from cold shock protein CspA) with fragmented genomic Escherichia coli DNA and cloned the repertoire of chimeric polypeptides for display on filamentous bacteriophage. Phage displaying folded polypeptides were selected by proteolysis; in most cases the protease-resistant chimeric polypeptides comprised genomic segments in their natural reading frames. Although the genomic segments appeared to have no sequence homologies with CspA, one of the originating proteins had the same fold as CspA, but another had a different fold. Four of the chimeric proteins were expressed as soluble polypeptides; they formed monomers and exhibited cooperative unfolding. Indeed, one of the chimeric proteins contained a set of very slowly exchanging amides and proved more stable than CspA itself. These results indicate that native-like proteins can be generated directly by combinatorial segment assembly from nonhomologous proteins, with implications for theories of the evolution of new protein folds, as well as providing a means of creating novel domains and architectures in vitro.
There is considerable evidence that proteins may have evolved by the assembly of nonhomologous genes; thus, in multidomain proteins, contiguous domains often have different architectures homologous to those from other proteins (1). Individual protein domains also may have evolved in the same manner, by assembly and/or exchange of small gene segments (2), leading to diversification of the domain architecture and even the generation of entirely new folds (3).
In principle, nonhomologous gene segments could have been joined by DNA recombination or by RNA splicing. Nonhomologous DNA recombination may occur as a result of exon shuffling (4) or can be induced by specific genetic mechanisms such as transpositional rearrangements (5, 6) and site-specific recombinations (7) but may also involve random deletions, insertions, and inversion of DNA fragments (8, 9). Nonhomologous gene segments also may have been recombined by RNA splicing (10–12), and it has been proposed that early in evolution the architecture of protein domains was generated by splicing together of small RNA gene segments (13, 14).
The evolution of protein domains through the recombination of smaller peptide segments is consistent with an average size of exons of 40 amino acid residues (15), which is about half the size of a small folded domain (1, 16), and with their conservation as blocks of coding sequences during exon shuffling (4, 15, 17). Experimentally, it has been shown that DNA shuffling of homologous genes can generate folded domains with improved properties (18–20). However, it is unlikely that such homologous recombinations can lead to the creation of entirely new protein folds, which are more likely to require the recombination of nonhomologous genes (3). We therefore have attempted to mimic this process by the combinatorial shuffling of small segments of different polypeptides in vitro. Specifically, we recombined DNA encoding the N-terminal half of the Escherichia coli cold shock protein A (CspA; ref. 21) with randomly fragmented genomic DNA from E. coli. The resulting repertoire of polypeptides was cloned for display on bacteriophage and selected by its ability to survive proteolysis. The sequences and biophysical properties of proteins selected from this repertoire were analyzed.
Materials and Methods
Vector Constructions.
The H102A mutant of barnase (22) was fused between the pelB leader peptide and the mature gene 3 protein (p3) of phage fd in a modified phagemid pHEN1 (23) to form the vector p22-12. Into p22-12, suitably amplified parts of the E. coli gene cspA (21) were cloned between the barnase and the p3 genes by using PstI and NotI restriction sites. In the resulting phagemid pC5-7, barnase is followed by the N-terminal 36 residues of CspA (the N-terminal Met mutated to Leu to accommodate a PstI site) and the DNA linker sequence GGG AGC TCA GGC GGC CGC AGA A (SacI and NotI restriction sites in italics) appended before the GAA codon for the first residue (Glu) of p3. In pC5-7, the barnase-Csp cassette is out of frame with the p3 gene. In the control vector pCsp/2, the barnase-Csp cassette is in frame with the p3 gene, but the first codon of the linker DNA constitutes an opal stop codon. Vectors for the cytoplasmic expression of soluble proteins were constructed by subcloning genes from the phagemids into the BamHI and HindIII sites of a modified QE30 vector (Qiagen, Chatsworth, CA) encoding a tetra-His tag. During subcloning by PCR, opal stop codons were converted into the Trp-encoding TGG triplet.
Library Construction.
Genomic DNA (ref. 24; 2 μg digested with SacI) from the E. coli strain TG1 (25) was amplified randomly in 30 PCR cycles (annealing at 30°C) with oligonucleotide SN6MIX (5′-GAG CCT GCA GAG CTC CGG NNN NNN-3′ at 40 pmol/ml). PCR products were extended in 30 cycles (annealing at 52°C) by using oligonucleotide XTND (5′-CGT GCG AGC CTG CAG AGC TCC GG-3′ at 4,000 pmol/ml). Products of around 140 bp were excised from an agarose gel and reamplified in 30 PCR cycles (annealing at 50°C) with oligonucleotide NOARG (5′-CGT GCG AGC CTG CAG AGC TCA GG-3′ at 500 pmol/ml).
