Abstract
The role that reticulate evolution (i.e., via lateral transfer, viral recombination and/or introgressive hybridization) has played in the origin and adaptation of individual taxa and even entire clades continues to be tested for all domains of life. Though falsified for some groups, the hypothesis of divergence in the face of gene flow is becoming accepted as a major facilitator of evolutionary change for many microorganisms, plants and animals. Yet, the effect of reticulate evolutionary change in certain assemblages has been doubted, either due to an actual dearth of genetic exchange among the lineages belonging to these clades or because of a lack of appropriate data to test alternative hypotheses. Marine organisms represent such an assemblage. In the past half-century, some evolutionary biologists interested in the origin and trajectory of marine organisms, particularly animals, have posited that horizontal transfer, introgression and hybrid speciation have been rare. In this review, we provide examples of such genetic exchange that have come to light largely as a result of analyses of molecular markers. Comparisons among these markers and between these loci and morphological characters have provided numerous examples of marine microorganisms, plants and animals that possess the signature of mosaic genomes.
Keywords: introgression, horizontal transfer, web of life, marine
1. Introduction
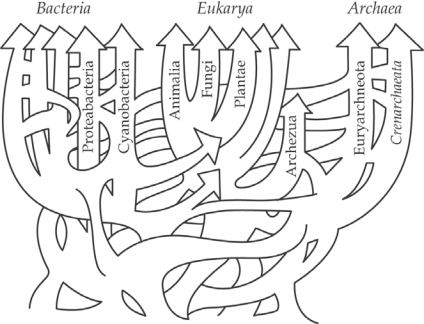
The occurrence of reticulate evolution (i.e., involving the processes of natural hybridization, horizontal transfer and viral recombination) is now well established as having affected the origin and adaptation of organisms from all of the domains of life (see [1–10] for reviews). The evaluation of available data sets, particularly those involving molecular markers, has thus led to the falsification of the hypothesis that most lineages have arisen and evolved in genetic isolation from other lineages (i.e., the allopatric model; see [6,7] for reviews). Instead, models of evolution, that incorporate divergence in the face of gene flow, have been repeatedly supported as more and more genomes have been examined in detail (e.g., [11–13]). Indeed, such is the phylogenetic extent of the genetic exchange that has been detected that we have argued [6,7] for the substitution of the tree-of-life metaphor with one best described as a web-of-life (Figure 1). Such a metaphor thus incorporates introgressive hybridization, lateral exchange and natural selection in the development of evolutionary lineages possessing mosaic genomes (i.e., genomes made up of elements from multiple evolutionary lineages; [6,7]).
Figure 1.
A representation of the history of biological diversification of all life, reflecting the role of introgressive hybridization and lateral exchange in the development of new lineages with mosaic genomes (from [8], as modified in [6]). Reprinted with permission from The American Association for the Advancement of Science [8].
In contrast to the above conclusions, some authors have suggested that the extent to which reticulate evolution has affected marine organisms is limited. For example, Arnold [5] agreed with the conclusion of Hubbs [14] that the available data supported “…the hypothesis that natural hybridization is less common in marine fishes.” This conclusion seems to be substantiated by the relative uniqueness of findings such as those reported by Roques et al. [15] in their paper on introgressive hybridization in redfish (genus Sebastes), which they referred to as “a rare marine example”. Yet, it is also possible that the rarity of such reports reflects a lack of data to test for genetic exchange, rather than an absence of such exchange. In discussing the dearth of examples of introgressive hybridization in entire clades of tropical birds, Grant and Grant [16] argued that the lack of examples from these groups might be due to cryptic morphological differences between species. Likewise, it is possible that in the marine realm reticulate evolution occurs at a similar frequency to that encountered for many non-marine clades, but the difficulty in collecting/observing the organisms has limited its detection [17]. Thus, the title of this review reflects the question of whether or not the marine realm reflects a “Final Frontier” in terms of testing for the role of reticulate evolution in the origin and trajectory of organismic lineages and assemblages.
In contrast to the rarity of reticulate evolution (whether biologically-based or due to lack of sampling) inferred for many marine clades, some researchers have invoked a major role for genetic exchange in the diversification of some taxa (e.g., corals; [18,19]). Furthermore, analyses of fossil records for marine organisms also support the contention that reticulate evolution has been a characteristic of certain assemblages across time as well as phylogenetic and geographic space [20,21]. Finally, if the evolutionary effect of reticulation is tested across the extreme taxonomic diversity present in this biome, including microorganisms, it is predicted that gene transfer would be seen as having a fundamentally important role in the evolution of marine environments. Thus, like many clades that reside in terrestrial and freshwater habitats, the evolution of marine organisms may also be better reflected by a web-of-life metaphor rather than the tree-of-life concept [6,7,17,22,23]. In this review, we cite marine examples of reticulate evolution in organisms as diverse as archaebacteria, bacteria, seaweed, eelgrass, coral, shrimp, tuna and fur seals. These examples are not exhaustive. Rather, they are included to reflect the breadth of organisms that have evolved in the face of gene flow. Specifically, we present evidence of a significant effect from genetic exchange on the population genetic structure, evolutionary diversity, and adaptive evolution across all major groups of marine organisms (Table 1).
Table 1.
Selected examples of marine organisms for which genetic exchange events (i.e., horizontal transfer, introgressive hybridization and/or hybrid speciation) have been inferred. Lineages that are either the donor or recipient of DNA sequences are included. The genus, species and common name (if available) for each example are given. In addition, whether the genetic exchange was characterized as horizontal transfer (as, for example, is the case in transfers between cyanobacteria and bacteriophages) or introgressive hybridization (i.e., involving sexual reproduction and backcrossing; 1) is noted. The term “Multiple” is given under the “Taxon” category to indicate the interaction of more than one lineage from the taxonomic group. For example, the categories of “Cyanobacteria” and “Bacteriophage” reflect analyses of at least 14 species/strains and ca. 100 divergent viral lineages, respectively. The type of analyses used (morphological analyses, population genetic surveys, genome sequence analyses and/or tests for phylogenetic discordance) to infer the exchange events are also indicated, along with the reference(s) that reported the findings. Note: the final example listed comes from fossil data and combines results from studies of crinoids and corals.
| Taxon | Common Name | Type of Genetic Exchange | Data | Reference(s) |
|---|---|---|---|---|
| Multiple | Cyanobacteria | Horizontal transfer | Genome sequence analyses, Phylogenetic discordance | [28,29,69–73] |
| Multiple | Bacteriophage | Horizontal transfer | Genome sequence analyses, Phylogenetic discordance | [72–74] |
| Multiple | Bacteria | Horizontal transfer | Genome sequence analyses, Phylogenetic discordance | [25] |
| Multiple | Archaebacteria | Horizontal transfer | Genome sequence analyses, Phylogenetic discordance | [25] |
| Spartina (Multiple) | Cordgrass | Introgressive hybridization, Hybrid speciation | Population genetic surveys, Phylogenetic discordance | [75–79] |
| Zostera (Multiple) | Eelgrass | Introgressive hybridization | Population genetic surveys | [34] |
| Fucus (Multiple) | Seaweed | Introgressive hybridization, Hybrid speciation | Population genetic surveys, Phylogenetic discordance | [80–84] |
| Pseudo-nitzschia pungens | Diatom | Introgressive hybridization | Morphological analyses, Population genetic surveys | [44–46] |
| Bythograea thermydron | Hydrothermal crab | Horizontal transfer (transposable elements) | Phylogenetic discordance | [32] |
| Ventiella sulfuris | Hydrothermal amphipod | Horizontal transfer (transposable elements) | Phylogenetic discordance | [32] |
| Maia brachydactila | Sea shoe | Horizontal transfer (transposable elements) | Phylogenetic discordance | [32] |
| Cancer pagurus | Crab | Horizontal transfer (transposable elements) | Phylogenetic discordance | [32] |
| Menippe (Multiple) | Stone crab | Introgressive hybridization | Population genetic surveys | [85–87] |
| Mysis (Multiple) | Opossum shrimp | Introgressive hybridization | Phylogenetic discordance | [48,50] |
| Diadema (Multiple) | Sea urchins | Introgressive hybridization | Population genetic surveys | [56] |
| Lytechinus (Multiple) | Sea urchins | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [57] |
| Strongylocentrotus (Multiple) | Sea urchins | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [53–55] |
| Asterias (Multiple) | Sea stars | Introgressive hybridization | Morphological analyses, Population genetic surveys, Phylogenetic discordance | [54,58,59] |
| Acrocnida brachiata | Brittle-star | Introgressive hybridization | Population genetic surveys | [88] |
| Alcyonium hibernicum | Coral | Hybrid speciation | Population genetic surveys, Phylogenetic discordance | [40] |
| Bellonella bocagei | Coral | Hybrid speciation | Population genetic surveys, Phylogenetic discordance | [40] |
| Pocillopora (Multiple) | Corals | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [41] |
| Acropora (Multiple) | Corals | Introgressive hybridization, Hybrid speciation | Morphological analyses, Population genetic surveys, Phylogenetic discordance | [19,35,89–94] |
| Littorina saxtilis | Snail | Introgressive hybridization | Morphological analyses | [95,96] |
| Mercenaria (Multiple) | Clams | Introgressive hybridization | Population genetic surveys | [97,98] |
| Macoma balthica | Clam | Introgressive hybridization | Population genetic surveys | [99,100] |
| Crassostrea virginica | American oyster | Introgressive hybridization | Population genetic surveys | [101,102] |
| Mytilus (Multiple) | Mussels | Introgressive hybridization | Morphological analyses, Population genetic surveys, Phylogenetic discordance | [103–114] |
| Bathymodiolus (Multiple) | Hydrothermal vent mussels | Introgressive hybridization | Morphological analyses, Population genetic surveys | [51,52] |
| Scophthalmus maximus | Turbot | Introgressive hybridization | Population genetic surveys | [115] |
| Clupea harengus | Atlantic herring | Introgressive hybridization | Population genetic surveys | [116] |
