Abstract
The NMDA receptor (NMDAR) subunit GluN1 is an obligatory component of NMDARs without a known functional homolog and is expressed in almost every neuronal cell type. The NMDAR system is a coincidence detector with critical roles in spatial learning and synaptic plasticity. Its coincidence detection property is crucial for the induction of hippocampal long-term potentiation (LTP). We have generated a mutant mouse model expressing a hypomorph of the Grin1N598R allele, which leads to a minority (about 10%) of coincidence detection-impaired NMDARs. Surprisingly, these animals revealed specific functional changes in the dentate gyrus (DG) of the hippocampal formation. Early LTP was expressed normally in area CA1 in vivo, but was completely suppressed at perforant path-granule cell synapses in the DG. In addition, there was a pronounced reduction in the amplitude of the evoked population spike in the DG. These specific changes were accompanied by behavioral impairments in spatial recognition, spatial learning, reversal learning, and retention. Our data show that minor changes in GluN1-dependent NMDAR physiology can cause dramatic consequences in synaptic signaling in a subregion-specific fashion despite the nonredundant nature of the GluN1 gene and its global expression.
According to Hebb's postulate, neurons require a molecular mechanism to detect synchronous activity in order to change the strength of synaptic connectivity (Hebb 1949). NMDA receptors (NMDARs) are molecular coincidence detectors, and selective NMDAR antagonists block the induction of long-term potentiation (LTP) in both the dentate gyrus (DG) and CA1 regions of the hippocampus (Bliss and Collingridge 1993; Martin et al. 2000). NMDARs have been long known for their role in spatial learning, but more recently have been implicated in other forms of cognitive function and dysfunction (Gruart et al. 2006; Whitlock et al. 2006; Castner and Williams 2007; Kristiansen et al. 2007; Wilson and Linster 2008).
Neuronal NMDARs are hetero-tetrameric ligand-gated ion channels typically comprised of two types of subunits. Two copies of the mandatory GluN1 subunit (or NR1 subunit [Collingridge et al. 2009] encoded by Grin1) are associated with two copies from the GluN2 family, GluN2A–D (or NR2A–D). The GluN1 subunit is expressed ubiquitously both spatially and temporally throughout the developing and adult brain. Global knockout mice models of the GluN1 subunit are postnatally lethal within hours after birth (Forrest et al. 1994; Li et al. 1994), and cell-specific GluN1 mice knockouts (Tsien et al. 1996; Nakazawa et al. 2002; McHugh et al. 2007; Niewoehner et al. 2007) have provided insights on how specific synapses and regional neuronal networks are dependent on NMDAR function.
The early postnatal lethality of the global GluN1 knockout is in contrast to the null mutants of the four AMPA receptor genes and other major synaptic proteins, such as αCaMKII (Silva et al. 1992a,b; Jia et al. 1996; Zamanillo et al. 1999; Meng et al. 2003). This can be at least partially explained by the absence of any close GluN1 homologs, which could functionally compensate for the absence of the GluN1 subunit. Recombinant expression studies defined the GluN1 subunit as a mandatory component of NMDARs. This constellation provides a specific opportunity to test whether different local neuronal subnetworks are affected differentially by mutant Grin1 alleles associated with subtle alterations of the functional properties of NMDARs.
GluN1 subunits with the N598R point mutation (GluN1R) yield functional NMDARs that are Mg2+ insensitive and Ca2+ impermeable (Burnashev et al. 1992; Mori et al. 1992). The Grin1N598R allele that codes for GluN1R subunits is a gain-of-function mutation that is dominant lethal, even in heterozygous and hemizygous lines (Single et al. 2000; Rudhard et al. 2003). NMDARs with GluN1R subunits do not act as coincidence detectors and, interestingly, mice expressing exclusively the GluN1R allele lack whisker-related pattern formation in the neonate brainstem (Rudhard et al. 2003).
To investigate the functional importance of GluN1 subunits with the N598R point mutation, we took advantage of the generation of a variant mutant line of mice (GluN1Rneo/+) expressing a minority (around 10%) of these mutant NMDARs. Even though the majority of the NMDARs are normal, all neurons expressing NMDARs will contain a subset of receptors carrying this mutation.