After digestion with SacI, the fragments were cloned into the vector pC5-7 and electroporated into TG1. About 60% of the recombinants contained monomeric inserts, and the remainder contained oligomers. Because of differences in the 3′ end of the PCR primers XTND and NOARG, 40% of clones with in-frame inserts contained a GGA-encoded Gly as part of the 3′ SacI site, and 60% contained the TGA-encoded opal stop codon at the same position. tRNATrp decodes TGA with an efficiency of up to 3% (26), leading to sufficient display of the barnase–chimera-p3 fusion on the phage but avoiding folding related, toxic effects. Only opal stop codon-containing clones were selected from the library, whereas, in their absence, almost exclusively chimeric gene fusions leading to a frameshift between the barnase and p3 genes were selected (unpublished data). Phage was prepared by using the helper phage KM13 (27), which encodes a trypsin-sensitive p3 mutant, to reduce contributions to infectivity from phage that do not display the fusion protein (28).
Selections.
For selections, 1010 colony-forming units (cfu) of phage were treated with 200 nM trypsin (specific for Arg or Lys in the P1 position) and 384 nM thermolysin (specific for aliphatic side chains in the P1′ position) in TBS-Ca buffer (25 mM Tris⋅HCl/137 mM NaCl/1 mM CaCl2, pH 7.4) for 10 min at 10°C. After proteolysis, phage was captured for 1 h with biotinylated C40A,C82A double-mutant of barstar (29, 30) immobilized on streptavidin-coated microtiter plate wells in 3% Marvel in PBS. Wells were washed 20 times with PBS (and once with 50 mM DTT in PBS for 5 min to elute phage containing proteolyzed p3 fusions held together solely by disulfide bridges). Phage was eluted at pH 2, neutralized, and propagated after reinfection.
ELISA.
Phage supernatants were screened by proteolysis in situ after capture on wells coated with barstar. Phage remaining bound after washes with PBS and DTT was detected in ELISA with an anti-M13 phage antibody–horseradish peroxidase conjugate (Pharmacia). Purified phages (1010 cfu per well) also were screened by proteolysis in solution, in which case proteases were inactivated with Pefabloc (Boehringer Mannheim) and EDTA before capture on immobilized barstar.
Protein Expression, Purification, and Analysis.
Proteins were expressed by induction of exponential bacterial cultures at 30°C and purified from the soluble fraction of the cytoplasm by using nitrilotriacetic acid–agarose (Qiagen). His-1g6 was purified after solubilization with 8 M urea in TBS and refolded by dialysis from 8 M, 4 M, 2 M, 1 M, 0.5 M, to 0 M urea in TBS. Proteins were purified further by gel filtration on a Superdex-75 column (Amersham Pharmacia). The molecular weight of proteolytic fragments was determined by using surface-enhanced laser desorption/ionization (Ciphergen, Palo Alto, CA).
CD was recorded as described (31). Thermodenaturation was fully reversible under the conditions used (10 μM protein in PBS, His-1c2 at 2 μM in 2.5 mM phosphate, pH 7). NMR experiments (32) were performed with protein at 1 mM in 20 mM phosphate/0.1 M NaCl buffer at pH 6.2 in 93% H2O/7% D2O or 99.9% D2O.
Protein homologs were identified in the blast 2.0 search of the E. coli gene products against Entrez's Molecular Modeling Database and the nr database (http://www.ncbi.nlm.nih.gov). Secondary structure predictions (33) were performed by using the default set-up at http://www.embl-heidelberg.de/predictprotein.
Results and Discussion
CspA forms a stable, five-stranded β-barrel of 70 residues (34), but the first three strands that are adjacent in the structure (Fig. 1A) are not capable of independent folding. We used this region (corresponding to the N-terminal 36-residue fragment) as a template to identify sequences from the E. coli genome that are able to create a folded protease-resistant domain, using proteolysis of phage-displayed proteins as a means of selection (27, 36, 37).