| Gadus morhua | Atlantic cod | Introgressive hybridization | Population genetic surveys | [117,118] |
| Platichthys flesus | European flounder | Introgressive hybridization | Population genetic surveys | [119] |
| Pleuronectes platessa | Plaice | Introgressive hybridization | Population genetic surveys | [119] |
| Thunnus (Multiple) | Tuna and Albacore | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [120–122] |
| Anguilla (Multiple) | Eels | Introgressive hybridization | Population genetic surveys | [123,124] |
| Sebastosomus (Multiple) | Rockfish | Introgressive hybridization | Population genetic surveys | [125] |
| Acanthochromis (Multiple) | Damselfish | Introgressive hybridization | Population genetic surveys | [126,127] |
| Plectropomus (Multiple) | Coral trout | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [60] |
| Acanthurus (Multiple) | Surgeonfish | Introgressive hybridization, Hybrid speciation | Population genetic surveys, Phylogenetic discordance | [61] |
| Chaetodon (Multiple) | Butterflyfish | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [128] |
| Sebastes (Multiple) | Redfish | Introgressive hybridization | Population genetic surveys | [15] |
| Salmonidae (Multiple) | Charr, Salmon | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [129,130] |
| Caretta caretta | Loggerhead turtle | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [62–64] |
| Lepidochelys (Multiple) | Kemp’s ridley and Olive ridley turtles | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [62–64] |
| Eretmochelys imbricata | Hawksbill turtle | Introgressive hybridization | Population genetic surveys, Phylogenetic discordance | [62–64] |
| Chelonia mydas | Green turtle | Introgressive hybridization | Population genetic surveys | [62] |
| Arctocephalus (Multiple) | Fur seals | Introgressive hybridization | Morphological analyses, Population genetic surveys | [65–68] |
| Eretmocrinus (Fossils) | Crinoids | Introgressive hybridization | Morphological analyses | [20] |
| Montastraea (Fossils) | Corals | Introgressive hybridization | Morphological analyses | [21] |
2. Horizontal Transfer
2.1. Horizontal Transfer and Adaptation in Marine Archaea, Bacteria and Cyanobacteria
Archaea and bacteria demonstrate evidence of extensive genetic exchange via horizontal gene transfers (Table 1). For example, DeLong [24] reported rDNA sequences characteristic of archaebacteria (i.e., Archaea) in a previously unknown environment for these organisms, that of oxygenated coastal surface waters. This observation generated the following hypothesis: Eubacteria, Archaea and Eukarya “…reside and compete in the ocean’s photic zone under the pervasive influence of light” [25]. This hypothesis leads to several predictions, one of which is that if Archaea and Eubacteria are to compete for light-limited resources, they must both possess genes involved in the utilization of photic energy. It is therefore significant that such genes have been isolated from Archaebacteria and Eubacteria (e.g., [25–27]). Specifically, photoproteins termed proteorhodopsins have been detected [26]. Furthermore, the detection of the shared genetic architecture for utilizing light energy has thus been ascribed to the lateral transfer of genes into the Archaea.
Frigaard et al. [25] also detected evidence for lateral transfer between planktonic bacteria and Archaea as well. This transfer was suggested to be adaptive in nature given that the proteorhodopsin genes isolated from euryarchaeotes were present in those isolates taken from photic, but not subphotic, regions of the water column. This finding was consistent with an adaptive scenario in which the organisms in the light-limited zones gained no benefit from the transfer of the photo-response genes, while those organisms living in the photic zone did [25]. From their findings, Frigaard et al. [25] made the general conclusion that “…lateral gene dispersal mechanisms, coupled with strong selection for proteorhodopsin in the light, have contributed to the distribution of these photoproteins among various [marine] members of all three of life’s domains.”
In addition to the above examples, cyanobacteria have also been shown to possess genetic components most likely resulting from horizontal transfer events. Shi and Falkowski [28]–in an examination of 682 loci–discovered widespread disagreement between phylogenies constructed for 13 cyanobacteria genomes. This discordance among phylogenetic hypotheses was apparently due to large-scale horizontal gene transfer. Indeed, of the 682 orthologs analyzed only 323 were placed into a “core set” whose evolutionary histories seemed congruent [28]. The majority of the loci thus appeared to have been potentially affected by genetic exchange. For example, it was hypothesized that the unique presence of the genetic architecture for nitrogen fixation originated from exchange with a heterotrophic prokaryotic lineage [28]. Likewise, Swingley et al. [29] also discovered evidence for the transfer of the genes (i.e., to produce chlorophyll d) that provided the cyanobacterial species, Acaryochloris marina, with the ability to utilize far-red light for photosynthesis. The close physical association of the cyanobacterium with other oxygenic phototrophs (e.g., Prochloron) and the selective benefit (to the recipients) of being outside the competitive milieu of organisms possessing chlorophyll a and/or b likely facilitated the acquiring of this function [29].
2.2. Horizontal Transfer and the Evolution of the Marine Protist, Micromonas
Worden et al. [30] suggested that members of the picoeukaryotic genus, Micromonas, could play a role as “sentinel organisms” for monitoring climate-change driven perturbations in oceanic systems. This potential utility as biogeochemical-indicator species is due to their distribution, and thus adaptation, across tropical to polar marine ecosystems.
Significantly, the differentiation and adaptive evolution of various isolates belonging to the Micromonas clade is likely the result of the combined action of horizontally-acquired genes and selection. In particular, a large fraction of the genes identified as “unique” to these eukaryotes lineages analyzed shared significant similarity with clades of prokaryotes. Furthermore, the two isolates examined by Worden et al. [30] were highly divergent at these loci reflecting this combination of reticulation and differential selection. Indeed, this pattern of divergence and acquisition (via horizontal transfer) reflects the evolutionarily dynamic nature of these protist lineages [30], and also indicates the potential for genetic exchange to underlie adaptive evolution [5–7].
2.3. Horizontal Transfer of Transposable Elements among Marine Invertebrates
The exchange of transposable elements (i.e., transposons) among distantly-related terrestrial organisms has been recognized for more than a decade (reviewed in [31]). For example, the relatively recent introduction of transposons known as “Type II class elements” was detected for the cosmopolitan invertebrate, Drosophila melanogaster. Likewise, a recent horizontal exchange of this class of element was inferred between D. melanogaster and D. willistoni, two species that last shared a common ancestor >50 million years ago [31].
Unlike the terrestrial lineages mentioned above, genomic information for marine invertebrates is relatively limited [32]. Tests for the horizontal exchange of transposable elements have been limited by this lack of genomic data. However, recent work has not only identified various Type II transposons, but has also identified instances of apparent horizontal exchange between phylogenetically-unrelated organisms that occur in close spatial proximity. Specifically, Casse et al. [32] identified mariner-like elements in the genomes of four different marine invertebrates (Table 1). Two of the species, Cancer pagurus and Maia brachydactila, are coastal crustaceans, while the remaining two species, Ventiella sulfuris and Bythograea thermydron are hydrothermal vent-associated organisms (an amphipod and crab, respectively [32]).
Though phylogenetically highly divergent lineages, the mariner-like elements isolated from the genomes of the coastal and the hydrothermal vent species demonstrated high levels of sequence similarity to the organism with which it was spatially associated [32]. In particular, the two hydrothermal organisms possessed elements that shared 99.5% similarity. Transposable elements isolated from the two coastal species likewise exhibited >99% sequence similarity. These findings led to the conclusion that, as for terrestrial invertebrates, horizontal transfer had resulted in the exchange of the Type II class elements between the unrelated, but sympatrically-distributed, coastal and hydrothermal vent species [32].
3. Introgressive Hybridization and Hybrid Speciation
3.1. Introgressive Hybridization in Marine Angiosperms
Table 1 lists three examples of marine plant clades that demonstrate the effect of introgressive hybridization and hybrid speciation (both homoploid–i.e., diploid hybrid derivative lineages–and polyploid). Of these, we will consider only the eelgrass, Zostera. This aquatic angiosperm, like other seagrasses, is an important constituent of the marine coastal communities in which it occurs. Though eelgrass genotypes have the capacity to reproduce asexually via rhizomatous growth, they also reproduce sexually, with their seeds having the ability to be transported over long distances by rafting on detached plants (e.g., up to 50km along the Northern European coast [33]).
Pollen flow among eelgrass populations appears limited (reviewed in [33]); however, gene flow via male gametes does occur at low frequencies. For example, in a sample of 28 Zostera “meadows” located both in the California Channel Islands and the adjacent mainland, Coyer et al. [34] detected genetic variation indicative of significant levels of gene flow, particularly for the coastal populations. Furthermore, their samples from the eelgrass sites included two species identified as Zostera marina and Z. pacifica. In addition to the genetic connectivity caused by clonal growth and sexual reproduction, these samples reflected admixtures of the two species’ genomes. Specifically, the assignment of genotypes to classes of Z. marina, Z. pacifica, or “introgressed”, utilizing microsatellite marker loci, detected hybrid/parental assemblages at the Santa Catalina, San Clemente and San Diego sites [34]. Each of these sites was suggested as a possible example of anthropogenically-mediated introgressive hybridization thus reflecting the extensive impact of humans on coastal environments. These results also led to the conclusion that introgressive hybridization may occur throughout the global distribution of Zostera populations, thereby contributing to the genetic variability in numerous eelgrass species [34].