Therefore, this mouse model is an ideal candidate to study the impact of subtle alterations of NMDAR function on different neuronal networks, such as those comprising the hippocampal formation.
Studies examining region-specific targeted disruption of GluN1 expression in subregions of the hippocampus have revealed subtle yet important contributions of this NMDAR subunit in synaptic plasticity and spatial learning and memory. CA1-restricted knockout of GluN1 expression in the hippocampus caused impaired spatial learning and memory as well as reduced CA1-LTP (Tsien et al. 1996). In the case of the disruption of GluN1 expression in the DG region of the hippocampus, more subtle behavioral impairments were apparent, including the inability to discriminate between two similar contexts (pattern separation) and deficits in spatial working memory despite normal LTP in the CA1 region (McHugh et al. 2007; Niewoehner et al. 2007).
Our GluN1Rneo/+ mice differ from the region-specific GluN1 mutant mice in that they express the mutant hypomorph at the same level in different subregions of the hippocampus. Interestingly, we found that this allele leads to substantial differences in short- and long-term plasticity between area CA1 and the DG of the hippocampus. The specific impairment in the DG was accompanied by impaired spatial recognition, spatial learning, reversal learning, and retention. Our data establish the possibility of a circuit-specific phenotype caused by a mutant variant of a globally expressed major nonredundant synaptic protein.
Results
Hypomorphic GluN1R expression in GluN1Rneo/+ mice
Initial expression analysis indicated that the GluN1Rneo allele is a null allele, due to the presence of the neo cassette in intron 18 (Fig. 1A). GluN1Rneo/Rneo mice did not survive beyond P0, as expected for animals that lack GluN1 subunits (Forrest et al. 1994; Li et al. 1994). In contrast, GluN1Rneo/+ mice reached adulthood, were fertile, and did not exhibit any obvious differences in mean body weight at 6 mo of age compared with their wild-type counterparts (GluN1+/+: 30.7 ± 0.5 g, n = 50; GluN1Rneo/+: 29.7 ± 0.4 g, n = 38; means ± SEM, t-test of mean adult body weight between both groups P > 0.2).
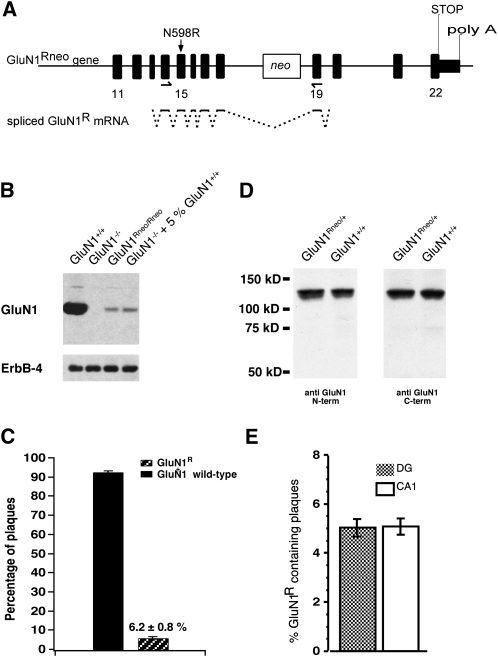
Figure 1.
GluN1Rneo/+ mice express NMDAR GluN1R subunits at low levels. (A) Genomic organization of the GluN1Rneo allele with neo cassette in intron 18. Arrows indicate PCR primers for mRNA quantification plaque assay. Splicing that leads to the GluN1R hypomorph is indicated for the PCR fragment. (B) Western blot from P0 brain membranes, probed with antibody against GluN1 C terminus. The weak GluN1Rneo/Rneo signal is comparable to that obtained from GluN1−/− preparations spiked with 5% GluN1+/+ material (sample-loading control: ErbB-4). (C) Relative GluN1R mRNA abundance in forebrain of adult GluN1Rneo/+ mice (n = 5), determined by a plaque assay. (D) Western blot on adult brain homogenate. Antibodies against GluN1 N or C terminus revealed full-length GluN1 subunits and no truncated species in GluN1Rneo/+. (E) Relative GluN1R mRNA abundance of GluN1R transcripts from individual CA1 pyramidal neurons or DG granule cells (n = 7 cells, each region), determined by plaque assay.