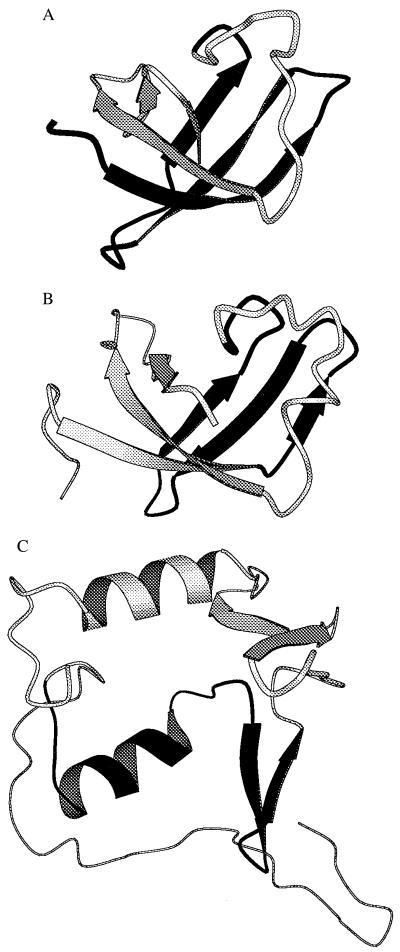
Figure 1.
Structures. Main-chain cartoons (35) from the structures of CspA [A, Protein Data Bank (PDB) file 1mjc], the S1 RNA-binding domain (SRD) (B, PDB file 1sro), and the first 110 residues from the Salmonella oligopeptide-binding protein (C, PDB file 1ola). In A, B, and C, respectively, we have highlighted in black residues 1–36 of CspA and the regions corresponding to the segment of 1b11 (residues 11–39 of SRD homologous to residues 369–397 of S1) and to the segment of 3a12 (residues 30–58 of the oligopeptide-binding domain homologous to residues 52–80 of the periplasmic transport protein).
The gene encoding the N-terminal CspA fragment was ligated to DNA fragments of around 120 bp that were created by random PCR amplification of genomic E. coli DNA. The fragments encode polypeptides of about 40 residues, which are expected to be too small to form globular domains (1, 16) on their own and which are similar in size to an average exon (15). The resulting chimeras were inserted between an N-terminal affinity tag (barnase) and the phage p3 protein of a phagemid, rescuing the phage with the protease-sensitive helper phage KM13 (27). After treatment with both trypsin and thermolysin, phages bearing chimeras that survived proteolysis were captured with the barnase ligand barstar, eluted at acid pH, and used to infect bacteria.
From 1010 phages (108 independent clones), 600 phages survived this first round of selection (5 × 106 phages in the absence of proteolysis), increasing to 2,000 and 4 × 104 after two and three rounds, respectively. Selected phages were grown up individually, bound to immobilized barstar, and treated in situ with trypsin and thermolysin, and proteolytic resistance was measured by detection of bound phage in ELISA. After two rounds, 6 of 192 (3%) phages retained >80% of their binding activity, increasing to 31 of 86 (36%) phages after three rounds. The sequences of 25 resistant phages from the third round revealed 11 different segments, and all could be identified (<1% nucleotide differences) within the E. coli genome (Table 1). For all 11 segments on phage there was an ORF from barnase to p3; for the majority of the segments (seven), the ORF was the same as that of the originating gene and, therefore, appends a segment of the originating protein to the CspA fragment. However, for some of the segments the ORF translates the originating gene out of frame or from the complementary strand, so as to append a segment of different character (Table 1).
Table 1.