3.2. Introgressive Hybridization and Hybrid Speciation in Corals
The literature concerning the role of reticulate evolution in the origin and diversification of some coral clades is extensive. For example, descriptions of widespread introgressive hybridization leading to an enrichment of genetic and morphological variation are replete for reef corals belonging to the genus Acropora [19]. In this regard, Hatta et al. [35] found high rates of experimental, interspecific fertilization between naturally hybridizing species of acroporids. In contrast, Knowlton et al. [36] and Levitan et al. [37] defined barriers to reproduction between different species and morphotypes of Montastraea corals, but with regional differences in the strength of isolation. In addition, Fukami et al. [38] detected a north to south hybridization gradient in these same Montastraea lineages using molecular and morphological analyses. This latter study provided evidence that introgression between these species occurred mostly in the northern portion of their distribution. Likewise, an analysis of the family Faviidae documented extensive paraphyly across numerous clades [39]. It was suggested that introgressive hybridization may have contributed to the observation of paraphyly. Specifically, Huang et al. [39] argued “Introgression…may have resulted in such disparity, where the gene tree does not resemble the species tree, and neither is well-correlated with morphological evolution…” Additional evidence for introgressive hybridization within this clade of corals has also come from the fossil record. Specifically, morphological variation across the fossil record of this coral genus led to the inference of introgressive hybridization during the Pleistocene period [21].
Examples of hybrid speciation and introgression affecting coral evolution have been found within the genera Alcyonium and Pocillopora as well. McFadden & Hutchinson [40] tested the hypothesis of hybrid speciation giving rise to members of two genera of European soft corals (i.e., Alcyonium and Bellonella). Specifically, they analyzed sequence variation in the rRNA internal transcribed spacer (“ITS-1”) region of the putative hybrid lineages, Alcyonium hibernicum and Bellonella bocagei. The following patterns of genetic variability did indeed support the hypothesis of hybrid derivation for each of these lineages: (1) A. hibernicum possessed a combination of two different sequence variants that were characteristic of divergent clades of soft corals, and (2) B. bocagei possessed divergent ITS-1 types inferred to be recombinants between the same two divergent sequence families [40].
Species belonging to the tropical eastern Pacific scleractinian coral genus, Pocillopora, are the dominant reef-building organisms in this region [41]. Unlike their congeners found throughout most of the genus’ geographical distribution, these species have shifted from internal brooding of larvae to free-spawning [42]. Interestingly, the transition in reproductive mode was correlated with introgressive hybridization among five species of these tropical corals. Like the study of Alcyonium and Bellonella, rRNA ITS sequence data were collected from Pocillopora individuals distributed in the tropical eastern Pacific [41]. These data revealed a sharing of sequence variants among Pocillopora damicornis, P. eydouxi, P. elegans, P. inflata and P. effusus. Not only did Combosch et al. [41] infer introgressive hybridization as the source of this genetic variation, but they also concluded that the gene flow was largely unidirectional, with P. damicornis acting as the recipient of the allelic variability. Thus, numerous coral assemblages, including Indo-Pacific acroporids, the European soft corals and the eastern Pacific Pocillopora clades, reflect signatures of reticulate evolution.
3.3. Introgressive Hybridization in Protists–The Diatom Genus Pseudo-nitzschia
Diatoms of the genus Pseudo-nitzschia are probably best known as the organisms responsible for ‘harmful algal blooms’ (HABs) during which large quantities of the neurotoxin, domoic acid is produced [43]. Domoic acid is the causative agent of ‘amnesic shellfish poisoning’ (ASP), a syndrome often caused (as the name suggests) by ingestion of shellfish laden with this neurotoxin [43]. Such poisoning events prior to the 1980’s had been ascribed mainly to toxin-producing dinoflagellate or cyanobacteria, taken up by shellfish that were subsequently eaten by humans [43]. However, a particular episode of poisoning in 1987 led to the discovery of Pseudo-nitzschia as the source of a low molecular weight amino acid (i.e., domoic acid) leading to ASP [43].
At least 12 species of Pseudo-nitzschia are now known to have the capacity to produce domoic acid (reviewed in [43,44]). Most of these species are now believed to be cosmopolitan [43,44], with genetic and morphological data indicative of multiple strains and/or varieties within some of the species [45,46]. For the present discussion it is significant that the evolution of multiple strains/varieties and their occurrence in sympatric associations have led to introgressive hybridization. This introgression has been documented using a variety of molecular and morphological markers. For example, D’Alelio et al. [45] detected admixtures of the Pseudo-nitzschia multistrata strains “A” and “B” (using the internal transcribed spacer region (ITS) of the ribosomal RNA genes) in samples from the Gulf of Naples. They thus found individual genomes defined as having A + B haplotypes. Because the individuals belonging to the A and B categories could not be defined on the basis of any other molecular or morphological parameter examined, it was concluded that this pattern of ITS admixture was due to the overlap of conspecific, but somewhat divergent, populations that had arisen either in situ or allopatrically [45].
Like the results from P. multistrata, analyses of P. pungens populations also detected patterns indicative of both divergence and introgression [46]. However, unlike P. multistrata, the P. pungens lineages were characterized by divergence in multiple genomic (i.e., ITS and chloroplast loci) and morphological characters [46]. Furthermore, the degree of divergence in the DNA and morphological characters allowed a relatively detailed definition of introgressive hybridization in a hybrid zone in the northeast Pacific. Casteleyn et al. [46] detected individuals exhibiting mixtures of morphological traits and chloroplast/nuclear haplotypes indicative of both first-generation and advanced-generation hybrids. As with all of the other examples given in this review, the detection of introgression within P. multistrata and P. pungens, suggests a broader base of genetic exchange among marine organisms than has been previously appreciated.
3.4. Introgressive Hybridization in Crustacea
3.4.1. Genus Tetraclita (Acorn Barnacles)
The predominant intertidal barnacle lineages in the northwestern Pacific Ocean belong to the genus Tetraclita. Because two of these “acorn barnacles” have been found to possess identical mitochondrial haplotypes, as well as very similar morphologies, they were recently reduced from specific to subspecific status. Tsang et al. [47] tested the genetic distinctiveness and the geographic pattern of genetic variation of these two subspecies (Tetraclita japonica japonica and T. j. formosana) using amplified fragment length polymorphisms. Tsang et al.’s [47] analysis led to the following series of observations and hypotheses: (1) warming in the oceans may have been the catalyst for poleward movement of some of the acorn barnacle lineages belonging to the genus Tetraclita; (2) T. j. formosana migrated to Japan and successfully colonized habitats there; (3) following this migration to Japan, and because of their relative scarcity, the T. j. formosana individuals mated frequently with the more numerous T. j. japonica barnacles; (4) the bouts of hybridization in Japan have resulted in numerous F1 and backcross hybrid individuals; (5) likewise, introgressive hybridization has also occurred in Okinawa; and (6) continued migration fueled by shifts in oceanic temperatures may lead to the genetic assimilation and thus disappearance of some of the Tetraclita lineages. Thus, although T. j. japonica and T. j. formosana were confirmed as genetically-differentiated lineages worthy of recognition, this distinctiveness may be lost if migration causes greater genetic admixture between these taxa [47].
3.4.2. Genus Mysis (Opossum Shrimp)
Opossum shrimp species belonging to the genus Mysis are distributed throughout aquatic habitats. Because they are found throughout the world’s marine and freshwater zones they have been used as a model system for testing hypotheses concerning the origin and evolutionary trajectories of geographically disjunct, but phylogenetically-related, zoogeographic elements [48]. Though questions concerning the processes that have affected current day distributions of such disjunct clades remain (e.g., [49]), studies of the opossum shrimp have defined phylogenetic signatures suggesting the role of reticulation in their evolutionary history.
Audzijonyte et al. [48] reconstructed the phylogenetic relationships among ca. 15 species of Mysis that possessed either circumarctic, northwest Atlantic, Continental or Caspian Sea distributions. The data that were used for the phylogenetic reconstructions included morphological characters and nuclear/mtDNA sequences. Instances suggestive of mtDNA introgression following divergence included (1) the Caspian Sea assemblage, (2) the circumarctic species, Mysis litoralis and M. oculata and (3) the continental species, M. salemaai and M. segerstralei [48]. In each case, the evidence for reticulate evolution came from phylogenetic discordance between evolutionary trees derived from different data sets.
A more recent analysis of morphological and genetic variation among Mysis species also resulted in the inference of post-divergence introgression. In this latter study, Audzijonyte and Väinölä [50] examined the divergence among the three circumpolar species, M. nordenskioldi, M. litoralis and M. oculata. Though previously difficult to separate, these three species were found to possess diagnostic combinations of morphological and genetic characteristics. However, Audzijonyte et al. [48] and Audzijonyte and Väinölä [50] also detected discordances. In particular, the three species were distinguishable using both morphology and nuclear loci, but M. litoralis and M. oculata formed an unresolved cluster based upon mtDNA variability. Once again, these data support the hypothesis of post-divergence, mtDNA introgression among Mysis lineages [50].
3.5. Introgressive Hybridization between Hydrothermal Vent Mussels of the Genus, Bathymodiolus
Like marine groups such as corals and cyanobacteria, mussels have a rich literature indicating extensive genetic exchange between divergent lineages (Table 1). For example, Arnold [5,7] has reviewed in detail work documenting the effect of introgression on the genetic structure of widely dispersed species of the genus, Mytilus. However, reticulate evolution is not limited to these near-shore taxa.
O’Mullan et al. [51] and Won et al. [52] reported genetic analyses of deep-sea hydrothermal vent mussels across an area of sympatry between the species, Bathymodiolus azoricus and B. puteoserpentis. Both studies detected introgressed individuals along a ridge in the area of overlap between the northern B. azoricus and southern B. puteoserpentis. In the first of the analyses, morphometric and genetic data (from both nuclear and mtDNA loci) “revealed a mixed population with gene frequencies and morphology that were broadly intermediate to those of the northern and southern species…” [51]. The spatially restricted nature of the hybrid individuals suggested the presence of selection against at least some of the hybrid genotypes. This latter hypothesis was supported by cytonuclear disequilibrium estimates [52]. In particular, Won et al. [52] discovered a pattern indicative of parental migration into the zone and restriction of the hybrids to the zone of overlap due to selectively disadvantageous interactions between genes inherited from the two species [52]. Notwithstanding the evidence for selection acting against some hybrid genotypes, recombination between the two hydrothermal vent mussel genomes–at least within the hybrid zone–was apparent.