However, close inspection of GluN1 protein expression levels of GluN1Rneo/Rneo mice indicated a low level of full-length protein originating from the GluN1Rneo allele. The protein level was ∼5% of that produced from the wild-type alleles (Fig. 1B). To quantify the expression of GluN1R full-length mRNA in adult GluN1Rneo/+ mice, we performed RT-PCR on total forebrain poly(A)+-RNA. The generated cDNA fragments were subcloned and the number of wild-type and GluN1R clones were identified using differential oligonucleotide hybridization. Our analysis suggested that 6.2 ± 0.8%, mean ± SEM (n = 5) of the total GluN1 population contained correctly spliced full-length GluN1R mRNA (Fig. 1C). Sequencing of GluN1R clones confirmed the presence of the N598R mutation and revealed no aberrant splicing. Initial immunoblot analysis of protein samples from adult GluN1Rneo/+ brains revealed no truncated GluN1 subunits using either N- or C-terminal-specific GluN1 antibodies, and this was still apparent when blot sensitivity was increased by overexposure (Fig. 1D).
We further investigated whether the hypomorphic allele is expressed at equal relative levels in different cell populations. Quantitative single cell RT-PCR from two hippocampal regions showed no difference in the relative abundance of GluN1R mRNA (CAl pyramidal cells: 5.1 ± 0.4%; DG granule cells: 5.0 ± 0.4%; mean ± SEM, n = 7 each region, two-tailed t-test of means, P > 0.9; Fig. 1E). We note that the GluN1R mRNA level found in these two cell populations is not significantly different from the average level in forebrain (two-tailed t-test of means, P > 0.2 for both cell populations compared with whole forebrain; Fig. 1C).
In summary, we conclude that the insertion of the neo cassette within intron 18 drastically reduced GluN1R expression, producing a hypomorphic GluN1R allele. Our GluN1Rneo/+ mice express two species of full-length GluN1 mRNA, GluN1 wild-type, and GluN1R in the ratio of about 95:5. Assuming two copies of the GluN1 subunit in mature NMDARs (Behe et al. 1995), a binomial analysis indicates that GluN1Rneo/+ mice should express about 90% of pure wild-type NMDARs, a very small amount of pure mutant (<0.4%; 6% mutant amounts to 0.36%), and about 10% of mixed receptors that carry one copy of wild-type GluN1+ together with one copy of mutant GluN1R subunit.
Thus, only a minority of NMDARs are affected in these animals. The minor alteration of the NMDAR system does not lead to any obvious change in gross neuroanatomy of adult animals (Supplemental Fig. S1A–F). More specifically, the morphology of the inhibitory hippocampal networks appeared unaltered. Immunohistochemistry for the glutamate decarboxylase GAD 67 revealed no obvious difference in the number and distribution of GAD 67-positive cells between wild-type and GluN1Rneo/+ mice in the DG (Supplemental Fig. S1G) or in CA1/CA3 (data not shown). This was confirmed by Western blots of GAD 67 protein (Supplemental Fig. S1H) and by counts of GAD 67 positive interneuron numbers in the two genotypes (three adult animals for each genotype, six hippocampal sections per animal): in the DG, wild-type 17 ± 2 cells/hippocampal section; GluN1Rneo/+ 18 ± 1 cells, mean ± SEM; in areas CA1/CA3, wild-type 115 ± 6 cells/hippocampal section; GluN1Rneo/+ 121 ± 12 cells. Furthermore, measurements of the level of a number of synaptic proteins (Supplemental Fig. S1H) and a screen for differential expression of a large number of genes failed to identify any obvious systematic molecular difference in the hippocampus between the two genotypes (Supplemental Fig. S1I). Yet, the mutant animals showed specific impairments in synaptic plasticity and spatial learning.