Sequences and origin of genomic segments
| Segment* | Sequence† | Genetic origin‡ | Protein origin§ |
|---|---|---|---|
| 1a7 (s) | GIATSAICDA QVIGEEPGQP TSTTCRFRSK FSAIAFPW | 8931–9041 in ECAE298, gatC | Minus strand |
| 1b11 (s, c) | GAAVRGNPQQ GDRVEGKIKS ITDFGIFIGL DGGIDGLVHL SDISW¶ | 6382–6514 in ECAE193, rpsA | 364–398 in RS1_ECOLI |
| 1c2 (s, c) | GRVISLTNEN GSHSVFSYDA LDRLVQQGGF DGRTORYHYD LTW | 2178–2303 in ECAE156, rhsD | 645–686 in RHSD_ECOLI |
| 1g6 (s, c) | GKSGVKTDYR ASASIACAYA GAGSSDSRRS FLCITRSESD GPW | 2694–2569 in ECAE116, rluA | Frameshift |
| 2f1 (s) | GAGTMAEEST DFPGVSRPQD MGGLGFWYRW NLGWMHDTLD YMKPHSW | 8558–8422 in ECAE419, glgB | 452–494 in GLGB_ECOLI |
| 2f3 (s, c) | GAGEPEIGAI MLFTAMDGSE MPGVIREING DSITVDFNHP PPW | 5431–5551 in ECAE113, slpA | 89–127 in FKBX_ECOLI |
| 2h2 (s) | GSAYNTNGLV QGDKYQIIGF PRFNQLTVYF HNLPW | 7955–7854 in ECAE475, yjbC | Minus strand |
| 3a12 (s) | GKAVGLPEIQ VIRDLFEGLV NQNEKGEIVP W | 1479–1568 in ECAE231, b1329 | 52–80 in MPPA_ECOLI |
| 1g7 | GWLKRKLNLK FNEASIAGCD ALLNAAW | 7290–7213 in ECAE217, b1191 | Frameshift |
| 1h12 | GCVPYTNFSL IYEGKCGMSG GRVEGKVIYE TQSTHKHSW | 12035–11927 in ECAE485, cadA | 334–367 in DCLY_ECOLI |
| 2e2 | GMWPLDMVNA IESGIGGTLG FLAAVIGPGT ILGKIMEVSW | 7398–7514 in ECAE324, dsdX | 45–83 in DSDX_ECOLI |
Segments retaining 80% barstar binding activity after proteolysis of phage in situ. Those retaining activity after proteolysis in solution are indicated by s in parentheses. Those purified as chimeric proteins are indicated by c in parentheses.
The sequence of the genomic segment as a C-terminal appendage to the N-terminal region of CspA (LQSGKMTGIV KWFNADKGFG FITPDDGSKD VFVHFSAGSS) is listed; sequences expressed in-frame with the originating gene are shown in italics.
The location of each segment within the E. coli genome is indicated by nucleotide numbers in the European Molecular Biology Laboratory (EMBL) database entry and name of the originating gene.
For those expressed in the same frame of the originating gene, the residue numbers of the corresponding protein and its identification number in the Swiss Protein Database are given.
A single base-pair deletion after the first 29 bp in the DNA insert of 1b11 renders the first 10 residues out of frame with the rspA gene.
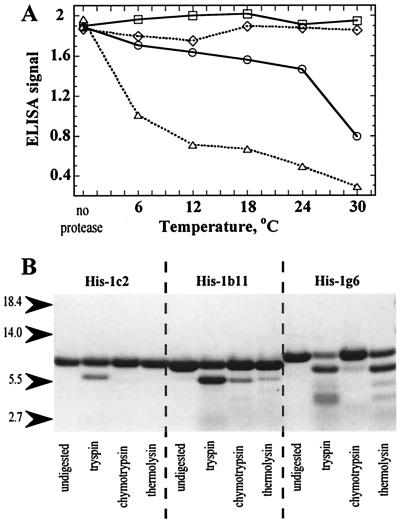
When tested for proteolytic stability in solution (Fig. 2A), 8 of the 11 unique phage clones retained >80% of their barstar-binding activity after trypsin/thermolysin treatment at 24°C (Table 1). The remaining phages were less well protected from proteolysis in solution than in situ. The eight chimeric genes were excised from the phage and cloned for expression in the bacterial cytoplasm with an N-terminal His-tag. Three of these proteins (His-1c2, His-2f3, His-1b11), all derived from segments expressed in the reading frame of the originating protein, could be purified from the soluble fraction of the cytoplasm via their His-tag. The remaining proteins formed inclusion bodies and were not studied further except for one (His-1g6), derived from a frameshift, which was refolded from inclusion bodies after solubilization in 8 M urea. Proteins His-1c2, His-2f3, and His-1g6 formed exclusively monomers on gel filtration, whereas His-1b11 formed 30% monomers with the remainder forming dimers. The monomeric proteins and the His-1b11 monomer were analyzed for resistance to proteolysis with trypsin, thermolysin, and chymotrypsin (Fig. 2B). In all cases, the N-terminal His-tag was excised through cleavage with trypsin at Arg-11 (absent in the phage but present in the soluble proteins at the junction with His-tag). His-1c2 otherwise was completely resistant, and the others were partly so.
Figure 2.
Proteolysis. (A) Proteolysis of selected phages. ELISA for barstar binding of phages 1c2 (□), 1b11 (○), 1g6 (⋄), and the half-barrel csp/2 (▵) before and after trypsin/thermolysin treatment at different temperatures. (B) Proteolysis of chimeric proteins. SDS-polyacrylamide gel of proteins His-1c2, His-1b11, and His-1g6 (40 μM) before and after treatment with trypsin, thermolysin, or chymotrypsin (40 nM) for 10 min at 20°C.