3.6. Introgressive Hybridization in Echinodermata
3.6.1. Sea Urchin Species
Numerous evolutionary studies involving various clades of sea urchins have defined reproductive barriers between congeners. For example, Levitan [53] reported the degree to which eggs from three species of Strongylocentrotus (i.e., droebachiensis, franciscanus, purpuratus) could be fertilized with either conspecific or heterospecific sperm. Levitan [53] found that eggs from females most easily fertilized with conspecific sperm (e.g., S. droebachiensis) were also less discriminating towards sperm from other species. Thus, Levitan [53] found a cline of reproductive isolation from S. droebachiensis (least isolated) to S. franciscanus (intermediate isolation) to S. purpuratus (highly isolated). Furthermore, Harper et al. [54], in an analysis of gene flow within and between species of Strongylocentrotus sea urchins, concluded that sympatric populations of different species exchanged genes at much lower frequencies than did populations of the same species separated by oceans.
In spite of reproductive barriers between sea urchin lineages, introgressive hybridization has been well documented between numerous species and subspecies (Table 1). Indeed, even the clades utilized to define reproductive isolation (e.g., Strongylocentrotus) contain genetic variation consistent with reticulate evolution. For example, both Addison and Hart [55] and Harper et al. [54] described extensive interspecific gene flow between S. droebachiensis and S. pallidus throughout the range of the former species (in the northwest Atlantic and Pacific oceans). Likewise, Lessios and Pearse [56] produced evidence of echinoid introgressive hybridization from a combined genetic and morphological analysis of Diadema paucispinum, D. savignyi and D. setosum. Specifically, these authors detected genotypes that suggested introgression between combinations of all three of the Diadema lineages. Finally, Zigler and Lessios [57] also reported variability at mitochondrial and nuclear loci demonstrative of introgression within the genus Lytechinus; intersubspecific and interspecific introgression was detected between Lytechinus variegatus variegatus/L. v. carolinus and L. variegatus/L. williamsi, respectively [57].
3.6.2. Genus Asterias (Sea Stars)
As with sea urchins of the genus Strongylocentrotus, Harper et al. [54] also detected patterns of genetic variation in sea stars reflective of long-distance gene flow within species, but a more limited effect from introgression between sympatric species. Yet, introgression does indeed affect the genetic structuring of Asterias species that co-occur and thus form hybrid zones. For example, both Harper & Hart [58] and Scheibling and Lauzon-Guay [59] reported morphological and/or mtDNA data demonstrating contemporary hybrid zones between Asterias forbesi/A. rubens and A. vulgaris/A. forbesi/A. rubens, respectively. Both of these above studies analyzed a zone of overlap in the northwest Atlantic. In the analysis by Harper and Hart [58], mtDNA and morphological data were collected and the resulting patterns of phenotypic and mtDNA variation were compared for several populations. The morphological characters suggested only two groups, reflective of A. forbesi and A. rubens phenotypes. However, the mtDNA sequence variability collected from the same populations was discordant with the morphology and suggested the presence of advanced-generation hybrids not detectable with quantitative (i.e., morphological) characters [58]. In contrast, a study based upon morphological characters diagnostic for A. vulgaris, A. forbesi and A. rubens did detect variation indicating a mosaic of “A. rubens” and “A. forbesi” phenotypes [59]. Furthermore, some individuals possessed morphological traits suggesting introgression involving A. vulgaris as well [59].
3.7. Introgressive Hybridization and Hybrid Speciation in Coral Reef Fishes Belonging to the Genera, Plectropomus and Acanthurus
Table 1 reflects the growing literature indicating the role of reticulate evolution within several different species complexes commonly known as coral reef fish. Two of these clades–Plectropomus and Acanthurus–exemplify the outcomes of introgressive hybridization and introgressive hybridization/hybrid speciation, respectively [60,61]. In particular, member lineages of these unrelated genera possess mosaic genomes and/or phenotypes reflecting contributions from multiple species. Furthermore, some of these hybrid lineages have been recognized as species.
van Herwerden et al. [60] defined the nuclear and mtDNA variation in two species of grouper, Plectropomus maculatus and P. leopardus, found along both the eastern and western Australian coastlines. The patterns of phylogenetic differentiation led these workers to infer both introgressive hybridization and incomplete lineage sorting as causal for discordances among the genetic markers. In particular, (1) the lack of reciprocal monophyly in mtDNA phylogenies for the eastern populations of the two species, but (2) the resolution of species-specific clades for the western samples, suggested an impact of introgression on the east coast lineages [60]. In contrast, incomplete lineage sorting was inferred as the cause of the discordances found among the nuclear-based phylogenies. Thus, one of three loci generated clades containing only one of the species. Two of the nuclear loci produced admixed clades containing samples from both P. maculatus and P. leopardus [60]. It is, however, possible that the discordance among the nuclear phylogenies, like those from the mtDNA, could also reflect the role of introgression.
A study of genetic variation in an area of sympatry in the eastern Indian Ocean between the coral reef surgeonfish, Acanthurus leucosternon and A. nigricans, also resolved patterns indicative of reticulate evolution (Figure 2). Marie et al. [61] collected DNA sequence data for three distinctive morphotypes, two reflecting A. leucosternon and A. nigricans and the third being a hypothesized hybrid species (i.e., “A. cf. leucosternon”). Sequence information was obtained from both mtDNA and nuclear loci. These data allowed simultaneous tests for introgression between A. leucosternon and A. nigricans, and the hybrid origin of A. cf. leucosternon. Both the introgression and hybrid speciation hypotheses were supported by the mtDNA data [61]. First, admixed clades of all three species were defined by the mtDNA sequence information (Figure 2). Indeed, the extent and directionality of introgression suggested concern that the A. leucosternon lineage might be lost from the region of overlap [61]. Second, mtDNA haplotypes characteristic of allopatric populations of both A. leucosternon and A. nigricans were detected in the sample of A. cf. leucosternon individuals (Figure 2); this is consistent with a hybrid origin for the “intermediate color patterns” possessed by A. cf. leucosternon [61].
Figure 2.
Genetic associations (based upon mtDNA sequence variation) between surgeonfish categorized as hybrids (“Acanthurus cf. leucosternon”), A. leucosternon or A. nigricans. The relative sizes of the circles reflect the number of individuals sharing a particular mtDNA haplotype. Bars on the lines connecting haplotypes indicate the number of substitutions differentiating them. Dashed lines surround the two major Acanthurus clades [61]. Reprinted with permission from Springer [61].
3.8. Introgressive Hybridization in Marine Turtles
Reticulate evolution in the form of introgressive hybridization has been well defined for numerous marine turtle taxa. For example, Karl et al. [62] used an analysis of both mtDNA and nuclear loci to test for the infrequent formation of hybrids among loggerhead, Kemp’s ridley, hawksbill and green sea turtles (Caretta caretta, Lepidochelys kempii, Eretmochelys imbricata and Chelonia mydas, respectively). Likewise, Bass et al. [63] detected divergent mtDNA haplotypes in Brazilian samples of hawksbill turtles that were identical, or nearly identical, to those found in loggerhead samples. Each of these analyses thus suggested the likelihood of low-frequency introgression in a number of marine turtle clades. Furthermore, the findings of Bass et al. [63] indicated the possibility of a large effect from introgressive hybridization on some lineages; 10 of 14 Brazilian “hawksbill” animals possessed mtDNA haplotypes most similar to loggerhead turtles.
Recently, Lara-Ruiz et al. [64] analyzed hawksbill populations from the state of Bahia in Brazil. This region contains > 90% of the E. imbricata nesting sites in Brazil. The high frequency of introgression in the Brazilian hawksbill samples suggested a decade earlier by Bass et al. [63] was confirmed by the much larger sample of 119 individuals. Over half of the turtles sampled (i.e., 67) possessed the expected mtDNA sequences characteristic of E. imbricata. Yet, of the remaining 52 individuals, 50 reflected introgression of mtDNA from C. caretta (loggerheads), while two possessed mtDNA from L. olivacea (i.e., the olive ridley lineage; [64]). Thus, introgressive hybridization among marine turtle lineages is taxonomically diverse and, in some cases, extensive in terms of the proportion of the population impacted.
3.9. Introgressive Hybridization in Fur Seals
Human-mediated environmental changes have been demonstrated to be catalysts for bouts of genetic exchange among a diverse array of organisms (see [6,7] for reviews). In this regard, introgressive hybridization among species of the fur seal genus, Arctocephalus reflects the role of anthropogenic processes [65,66]. Analyses of various fur seal populations thus suggested that the observed introgression was at least partly caused by the extinction (or near-extinction) of populations of Arctocephalus gazella, A. tropicalis and A. forsteri (Antarctic, subantarctic and New Zealand fur seals, respectively) due to human harvesting. The introgression among the fur seals was hypothesized to have occurred due to the recolonization of islands by multiple species occupied formerly by a single taxon [65,66].