GluN1Rneo/+ mice exhibit impaired in vivo synaptic plasticity in the DG
In vivo electrophysiological analysis within distinct subregions is an elegant strategy that permits both evaluation of synaptic plasticity together with network properties, where the contribution of the native neuronal circuitry is intact (Bliss et al. 2000; Jones et al. 2001; Jedlicka et al. 2009). Using this approach, we first investigated aspects of short- and long-term plasticity in the hippocampus (Fig. 2). In vivo LTP in CA1 following tetanic stimulation of Schaffer collaterals was indistinguishable between GluN1Rneo/+ mice and controls (potentiation 55–60 min after induction relative to baseline measured in the 5 min before induction; GluN1Rneo/+: 18.7 ± 7.1%, n = 5; GluN1+/+: 18.8 ± 4.7%, n = 5; mean ± SEM; Fig. 2A).
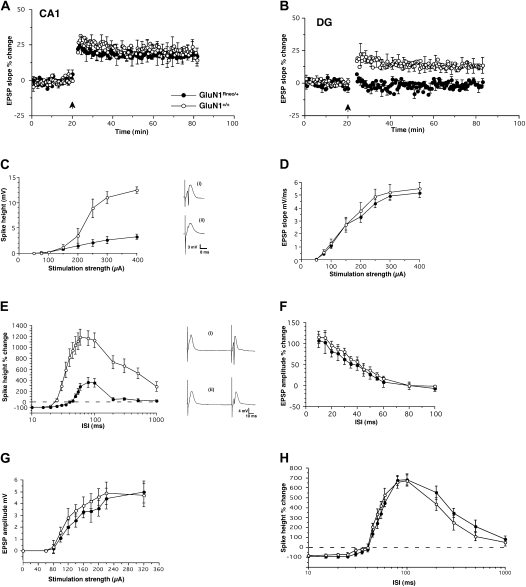
Figure 2.
Altered electrophysiological properties in the hippocampus of GluN1Rneo/+ mice, in vivo. (A) Unaltered LTP in area CA1 after tetanic stimulation (n = 5, each genotype; ●, GluN1Rneo/+; ○, GluN1+/+). (B) Absence of LTP in the DG in GluN1Rneo/+ animals (n = 9) after tetanic stimulation of the perforant path, in contrast to wild-type GluN1+/+ (n = 8). (C) Stimulus-response curves for the population spike in the DG. Note strongly reduced population spike amplitude of GluN1Rneo/+ animals compared with GluN1+/+ wild-type mice (n = 7, each). Insets: Representative EPSP traces are included on the right (i) GluN1+/+ and (ii) GluN1Rneo/+; scales: 8 msec, 3 mV. (D) Stimulus-response curves for field EPSP in the DG are similar in GluN1Rneo/+ and GluN1+/+ (n = 7, each). (E) DG paired-pulse interaction. Note prolonged paired-pulse inhibition of population spike at short intervals and reduced facilitation at long intervals in GluN1Rneo/+ animals compared with GluN1+/+ wild-type mice, measured as percent change of the population spike height (n = 10, each). ISI corresponds to interstimulus interval. Representative traces are included on the right (i) GluN1+/+ and (ii) GluN1Rneo/+; scales: 10 msec, 4 mV. (F) Subthreshold stimulation resulted in identical paired-pulse facilitation of the EPSP in both genotypes, in the DG (n = 7, each). (G) Input/output curves of EPSP amplitude in the CA1 region from both genotypes (n = 5, each). (H) CA1 paired-pulse interaction. The inhibition and facilitation of the population spike was similar in both genotypes in area CA1 (n = 4, each).
In contrast, the two genotypes showed pronounced differences in the amplitude of LTP in the DG (Fig. 2B). LTP induced by tetanic stimulation of perforant path fibers was completely absent in GluN1Rneo/+ mice (potentiation GluN1Rneo/+: −0.88 ± 2.7%, n = 9; GluN1+/+: 12.6 ± 4.6%, n = 8, P < 0.001, two-tailed t-test comparing means 40–60 min post-tetanus). Our measurements were limited to a 60-min post-induction period and demonstrate that early LTP in GluN1Rneo/+ animals is normal in the CA1 region, but abolished in the DG. Given the absence of any sign of decay of LTP in the CA1 region or of recovery in the DG at 60 min post-induction, it is probable that late LTP follows the same pattern as early LTP.
Granule cell firing properties are altered in the DG of GluN1Rneo/+ mice
Further investigation of population responses of DG granule cells to synaptic stimulation revealed substantially reduced granule cell firing, as assessed by the input/output function for the population spike, despite indistinguishable field EPSP responses (Fig. 2C,D). There was no obvious broadening of the population spike, making it unlikely that the reduced spike height was secondary to a more asynchronous firing of the granule cells.