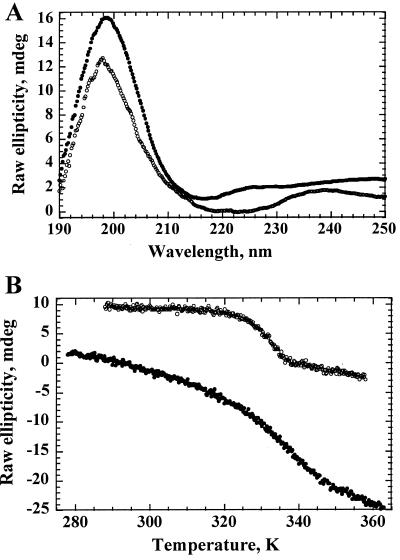
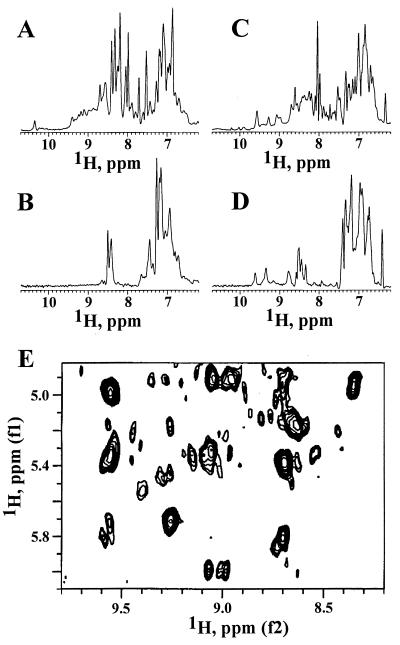
All four proteins had CD spectra (Fig. 3A) with a single trough between 215 nm and 225 nm characteristic for proteins rich in β structure (38). All showed cooperative folding characteristics with sigmoidal melting curves (Fig. 3B) and midpoints of unfolding transition between 48°C and 62°C (Table 2). The chemical shift dispersion of many amide protons to values downfield of 9 ppm (Fig. 4 A and C) and of methyl group protons to values around 0 ppm in the NMR spectra of His-2f3 and His-1c2 are further characteristics of folded proteins (41). Furthermore, downfield chemical shifts of Cα protons to values between 5 and 6 ppm, as seen in the NMR spectrum of His-1c2 (Fig. 4E), are observed frequently in β-sheet-containing proteins such as the immunoglobulins (42).
Figure 3.
CD and thermodenaturation. (A) CD. Spectra of His-1c2 (upper trace) and His-2f3 (lower trace) were recorded at 20°C. (B) Thermodenaturation. Ellipticity of His-1c2 (at 205 nm; upper trace) and His-2f3 (at 223 nm; lower trace) was measured at various temperatures.
Table 2.
Biophysical parameters of proteins
| Protein* | Tm, °C | ΔG,† kcal/mol | Molecular mass, Da |
|---|---|---|---|
| His-Csp | 59.8 | 3.6 | 8,565 |
| His-Csp/2‡ | No expression | 5,854 | |
| His-1b11 | 57.1 | 2.0 | 10,722 |
| His-1c2 | 54.8 | 5.3 | 10,972 |
| His-1g6 | 48.4 | 2.4 | 10,485 |
| His-2f3 | 61.4 | 1.8 | 10,582 |
The sequences of the chimeric proteins are as described in Table 1 with appended tags MRGSHHHHGSR (N terminus) and AQAEA (C terminus).
The conformational stability ΔG at a temperature T was calculated by using the Gibbs–Helmholtz equation ΔG(T) = ΔHm(1 − T/Tm) − ΔCp [(Tm − T) + ln(T/Tm)], while inferring the midpoint of thermal unfolding (Tm) and the enthalpy change for unfolding (ΔHm) at the Tm from the denaturation curve (30) and assuming for ΔCp (the difference in heat capacity between unfolded and folded conformation at constant pressure) a value of 12 cal per residue (40).
The His-Csp/2 protein, which comprises the N-terminal half of CspA (LQSGKMTGIV KWFNADKGFG FITPDDGSKD VFVHFSAW) plus the terminal tags, was found in neither the soluble nor the insoluble fraction of the cytoplasm and is presumed to have been degraded within the cell.