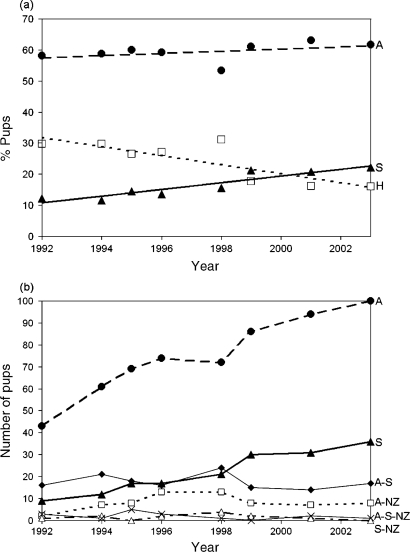
In their analysis of the genetic structure of Macquarie Island, Lancaster et al. [65] collected mtDNA and nuclear sequence variation for 1007 pups sampled over an eight year period (Figure 3). Though A. gazella genotypes predominated, hybrids among all three species were also detected, with the percentage of hybrid pups averaging ca. 23% and varying from 17–30% across years (Figure 3). Four hybrid categories were identifiable from the Macquarie Island samples. These included: (1) A. gazella x A. tropicalis; (2) A. gazella x A. forsteri, (3) A. tropicalis x A. forsteri and (4) A. gazella x A. tropicalis x A. forsteri (Figure 3) [65]. Though lower reproductive success was detected for some of the hybrid classes, the presence of “multiple mating strategies” have led to the establishment of this genetically diverse hybrid zone among the three fur seal taxa [67,68]. Likewise, Kingston and Gwilliam [66] also detected introgression on Iles Crozet, but only between subantarctic and Antarctic fur seals. Furthermore, their estimated frequencies of hybridization were lower than that of Lancaster et al. [65]; these authors estimated the frequency of F1 hybrids at 1% of the total population and 1.6% of the pups. They also concluded that at least 2.4% (and possibly as much as 4%) of the population consisted of backcross progeny [66].
Figure 3.
Change over time in a) the percentage of Antarctic (“A”), subantarctic (“S”) and hybrid (“H”) fur seal pups and b) the numbers of A, S and H pups, including values for the four hybrid classes (i.e., Antarctic x subantarctic, “A-S”; Antarctic x New Zealand, “A-NZ”; subantarctic x New Zealand, “S-NZ”; Antarctic x subantarctic x New Zealand, “A-S-NZ”; from [65]). Reproduced with permission from Wiley-Blackwell [65].
4. Conclusions and Future Directions
We believe that the above examples (and those given in previous reviews, see [17]) indicate that reticulate evolution (as reflected by introgressive hybridization, hybrid speciation and horizontal transfer events) is not limited to a few categories of marine organisms. Indeed, though reproductive isolation is a key factor in the speciation process–reducing genetic exchange in areas of geographic overlap between closely related organisms–divergence of marine lineages in the face of gene flow (i.e., sympatric or parapatric divergence) is indicated for a wide array of organisms (Table 1). Thus, genetic exchange and evolutionary diversification appear to reflect simultaneous processes during many radiations of marine clades. Furthermore, not only has genetic exchange occurred, but also this exchange has been associated with the origin of new lineages and, in some cases, the transfer of adaptations leading to the invasion of new habitats (e.g., Archaebacteria).
All of the above observations belie the conclusion that genetic exchange involving marine assemblages is relatively rare compared to that observed for terrestrial organisms. In addition, the origins of novel lineages and/or adaptations via hybridization and horizontal transfer indicate the potential evolutionary significance of exchange between divergent marine lineages. Yet, questions remain. Most importantly it remains to be seen whether there is a relationship between the clade to which an organism belongs and the potential for genetic exchange? The relevant data suggest that the genomic architecture, adaptive potential and thus ecological and evolutionary diversification of prokaryotic organisms in the marine realm, like their terrestrial counterparts, have been greatly affected by gene acquisition via horizontal gene transfer. Furthermore, some might argue that plant clades would also demonstrate a high proportion of reticulate events. But, what of animal lineages? As already mentioned, some authors (including MLA) have previously argued against a prominent role for introgressive hybridization among marine animals. Indeed, there are still few data sets to test for genetic exchange among such groups–though those that exist uniformly detect patterns that reject the strictly allopatric model of divergence and the application of the biological species concept as a robust descriptor of evolutionary pattern and process. Thus, as with most evolutionary hypotheses, those that predict the frequency, phylogenetic distribution and effect of genetic exchange on adaptive change and speciation in marine organisms necessitate additional, detailed studies of the genetic/genomic constitution of diverse lineages of prokaryotes and eukaryotes.
Acknowledgments
MLA was supported by NSF grant DEB-0345123. MLA also wishes to thank his hosts R. Sukumar and H.S. Suresh for the opportunity to visit the Indian Institute of Science (Bangalore, India), during which time much of this review was written. We would also like to thank M. Adreani, D. Ferrell, D. Levitan, C. Riginos and C. terHorst for helpful comments on this manuscript.
References
- 1.Anderson E. Introgressive Hybridization. John Wiley and Sons, Inc; New York, NY, USA: 1949. [Google Scholar]
- 2.Anderson E, Stebbins GL., Jr Hybridization as an evolutionary stimulus. Evolution. 1954;8:378–388. [Google Scholar]
- 3.Grant V. Plant Speciation. Columbia University Press; New York, NY, USA: 1981. [Google Scholar]
- 4.Dowling TE, DeMarais BD. Evolutionary significance of introgressive hybridization in cyprinid fishes. Nature. 1993;362:444–446. [Google Scholar]
- 5.Arnold ML. Natural Hybridization and Evolution. Oxford University Press; Oxford, UK: 1997. [Google Scholar]
- 6.Arnold ML. Evolution through Genetic Exchange. Oxford University Press; Oxford, UK: 2006. [Google Scholar]
- 7.Arnold ML. Reticulate Evolution and Humans: Origins and Ecology. Oxford University Press; Oxford, UK: 2008. [Google Scholar]
- 8.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 9.Ochman H, Lerat E, Daubin V. Examining bacterial species under the specter of gene transfer and exchange. Proc. Natl. Acad. Sci. USA. 2005;102:6595–6599. doi: 10.1073/pnas.0502035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold ML, Meyer A. Natural hybridization in primates: One evolutionary mechanism. Zoology. 2006;109:261–276. doi: 10.1016/j.zool.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. USA. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dambroski HR, Linn C, Jr, Berlocher SH, Forbes AA, Roelofs W, Feder JL. The genetic basis for fruit odor discrimination in Rhagoletis flies and its significance for sympatric host shifts. Evolution. 2005;59:1953–1964. [PubMed] [Google Scholar]
- 13.Vana G, Westover KM. Origin of the 1918 Spanish influenza virus: A comparative genomic analysis. Mol. Phylogenet. Evol. 2008;47:1100–1110. doi: 10.1016/j.ympev.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Hubbs CL. Hybridization between fish species in nature. Syst. Zool. 1955;4:1–20. [Google Scholar]
- 15.Roques S, Sévigny J-M, Bernatchez L. Evidence for broadscale introgressive hybridization between two redfish (genus Sebastes) in the North-west Atlantic: A rare marine example. Mol. Ecol. 2001;10:149–165. doi: 10.1046/j.1365-294x.2001.01195.x. [DOI] [PubMed] [Google Scholar]
- 16.Grant PR, Grant BR. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. [DOI] [PubMed] [Google Scholar]
- 17.Gardner JPA. Hybridization in the sea. Adv. Mar. Biol. 1997;31:1–78. [Google Scholar]
- 18.Veron JEN. Corals in Space and Time The Biogeography and Evolution of the Scleractinia. University of New South Wales Press; Sydney, Australia: 1995. [Google Scholar]
- 19.Willis BL, van Oppen MJH, Miller DJ, Vollmer SV, Ayre DJ. The role of hybridization in the evolution of reef corals. Ann. Rev. Ecol. Evol. Syst. 2006;37:489–517. [Google Scholar]
- 20.Ausich WI, Meyer DL. Hybrid crinoids in the fossil record (Early Mississippian, Phylum Echinodermata) Paleobiology. 1994;20:362–367. [Google Scholar]
- 21.Budd AF, Pandolfi JM. Overlapping species boundaries and hybridization within the Montastraea “annularis” reef coral complex in the Pleistocene of the Bahama Islands. Paleobiology. 2004;30:396–425. [Google Scholar]
- 22.Arnold ML, Larson EJ. Evolution’s new look. Wilson Quart. 2004 Autumn;:60–72. [Google Scholar]
- 23.Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl. Acad. Sci. USA. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DeLong EF. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frigaard N-U, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- 26.Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 27.Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 28.Shi T, Falkowski PG. Genome evolution in cyanobacteria: The stable core and the variable shell. Proc. Natl. Acad. Sci. USA. 2008;105:2510–2515. doi: 10.1073/pnas.0711165105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swingley WD, Chen M, Cheung PC, Conrad AL, Dejesa LC, Hao JC, Honchak BM, Karbach LE, Kurdoglu A, Lahiri S, Mastrian SD, Miyashita H, Page L, Ramakrishna P, Satoh S, Sattley WM, Shimada Y, Taylor HL, Tomo T, Tsuchiya T, Wang ZT, Raymond J, Mimuro M, Blankenship RE, Touchman JW. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc. Natl. Acad. Sci. USA. 2008;105:2005–2010. doi: 10.1073/pnas.0709772105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worden AZ, Lee J-H, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, Foulon E, Grimwood J, Gundlach H, Henrissat B, Napoli C, McDonald SM, Parker MS, Rombauts S, Salamov A, von Dassow P, Badger JH, Coutinho PM, Demir E, Dubchak I, Dubchak C, Eikrem W, Gready JE, John U, Lanier W, Lindquist EA, Lucas S, Mayer KFX, Moreau H, Not F, Otillar R, Panaud O, Pangilinan J, Paulsen I, Piegu B, Poliakov A, Robbens S, Schmutz J, Toulza E, Wyss T, Zelensky A, Zhou K, Armbrust EV, Bhattacharya D, Goodenough UW, van de Peer Y, Grigoriev IV. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 31.Kidwell MG. Lateral transfer in natural populations of eukaryotes. Ann. Rev. Genet. 1993;27:235–256. doi: 10.1146/annurev.ge.27.120193.001315. [DOI] [PubMed] [Google Scholar]
- 32.Casse N, Bui QT, Nicolas V, Renault S, Bigot Y, Laulier M. Species sympatry and horizontal transfers of Mariner transposons in marine crustacean genomes. Mol. Phylogenet. Evol. 2006;40:609–619. doi: 10.1016/j.ympev.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Reusch TBH. Microsatellites reveal high population connectivity in eelgrass (Zostera marina) in two contrasting coastal areas. Limnol. Oceanogr. 2002;47:78–85. [Google Scholar]
- 34.Coyer JA, Miller KA, Engle JM, Veldsink J, Cabello-Pasini A, Stam WT, Olsen JL. Eelgrass meadows in the California Channel Islands and adjacent coast reveal a mosaic of two species, evidence for introgression and variable clonality. Ann. Bot. 2008;101:73–87. doi: 10.1093/aob/mcm288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatta M, Fukami H, Wang W, Omori M, Shimoike K, Hayashibara T, Ina Y, Sugiyama T. Reproductive and genetic evidence for a reticulate evolutionary history of mass-spawning corals. Mol. Biol. Evol. 1999;16:1607–1613. doi: 10.1093/oxfordjournals.molbev.a026073. [DOI] [PubMed] [Google Scholar]
- 36.Knowlton N, Weil E, Wright LA, Guzman HM. Sibling species in Montastraea annularis, coral bleaching, and the coral climate record. Science. 1992;255:330–333. doi: 10.1126/science.255.5042.330. [DOI] [PubMed] [Google Scholar]
- 37.Levitan DR, Fukami H, Jara J, Kline D, McGovern TM, McGhee KE, Swanson CA, Knowlton N. Mechanisms of reproductive isolation among sympatric broadcast-spawning corals of the Montastraea annularis species complex. Evolution. 2004;58:308–323. [PubMed] [Google Scholar]
- 38.Fukami H, Budd AF, Levitan DR, Jara J, Kersanach R, Knowlton N. Geographic differences in species boundaries among members of the Montastraea annularis complex based on molecular and morphological markers. Evolution. 2004;58:324–337. [PubMed] [Google Scholar]
- 39.Huang D, Meier R, Todd PA, Chou LM. More evidence for pervasive paraphyly in scleractinian corals: Systematic study of Southeast Asian Faviidae (Cnidaria; Scleractinia) based on molecular and morphological data. Mol. Phylogenet. Evol. 2009;50:102–116. doi: 10.1016/j.ympev.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 40.McFadden CS, Hutchinson MB. Molecular evidence for the hybrid origin of species in the soft coral genus Alcyonium (Cnidaria: Anthozoa: Octocorallia) Mol. Ecol. 2004;13:1495–1505. doi: 10.1111/j.1365-294X.2004.02167.x. [DOI] [PubMed] [Google Scholar]
- 41.Combosch DJ, Guzman HM, Schuhmacher H, Vollmer SV. Interspecific hybridization and restricted trans-Pacific gene flow in the tropical eastern Pacific. Pocillopora. Mol. Ecol. 2008;17:1304–1312. doi: 10.1111/j.1365-294X.2007.03672.x. [DOI] [PubMed] [Google Scholar]
- 42.Glynn PW, Gassman NJ, Eakin CM, Cortes J, Smith DB, Guzman HM. Reef coral reproduction in the eastern Pacific: Costa Rica, Panama, and Galapagos Islands (Ecuador) I. Pocilloporiidae. Mar. Biol. 1991;109:355–368. [Google Scholar]
- 43.Bates SS. Domoic-Acid-producing diatoms: Another genus added! J. Phycol. 2000;36:978–985. [Google Scholar]
- 44.Hasle GR. Are most domoic acid-producing species of the diatom genus Pseudo-nitzschia cosmopolites? Harmful Algae. 2002;1:137–146. [Google Scholar]
- 45.D’Alelio D, Amato A, Kooistra WHCF, Procaccini G, Casotti R, Montresor M. Internal transcribed spacer polymorphism in Pseudo-nitzschia multistriata (Bacillariophyceae) in the Gulf of Naples: Recent divergence or intraspecific hybridization? Protist. 2009;160:9–20. doi: 10.1016/j.protis.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Casteleyn G, Adams NG, Vanormelingen P, Debeer A-E, Sabbe K, Vyverman W. Natural hybrids in the marine diatom Pseudo-nitzschia pungens (Bacillariophyceae): Genetic and morphological evidence. Protist. 2009;160:343–354. doi: 10.1016/j.protis.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Tsang LM, Chan BKK, Ma KY, Chu KH. Genetic differentiation, hybridization and adaptive divergence in two subspecies of the acorn barnacle Tetraclita japonica in the northwestern Pacific. Mol. Ecol. 2008;17:4151–4163. doi: 10.1111/j.1365-294x.2008.03907.x. [DOI] [PubMed] [Google Scholar]
- 48.Audzijonyte A, Damgaard J, Varvio S-L, Vainio JK, Väinölä R. Phylogeny of Mysis (Crustacea, Mysida): History of continental invasions inferred from molecular and morphological data. Cladistics. 2005;21:575–596. doi: 10.1111/j.1096-0031.2005.00081.x. [DOI] [PubMed] [Google Scholar]
- 49.Vainio JK, Väinölä R. Refugial races and postglacial colonization history of the freshwater amphipod Gammarus lacustris in northern Europe. Biol. J. Linn. Soc. 2003;79:523–542. [Google Scholar]
- 50.Audzijonyte A, Väinölä R. Mysis nordenskioldi n. sp. (Crustacaea, Mysida), a circumpolar coastal mysid separated from the NE Pacific M. litoralis (Banner, 1948) Polar Biol. 2007;30:1137–1157. [Google Scholar]
- 51.O’Mullan GD, Maas PAY, Lutz RA, Vrijenhoek RC. A hybrid zone between hydrothermal vent mussels (Bivalvia: Mytilidae) from the Mid-Atlantic Ridge. Mol. Ecol. 2001;10:2819–2831. doi: 10.1046/j.0962-1083.2001.01401.x. [DOI] [PubMed] [Google Scholar]
- 52.Won Y, Hallam SJ, O’Mullan GD, Vrijenhoek RC. Cytonuclear disequilibrium in a hybrid zone involving deep-sea hydrothermal vent mussels of the genus Bathymodiolus. Mol. Ecol. 2003;12:3185–3190. doi: 10.1046/j.1365-294x.2003.01974.x. [DOI] [PubMed] [Google Scholar]
- 53.Levitan DR. The relationship between conspecific fertilization success and reproductive isolation among three congeneric sea urchins. Evolution. 2002;56:1599–1609. doi: 10.1111/j.0014-3820.2002.tb01472.x. [DOI] [PubMed] [Google Scholar]
- 54.Harper FM, Addison JA, Hart MW. Introgression versus immigration in hybridizing high-dispersal echinoderms. Evolution. 2007;61:2410–2418. doi: 10.1111/j.1558-5646.2007.00200.x. [DOI] [PubMed] [Google Scholar]
- 55.Addison JA, Hart MW. Colonization, dispersal, and hybridization influence phylogeography of north Atlantic sea urchins (Strongylocentrotus droebachiensis) Evolution. 2005;59:532–543. [PubMed] [Google Scholar]
- 56.Lessios HA, Pearse JS. Hybridization and introgression between Indo-Pacific species of Diadema. Mar. Biol. 1996;126:715–723. [Google Scholar]
- 57.Zigler KS, Lessios HA. Speciation on the coasts of the New World: Phylogeography and the evolution of bindin in the sea urchin genus Lytechinus. Evolution. 2004;58:1225–1241. doi: 10.1111/j.0014-3820.2004.tb01702.x. [DOI] [PubMed] [Google Scholar]
- 58.Harper FM, Hart MW. Morphological and phylogenetic evidence for hybridization and introgression in a sea star secondary contact zone. Invert. Biol. 2007;126:373–384. [Google Scholar]
- 59.Scheibling RE, Lauzon-Guay J-S. Feeding aggregations of sea stars (Asterias spp. and Henricia sanguinolenta) associated with sea urchin (Strongylocentrotus droebachiensis) grazing fronts in Nova Scotia. Mar. Biol. 2007;151:1175–1183. [Google Scholar]
- 60.van Herwerden L, Choat JH, Dudgeon CL, Carlos G, Newman SJ, Frisch A, van Oppen M. Contrasting patterns of genetic structure in two species of the coral trout Plectropomus (Serranidae) from east and west Australia: Introgressive hybridisation or ancestral polymorphisms. Mol. Phylogenet. Evol. 2006;41:420–435. doi: 10.1016/j.ympev.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 61.Marie AD, van Herwerden L, Choat JH, Hobbs J-PA. Hybridization of reef fishes at the Indo-Pacific biogeographic barrier: A case study. Coral Reefs. 2007;26:841–850. [Google Scholar]