Figure 4.
NMR. One-dimensional 1H NMR spectra of His-2f3 in H2O (A) and after incubation for 24 h at 25°C in D2O (B) were recorded at 25°C, one-dimensional 1H NMR spectra of His-1c2 in H2O (C) and after incubation for 24 h were recorded at 25°C in D2O (D) at 30°C, and the two-dimensional 1H nuclear Overhauser effect spectroscopy spectrum of His-1c2 (E) in H2O was recorded at 30°C.
The conformational stability (ΔG) or folding energy of His-1b11, His-2f3, and His-1g6 ranged between 1.8 and 2.4 kcal/mol (Table 2). These values are similar to the most stable, de novo designed β structure protein, betadoublet (2.5 kcal/mol; ref. 43), but lower than most natural proteins (5–15 kcal/mol; ref. 44). The His-1c2 protein had a much higher stability (5.3 kcal/mol), similar to the most stable de novo designed four helix bundles (45) and to some natural proteins (44). Indeed, His-1c2 was 1.7 kcal/mol more stable than His-CspA (Table 2). Amide exchange in His-1c2 was slow, allowing the observation of many amide protons in a one-dimensional 1H NMR spectrum after 24 h at 25°C in D2O (Fig. 4D). The group of amide signals between 8.7 and 10 ppm was detectable even 3 weeks later at about 40% of their original intensity. By contrast, for His-2f3, no amide protons were observed after 24 h in D2O (Fig. 4B). The greater resistance of His-1c2 to proteolysis compared with the other chimeric proteins correlates with its greater conformational stability rather than its melting temperature (Table 2). Indeed, selection conditions involving proteolysis for longer times (at temperatures ≪Tm) would be expected to favor the selection of proteins of higher conformational stability.
Earlier work has shown that folded hybrid domains can be created from homologous proteins (18–20). With our hybrid domains we were unable to detect any homologies between CspA and any of the donor proteins by sequence comparisons alone. However, we did detect a structural similarity with CspA for one of the donor segments. The structure of the 30S ribosomal subunit protein S1 (the origin of 1b11) appears to comprise a five-stranded β-barrel like that of CspA (as deduced from the known structure (46) of the S1 RNA-binding domain (SRD) of polynucleotide phosphorylase and its sequence homology with S1). The segment of 1b11 is derived from the four adjacent β-strands at the N-terminal portion of the β-barrel (Fig. 1B). By contrast, the structure of the periplasmic transport protein (the origin of segment 3a12) is evidently an α/β protein rather than a β-barrel (as deduced from the known structure of the Salmonella oligopeptide-binding protein (47) and its sequence homologies with the periplasmic transport protein). The segment of 3a12 is derived from a region homologous to a helix and two short antiparallel β-strands (Fig. 1C). Thus sequences derived from the same region of the same fold are juxtaposed in the chimeric protein 1b11, whereas sequences from different folds are juxtaposed in the chimeric protein 3a12.
We do not know whether the original architecture of the CspA fragment and the selected segments has been retained or transmuted in the chimeric proteins, and only the determination of their structures will show whether their folds resemble the fold of CspA. So far, the spectroscopic analyses of the four isolated proteins (Figs. 3A and 4) and secondary structure predictions of all 11 chimeric proteins (36% β-strand and 1.6% helix for the selected segments within the chimeras) suggest that the chimeric proteins fold predominantly into β structure (which does not necessarily correspond to their secondary structure as adopted in the different context of the original protein). It appears that our template (derived from three contiguous β-strands) has acted as a bait for sequences able to complete the structure of small stable domains, in this case most readily by folding into β structure. This pathway may have favored sequences derived from regions of β structure of existing proteins, but not to the exclusion of others, including those derived from out-of-frame readings.
Our results are entirely consistent with proposals that the architecture of proteins was created by the shuffling of polypeptide segments (48, 49). Furthermore, the results indicate that the combination of segments from modern proteins also is capable of generating new proteins. Thus, the generation of new proteins may be expected to be a continuing process; indeed, it may be possible to detect traces of such recombinations by genome analysis (50).
Our data also show that in-frame recombinations are more likely to generate stable proteins. This result highlights an advantage of exons in the evolution of protein architecture, because nonhomologous recombination of DNA or RNA should be aided by the arrangements of genes as exons and introns and in-frame recombinations should be aided by the predominant use of splice junctions in the same reading frame. However, that out-of-frame recombinations can generate stable proteins also may be important in protein evolution. Other reading frames have the potential to contribute sequence of a different character and also may represent a source of great sequence diversity; thus, the accumulation of silent mutations in one reading frame will be revealed on changing frames.