- 62.Karl SA, Bowen BW, Avise JC. Hybridization among the ancient mariners: Characterization of marine turtle hybrids with molecular genetic assays. J. Hered. 1995;86:262–268. doi: 10.1093/oxfordjournals.jhered.a111579. [DOI] [PubMed] [Google Scholar]
- 63.Bass AL, Good DA, Bjorndal KA, Richardson JI, Hillis Z-M, Horrocks JA, Bowen BW. Testing models of female reproductive migratory behaviour and population structure in the Caribbean hawksbill turtle, Eretmochelys imbricata, with mtDNA sequences. Mol. Ecol. 1996;5:321–328. [PubMed] [Google Scholar]
- 64.Lara-Ruiz P, Lopez GG, Santos FR, Soares LS. Extensive hybridization in hawksbill turtles (Eretmochelys imbricata) nesting in Brazil revealed by mtDNA analyses. Conserv. Genet. 2006;7:773–781. [Google Scholar]
- 65.Lancaster ML, Gemmel NJ, Negro S, Goldsworthy S, Sunnucks P. Ménage à trois on Macquarie Island: Hybridization among three species of fur seal (Arctocephalus spp.) following historical population extinction. Mol. Ecol. 2006;15:3681–3692. doi: 10.1111/j.1365-294X.2006.03041.x. [DOI] [PubMed] [Google Scholar]
- 66.Kingston JJ, Gwilliam J. Hybridization between two sympatrically breeding species of fur seal at Iles Crozet revealed by genetic analysis. Conserv. Genet. 2007;8:1133–1145. [Google Scholar]
- 67.Lancaster ML, Bradshaw CJA, Goldsworthy S, Sunnucks P. Lower reproductive success in hybrid fur seal males indicates fitness costs to hybridization. Mol. Ecol. 2007;16:3187–3197. doi: 10.1111/j.1365-294X.2007.03339.x. [DOI] [PubMed] [Google Scholar]
- 68.Lancaster ML, Goldsworthy S, Sunnucks P. Multiple mating strategies explain unexpected genetic mixing of New Zealand fur seals with two congenerics in a recently recolonized population. Mol. Ecol. 2007;16:5267–5276. doi: 10.1111/j.1365-294X.2007.03586.x. [DOI] [PubMed] [Google Scholar]
- 69.Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, Duprat S, Galperin MY, Koonin EV, Le Gall F, Makarova KS, Ostrowski M, Oztas S, Robert C, Rogozin IB, Scanlan DJ, de Marsac NT, Weissenbach J, Wincker P, Wolf YI, Hess WR. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl. Acad. Sci. USA. 2003;100:10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palenik B, Brahamsha B, Larimer FW, Land M, Hauser L, Chain P, Lamerdin J, Regala W, Allen EE, McCarren J, Paulsen I, Dufresne A, Partensky F, Webb EA, Waterbury J. The genome of a motile marine Synechococcus. Nature. 2003;424:1037–1042. doi: 10.1038/nature01943. [DOI] [PubMed] [Google Scholar]
- 71.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, Johnson ZI, Land M, Lindell D, Post AF, Regala W, Shah M, Shaw SL, Steglich C, Sullivan MB, Ting CS, Tolonen A, Webb EA, Zinser ER, Chisholm SW. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 72.Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Millard A, Clokie MRJ, Shub DA, Mann NH. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc. Natl. Acad. Sci. USA. 2004;101:11007–11012. doi: 10.1073/pnas.0401478101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Filée J, Tétart F, Suttle CA, Krisch HM. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc. Natl. Acad. Sci. USA. 2005;102:12471–12476. doi: 10.1073/pnas.0503404102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Daehler CC, Strong DR. Hybridization between introduced smooth cordgrass (Spartina alterniflora; Poaceae) and native California cordgrass (S. foliosa) in San Francisco Bay, California, USA. Amer. J. Bot. 1997;84:607–611. [PubMed] [Google Scholar]
- 76.Anttila CK, King RA, Ferris C, Ayres DR, Strong DR. Reciprocal hybrid formation of Spartina in San Francisco Bay. Mol. Ecol. 2000;9:765–770. doi: 10.1046/j.1365-294x.2000.00935.x. [DOI] [PubMed] [Google Scholar]
- 77.Baumel A, Ainouche ML, Bayer RJ, Ainouche AK, Misset MT. Molecular phylogeny of hybridizing species from the genus Spartina Schreb. (Poaceae) Mol. Phylogenet. Evol. 2002;22:303–314. doi: 10.1006/mpev.2001.1064. [DOI] [PubMed] [Google Scholar]
- 78.Baumel A, Ainouche ML, Misset MT, Gourret J-P, Bayer RJ. Genetic evidence for hybridization between the native Spartina maritima and the introduced Spartina alterniflora (Poaceae) in south-west France: Spartina x neyrautii re-examined. Plant Syst. Evol. 2003;237:87–97. [Google Scholar]
- 79.Ayres DR, Grotkopp E, Zaremba K, Sloop CM, Blum MJ, Bailey JP, Anttila CK, Strong DR. Hybridization between invasive Spartina densiflora (Poaceae) and native S. foliosa in San Francisco Bay, California, USA. Amer. J. Bot. 2008;95:713–719. doi: 10.3732/ajb.2007358. [DOI] [PubMed] [Google Scholar]
- 80.Coyer JA, Peters AF, Hoarau G, Stam WT, Olsen JL. Hybridization of the marine seaweeds, Fucus serratus and Fucus evanescens (Heterokontophyta: Phaeophyceae) in a 100-year-old zone of secondary contact. Proc. R. Soc. Lond. B. 2002;269:1829–1834. doi: 10.1098/rspb.2002.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wallace AL, Klein AS, Mathieson AC. Determining the affinities of salt marsh fucoids using microsatellite markers: Evidence of hybridization and introgression between two species of Fucus (Phaeophyta) in a marine estuary. J. Phycol. 2004;40:1013–1027. [Google Scholar]
- 82.Coyer JA, Hoarau G, Oudot-Le Secq M-P, Stam WT, Olsen JL. A mtDNA-based phylogeny of the brown algal genus Fucus (Heterokontophyta: Phaeophyta) Mol. Phylogenet. Evol. 2006;39:209–222. doi: 10.1016/j.ympev.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 83.Coyer JA, Hoarau G, Pearson GA, Serrão EA, Stam WT, Olsen JL. Convergent adaptation to a marginal habitat by homoploid hybrids and polyploidy ecads in the seaweed genus Fucus. Biol. Let. 2006;2:405–408. doi: 10.1098/rsbl.2006.0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coyer JA, Hoarau G, Stam WT, Olsen JL. Hybridization and introgression in a mixed population of the intertidal seaweeds Fucus evanescens and F. serratus. J. Evol. Biol. 2007;20:2322–2333. doi: 10.1111/j.1420-9101.2007.01411.x. [DOI] [PubMed] [Google Scholar]
- 85.Bert TM. Speciation in western Atlantic stone crabs (genus Menippe): The role of geological processes and climatic events in the formation and distribution of species. Mar. Biol. 1986;93:157–170. [Google Scholar]
- 86.Bert TM, Harrison RG. Hybridization in western Atlantic stone crabs (genus Menippe): Evolutionary history and ecological context influence species interactions. Evolution. 1988;42:528–544. doi: 10.1111/j.1558-5646.1988.tb04158.x. [DOI] [PubMed] [Google Scholar]
- 87.Bert TM, McCarthy KJ, Cruz-Lopez H, Bogdanowicz S. Character discriminatory power, character-set congruence, and the classification of individuals from hybrid zones: An example using stone crabs (Menippe) Evolution. 1996;50:655–671. doi: 10.1111/j.1558-5646.1996.tb03876.x. [DOI] [PubMed] [Google Scholar]
- 88.Muths D, Davoult D, Gentil F, Jollivet D. Incomplete cryptic speciation between intertidal and subtidal morphs of Acrocnida brachiata (Echinodermata: Ophiuroidea) in the northeast Atlantic. Mol. Ecol. 2006;15:3303–3318. doi: 10.1111/j.1365-294X.2006.03000.x. [DOI] [PubMed] [Google Scholar]
- 89.Wallace CC, Willis BL. Systematics of the coral genus Acropora: Implications of new biological findings for species concepts. Ann. Rev. Ecol. Syst. 1994;25:237–262. [Google Scholar]
- 90.van Oppen MJH, McDonald BJ, Willis B, Miller DJ. The evolutionary history of the coral genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: Reticulation, incomplete lineage sorting, or morphological convergence? Mol. Biol. Evol. 2001;18:1315–1329. doi: 10.1093/oxfordjournals.molbev.a003916. [DOI] [PubMed] [Google Scholar]
- 91.Márquez LM, van Oppen MJH, Willis BL, Reyes A, Miller DJ. The highly cross-fertile coral species, Acropora hyacinthus and Acropora cytherea, constitute statistically distinguishable lineages. Mol. Ecol. 2002;11:1339–1349. doi: 10.1046/j.1365-294x.2002.01526.x. [DOI] [PubMed] [Google Scholar]
- 92.van Oppen MJH, Willis BL, van Rheede T, Miller DJ. Spawning times, reproductive compatibilities and genetic structuring in the Acropora aspera group: Evidence for natural hybridization and semi-permeable species boundaries in corals. Mol. Ecol. 2002;11:1363–1376. doi: 10.1046/j.1365-294x.2002.01527.x. [DOI] [PubMed] [Google Scholar]
- 93.Vollmer SV, Palumbi SR. Hybridization and the evolution of reef coral diversity. Science. 2002;296:2023–2025. doi: 10.1126/science.1069524. [DOI] [PubMed] [Google Scholar]
- 94.Vollmer SV, Palumbi SR. Restricted gene flow in the Caribbean staghorn coral Acropora cervicornis: Implications for the recovery of endangered reefs. J. Hered. 2007;98:40–50. doi: 10.1093/jhered/esl057. [DOI] [PubMed] [Google Scholar]
- 95.Rolán-Alvarez E, Johannesson K, Erlandsson J. The maintenance of a cline in the marine snail Littorina saxtilis: The role of home site advantage and hybrid fitness. Evolution. 1997;51:1838–1847. doi: 10.1111/j.1558-5646.1997.tb05107.x. [DOI] [PubMed] [Google Scholar]