Our results suggest further that any segment of sequence in the genome may have the potential to produce folded domains with several other segments in the same genome. We do not know how frequently such recombinations would take place in nature, because these would be expected to depend on many factors, including the number of segments and the distance between each pair of segments on a chromosome and the presence of small regions of homology. However, our data indicate that, in the case of the N-terminal half of CspA, new polypeptides capable of folding are to be expected at a rate of no less than 10−7 for each nonhomologous recombination with another gene segment of similar size [at least four to eight new folded proteins (Table 1) were rescued from, at most, 108 independent recombinations]. This frequency is likely to vary greatly, depending on the lengths and sequences of the reshuffled gene segments.
In addition to the implications for protein evolution, our results also may lead to novel approaches for making proteins in vitro (51, 52); for example, our strategy provides a means of presenting the same segment of sequence in a variety of structural contexts. Furthermore, the use of segments from humans may allow the creation of novel human hybrid proteins with limited immunogenicity for therapeutic intervention.
Acknowledgments
We are indebted to G. Grigg, R. Holliday, A. Lesk, and C. Chothia for their comments on the manuscript, C. Johnson for advice on the thermodynamic analysis, and I. Lavenir for the mass spectrometer analysis.
Abbreviation
- CspA
cold shock protein A
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170145497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170145497
References
- 1.Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 2.Blake C C F. Nature (London) 1978;273:267. [Google Scholar]
- 3.Bogarad L D, Deem M W. Proc Natl Acad Sci USA. 1999;96:2591–2595. doi: 10.1073/pnas.96.6.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilbert W. Nature (London) 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 5.McClintock B. Cold Spring Harbor Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Craig N L. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2339–2362. [Google Scholar]
- 7.Nash H A. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2363–2376. [Google Scholar]
- 8.Krawiec S, Riley M. Microbiol Rev. 1990;54:502–539. doi: 10.1128/mr.54.4.502-539.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth J R, Benson N J, Galitski T, Haack K, Lawrence J G, Miesel L. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 2256–2276. [Google Scholar]
- 10.Sharp P A. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 11.Lai M M C. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chetverin A B, Chetverina H V, Demidenko A A, Ugarov V I. Cell. 1997;88:503–513. doi: 10.1016/S0092-8674(00)81890-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert W. Nature (London) 1986;319:618. [Google Scholar]
- 14.Darnell J E, Doolittle W F. Proc Natl Acad Sci USA. 1986;83:1271–1275. doi: 10.1073/pnas.83.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blake C. Nature (London) 1983;306:535–537. doi: 10.1038/306535a0. [DOI] [PubMed] [Google Scholar]
- 16.Chothia C, Hubbard T, Brenner S, Barns H, Murzin A. Annu Rev Biophys Biomol Struct. 1997;26:597–627. doi: 10.1146/annurev.biophys.26.1.597. [DOI] [PubMed] [Google Scholar]
- 17.Stoltzfus A, Spencer D F, Zuker M, Logsdon J M, Jr, Doolittle W F. Science. 1994;265:202–207. doi: 10.1126/science.8023140. [DOI] [PubMed] [Google Scholar]
- 18.Crameri A, Raillard S A, Bermudez E, Stemmer W P. Nature (London) 1998;391:288–291. doi: 10.1038/34663. [DOI] [PubMed] [Google Scholar]
- 19.Ostermeier M, Nixon A E, Shim J H, Benkovic S J. Proc Natl Acad Sci USA. 1999;96:3562–3567. doi: 10.1073/pnas.96.7.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altamirano M M, Blackburn J M, Aguayo C, Fersht A R. Nature (London) 2000;403:617–622. doi: 10.1038/35001001. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein J, Pollitt N S, Inouye M. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meiering E M, Serrano L, Fersht A R. J Mol Biol. 1992;225:585–589. doi: 10.1016/0022-2836(92)90387-y. [DOI] [PubMed] [Google Scholar]
- 23.Hoogenboom H R, Griffiths A D, Johnson K S, Chiswell D J, Hudson P, Winter G. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1995. p. 2.4.1. [Google Scholar]
- 25.Gibson T J. Ph.D. thesis. Cambridge, U.K.: Univ. of Cambridge; 1984. [Google Scholar]
- 26.Eggertsson G, Söll D. Microbiol Rev. 1988;52:354–374. doi: 10.1128/mr.52.3.354-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kristensen P, Winter G. Folding Des. 1997;3:321–328. doi: 10.1016/S1359-0278(98)00044-3. [DOI] [PubMed] [Google Scholar]
- 28.Jestin J L, Kristensen P, Winter G. Angew Chem Int Ed Engl. 1999;38:1124–1127. doi: 10.1002/(SICI)1521-3773(19990419)38:8<1124::AID-ANIE1124>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 29.Hartley R W. Biochemistry. 1993;32:5978–5984. doi: 10.1021/bi00074a008. [DOI] [PubMed] [Google Scholar]
- 30.Lubienski M J, Bycroft M, Jones D N M, Fersht A R. FEBS Lett. 1993;332:81–87. doi: 10.1016/0014-5793(93)80489-h. [DOI] [PubMed] [Google Scholar]
- 31.Davies J, Riechmann L. FEBS Lett. 1995;377:92–96. doi: 10.1016/0014-5793(95)01313-x. [DOI] [PubMed] [Google Scholar]
- 32.Riechmann L, Holliger P. Cell. 1997;90:351–360. doi: 10.1016/s0092-8674(00)80342-6. [DOI] [PubMed] [Google Scholar]
- 33.Rost B, Sander C. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindelin H, Maraheil M A, Heinemann U. Proc Natl Acad Sci USA. 1994;91:5119–5123. doi: 10.1073/pnas.91.11.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 36.Sieber V, Plückthun A, Schmid F X. Nat Biotechnol. 1998;16:955–960. doi: 10.1038/nbt1098-955. [DOI] [PubMed] [Google Scholar]
- 37.Finucane M D, Tuna M, Lees J H, Woolfson D N. Biochemistry. 1999;38:11604–11612. doi: 10.1021/bi990765n. [DOI] [PubMed] [Google Scholar]
- 38.Greenfield N, Fasman G D. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- 39.Agashe V R, Udgaonkar J B. Biochemistry. 1995;34:3286–3299. doi: 10.1021/bi00010a019. [DOI] [PubMed] [Google Scholar]
- 40.Edelhoch H, Osborne J C., Jr Adv Protein Chem. 1976;30:183–250. doi: 10.1016/s0065-3233(08)60480-5. [DOI] [PubMed] [Google Scholar]
- 41.Wüthrich K. In: NMR of Proteins and Nucleic Acids. Wüthrich K, editor. New York: Wiley; 1986. pp. 26–39. [Google Scholar]
- 42.Riechmann L, Davies J. J Biomol NMR. 1995;6:141–152. doi: 10.1007/BF00211778. [DOI] [PubMed] [Google Scholar]
- 43.Quinn T P, Tweedy N B, Williams R W, Richardson J S, Richardson D C. Proc Natl Acad Sci USA. 1994;91:8747–8751. doi: 10.1073/pnas.91.19.8747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pace C N. Trends Biochem Sci. 1990;15:14–17. doi: 10.1016/0968-0004(90)90124-t. [DOI] [PubMed] [Google Scholar]
- 45.Kamtekar S, Schiffer J M, Xiong H, Babik J M, Hecht M. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 46.Bycroft M, Hubbard T J, Proctor M, Freund S M, Murzin A G. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 47.Tame J R, Murshudov G N, Dodson E J, Neil T K, Dodson G G, Higgins C F, Wilkinson A J. Science. 1994;264:1578–1581. doi: 10.1126/science.8202710. [DOI] [PubMed] [Google Scholar]
- 48.Doolittle R F, Bork P. Sci Am. 1993;269(4):34–40. doi: 10.1038/scientificamerican1093-50. [DOI] [PubMed] [Google Scholar]
- 49.Gilbert W, De Souza S J, Long M. Proc Natl Acad Sci USA. 1997;94:7698–7703. doi: 10.1073/pnas.94.15.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whisstock J C, Irving J A, Bottomley S P, Pike R N, Lesk A M. Proteins. 1999;36:31–41. doi: 10.1002/(sici)1097-0134(19990701)36:1<31::aid-prot3>3.3.co;2-h. [DOI] [PubMed] [Google Scholar]
- 51.Hecht M. Proc Natl Acad Sci USA. 1994;91:8729–8730. doi: 10.1073/pnas.91.19.8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Regan L. Structure. 1998;6:1–4. doi: 10.1016/s0969-2126(98)00001-x. [DOI] [PubMed] [Google Scholar]