- 96.Cruz R, Vilas C, Mosquera J, García C. The close relationship between estimated divergent selection and observed differentiation supports the selective origin of a marine snail hybrid zone. J. Evol. Biol. 2004;17:1221–1229. doi: 10.1111/j.1420-9101.2004.00802.x. [DOI] [PubMed] [Google Scholar]
- 97.Bert TM, Hesselman DM, Arnold WS, Moore WS, Cruz-Lopez H, Marelli DC. High frequency of gonadal neoplasia in a hard clam (Mercenaria spp.) hybrid zone. Mar. Biol. 1993;117:97–104. [Google Scholar]
- 98.Bert TM, Arnold WS. An empirical test of predictions of two competing models for the maintenance and fate of hybrid zones: Both models are supported in a hard-clam hybrid zone. Evolution. 1995;49:276–289. doi: 10.1111/j.1558-5646.1995.tb02240.x. [DOI] [PubMed] [Google Scholar]
- 99.Strelkov P, Nikula R, Väinölä R. Macoma balthica in the White and Barents Seas: Properties of a widespread marine hybrid swarm (Mollusca: Bivalvia) Mol. Ecol. 2007;16:4110–4127. doi: 10.1111/j.1365-294X.2007.03463.x. [DOI] [PubMed] [Google Scholar]
- 100.Riginos C, Cunningham CW. Hybridization in postglacial marine habitats. Mol. Ecol. 2007;16:3971–3972. doi: 10.1111/j.1365-294X.2007.03505.x. [DOI] [PubMed] [Google Scholar]
- 101.Hare MP, Avise JC. Molecular genetic analysis of a stepped multilocus cline in the American oyster (Crassostrea virginica) Evolution. 1996;50:2305–2315. doi: 10.1111/j.1558-5646.1996.tb03618.x. [DOI] [PubMed] [Google Scholar]
- 102.Murray MC, Hare MP. A genomic scan for divergent selection in a secondary contact zone between Atlantic and Gulf of Mexico oysters, Crassostrea virginica. Mol. Ecol. 2006;15:4229–4242. doi: 10.1111/j.1365-294X.2006.03060.x. [DOI] [PubMed] [Google Scholar]
- 103.Skibinski DOF, Ahmad M, Beardmore JA. Genetic evidence for naturally occurring hybrids between Mytilus edulis and Mytilus galloprovincialis. Evolution. 1978;32:354–364. doi: 10.1111/j.1558-5646.1978.tb00651.x. [DOI] [PubMed] [Google Scholar]
- 104.Rawson PD, Hilbish TJ. Asymmetric introgression of mitochondrial DNA among European populations of blue mussels (Mytilus spp.) Evolution. 1998;52:100–108. doi: 10.1111/j.1558-5646.1998.tb05142.x. [DOI] [PubMed] [Google Scholar]
- 105.Daguin C, Bonhomme F, Borsa P. The zone of sympatry and hybridization of Mytilus edulis and M. galloprovincialis, as described by intron length polymorphism at locus mac-1. Heredity. 2001;86:342–354. doi: 10.1046/j.1365-2540.2001.00832.x. [DOI] [PubMed] [Google Scholar]
- 106.Riginos C, Sukhdeo K, Cunningham CW. Evidence for selection at multiple allozyme loci across a mussel hybrid zone. Mol. Biol. Evol. 2002;19:347–351. doi: 10.1093/oxfordjournals.molbev.a004088. [DOI] [PubMed] [Google Scholar]
- 107.Bierne N, Borsa P, Daguin C, Jollivet D, Viard F, Bonhomme F, David P. Introgression patterns in the mosaic hybrid zone between Mytilus edulis and M. galloprovincialis. Mol. Ecol. 2003;12:447–461. doi: 10.1046/j.1365-294x.2003.01730.x. [DOI] [PubMed] [Google Scholar]
- 108.Gilg MR, Hilbish TJ. Patterns of larval dispersal and their effect on the maintenance of a blue mussel hybrid zone in southwestern England. Evolution. 2003;57:1061–1077. doi: 10.1111/j.0014-3820.2003.tb00316.x. [DOI] [PubMed] [Google Scholar]
- 109.Riginos C, Hickerson MJ, Henzler CM, Cunningham CW. Differential patterns of male and female mtDNA exchange across the Atlantic Ocean in the blue mussel, Mytilus edulis. Evolution. 2004;58:2438–2451. doi: 10.1111/j.0014-3820.2004.tb00873.x. [DOI] [PubMed] [Google Scholar]
- 110.Toro J, Innes DJ, Thompson RJ. Genetic variation among life-history stages of mussels in a Mytilus edulis—M. trossulus hybrid zone. Mar. Biol. 2004;145:713–725. [Google Scholar]
- 111.Riginos C, Cunningham CW. Local adaptation and species segregation in two mussel (Mytilus edulis x Mytilus trossulus) hybrid zones. Mol. Ecol. 2005;14:381–400. doi: 10.1111/j.1365-294X.2004.02379.x. [DOI] [PubMed] [Google Scholar]
- 112.Borsa P, Daguin C, Bierne N. Genomic reticulation indicates mixed ancestry in southern-hemisphere Mytilus spp. mussels. Biol. J. Linn. Soc. 2007;92:747–754. [Google Scholar]
- 113.Coghlan B, Gosling E. Genetic structure of hybrid mussel populations in the west of Ireland: Two hypotheses revisited. Mar. Biol. 2007;150:841–852. [Google Scholar]
- 114.Shields JL, Barnes P, Heath DD. Growth and survival differences among native, introduced and hybrid blue mussels (Mytilus spp.): Genotype, environments and interaction effects. Mar. Biol. 2008;154:919–928. [Google Scholar]
- 115.Nielsen EE, Nielsen PH, Meldrup D, Hansen MM. Genetic population structure of turbot (Scophthalmus maximus L.) supports the presence of multiple hybrid zones for marine fishes in the transition zone between the Baltic Sea and the North Sea. Mol. Ecol. 2004;13:585–595. doi: 10.1046/j.1365-294x.2004.02097.x. [DOI] [PubMed] [Google Scholar]
- 116.Bekkevold D, Andre C, Dahlgren TG, Clausen LAW, Torstensen E, Mosegaard H, Carvalho GR, Christensen TB, Norlinder E, Ruzzante DE. Environmental correlates of population differentiation in Atlantic herring. Evolution. 2005;59:2656–2668. [PubMed] [Google Scholar]
- 117.Nielsen EE, Hansen MM, Ruzzante DE, Meldrup D, Grønkjær P. Evidence of a hybrid-zone in Atlantic cod (Gadus morhua) in the Baltic and the Danish Belt Sea revealed by individual admixture analysis. Mol. Ecol. 2003;12:1497–1508. doi: 10.1046/j.1365-294x.2003.01819.x. [DOI] [PubMed] [Google Scholar]
- 118.Nielsen EE, Grønkjær P, Meldrup D, Paulsen H. Retention of juveniles within a hybrid zone between North Sea and Baltic Sea Atlantic cod (Gadus morhua) Can. J. Fish. Aquat. Sci. 2005;62:2219–2225. [Google Scholar]
- 119.Hemmer-Hansen J, Nielsen EEG, Grønkjær P, Loeschcke V. Evolutionary mechanisms shaping the genetic population structure of marine fishes; lessons from the European flounder (Platichthys flesus L.) Mol. Ecol. 2007;16:3104–3118. doi: 10.1111/j.1365-294X.2007.03367.x. [DOI] [PubMed] [Google Scholar]
- 120.Chow S, Kishino H. Phylogenetic relationships between tuna species of the genus Thunnus (Scombridae: Teleostei): Inconsistent implications from morphology, nuclear and mitochondrial genomes. J. Mol. Evol. 1995;41:741–748. doi: 10.1007/BF00173154. [DOI] [PubMed] [Google Scholar]
- 121.Alvarado Bremer JR, Naseri I, Ely B. Orthodox and unorthodox phylogenetic relationships among tunas revealed by the nucleotide sequence analysis of the mitochondrial DNA control region. J. Fish Biol. 1997;50:540–554. [Google Scholar]
- 122.Alvarado Bremer JR, Viñas J, Mejuto J, Ely B, Pla C. Comparative phylogeography of Atlantic bluefin tuna and swordfish: The combined effects of vicariance, secondary contact, introgression, and population expansion on the regional phylogenies of two highly migratory pelagic fishes. Mol. Phylogenet. Evol. 2005;36:169–187. doi: 10.1016/j.ympev.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 123.Avise JC, Nelson WS, Arnold J, Koehn RK, Williams GC, Thorsteinsson V. The evolutionary genetic status of Icelandic eels. Evolution. 1990;44:1254–1262. doi: 10.1111/j.1558-5646.1990.tb05229.x. [DOI] [PubMed] [Google Scholar]
- 124.Albert V, Jónsson B, Bernatchez L. Natural hybrids in Atlantic eels (Anguilla anguilla, A. rostrata): Evidence for successful reproduction and fluctuating abundance in space and time. Mol. Ecol. 2006;15:1903–1916. doi: 10.1111/j.1365-294X.2006.02917.x. [DOI] [PubMed] [Google Scholar]
- 125.Burford MO, Bernardi G. Incipient speciation within a subgenus of rockfish (Sebastosomus) provides evidence of recent radiations within an ancient species flock. Mar. Biol. 2008;154:701–717. [Google Scholar]
- 126.Planes S, Doherty PJ. Genetic and color interactions at a contact zone of Acanthochromis polyacanthus: A marine fish lacking pelagic larvae. Evolution. 1997;51:1232–1243. doi: 10.1111/j.1558-5646.1997.tb03970.x. [DOI] [PubMed] [Google Scholar]
- 127.van Herwerden L, Doherty PJ. Contrasting genetic structure across two hybrid zones of a tropical reef fish, Acanthochromis polyacanthus (Bleeker 1855) J. Evol. Biol. 2006;19:239–252. doi: 10.1111/j.1420-9101.2005.00969.x. [DOI] [PubMed] [Google Scholar]
- 128.McMillan WO, Weigt LA, Palumbi SR. Color pattern evolution, assortative mating, and genetic differentiation in brightly colored butterflyfishes (Chaetodontidae) Evolution. 1999;53:247–260. doi: 10.1111/j.1558-5646.1999.tb05350.x. [DOI] [PubMed] [Google Scholar]
- 129.Crespi BJ, Fulton MJ. Molecular systematics of Salmonidae: Combined nuclear data yields a robust phylogeny. Mol. Phylogenet. Evol. 2004;31:658–679. doi: 10.1016/j.ympev.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 130.Campos JL, Posada D, Morán P. Introgression and genetic structure in northern Spanish Atlantic salmon (Salmo salar L.) populations according to mtDNA data. Conserv. Genet. 2008;9:157–169. [Google Scholar]