Analysis of paired-pulse interactions revealed a similar divergence between synaptic responses and the discharge of granule cells in the two genotypes (Fig. 2E,F). The paired-pulse ratio of the field EPSP over a range of interstimulus intervals (ISI) was unaltered (Fig. 2F). In contrast, when we examined paired-pulse inhibition and facilitation of the population spike, we found a marked increase in the duration of inhibition in GluN1Rneo/+ mice compared with controls, up to 35 msec, compared with 20 msec in controls. There was also a profound decrease in facilitation at longer ISIs (Fig. 2E); the maximal potentiation was found at ISIs between 60 and 100 msec, with an increase of 1185 ± 147% for wild-type and 360 ± 89% (mean ± SEM) for mutant animals. There was no difference between the EPSP amplitude input/output curves between the genotypes in area CA1 (Fig. 2G). In contrast to the DG, paired-pulse interactions in area CA1 were not altered in GluN1Rneo/+ mice (Fig. 2H).
In summary, our electrophysiological recordings revealed a substantial impairment of short- and long-term plasticity in the DG as well as altered granule cell firing, whereas plasticity in area CA1 appeared normal, despite the identical expression levels of the mutant allele in these two subregions of the hippocampus.
GluN1Rneo/+ mice show impaired spatial learning
Our electrophysiological observations in the DG prompted us to examine whether these localized changes influence the ability of GluN1Rneo/+ animals to perform a hippocampus-dependent behavioral task such as spatial learning. NMDARs containing GluN1R subunits are impaired in their coincidence-detection properties (Burnashev et al. 1992; Single et al. 2000; Rudhard et al. 2003). This raises the question as to whether the minimal but potentially critical disruption of the NMDAR-mediated signaling system in GluN1Rneo/+ animals affects spatial learning or the retention of spatial information.
We first examined the ability of GluN1Rneo/+ mice to learn the hidden platform reference memory version of the watermaze task (Morris et al. 1982; Schenk and Morris 1985), beginning with pretraining that consisted of navigation to a visible cue in the absence of extramaze cues. The escape latency to reach the platform was reduced during training in both GluN1+/+ (analysis of variance, ANOVA, escape latency across days of training, F(5,72) = 38.9, P < 0.001) and GluN1Rneo/+ mice (ANOVA, F(5,66) = 13.4, P < 0.001). No significant difference between the genotypes was observed at the completion of training (day 6, ANOVA P > 0.33; Fig. 3C), indicating that any impairment in this test can be fully overcome with training.
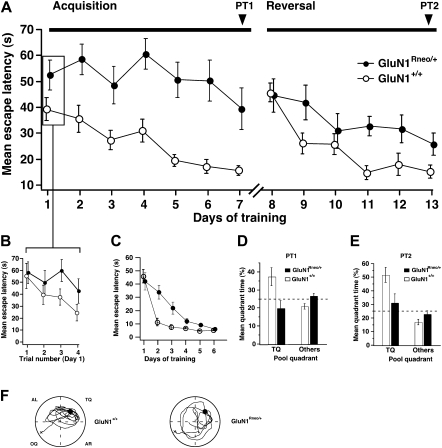
Figure 3.
Impaired spatial learning of GluN1Rneo/+ mice. (A,B) GluN1Rneo/+ (n = 12) and wild-type GluN1+/+ littermates (n = 13) were trained to a fixed hidden platform position in a watermaze for seven consecutive days (acquisition, four trials a day), followed by 6 d, to a platform position in the opposite pool quadrant (reversal, after 21-d interval). (B) Identical escape latencies for both genotypes at the beginning of the experiment, as shown by the results from individual trials on training day 1. (C) (○) Wild-type GluN1+/+, (n = 13) and (●) GluN1Rneo/+ mice, (n = 12) were trained for 6 d with four trials per day to locate the platform marked by a local visual cue, prior to the training for the hidden platform. The two groups performed equivalently well at day 1 (ANOVA, P > 0.65), and by training day 6 (ANOVA, P > 0.33). The differences between the groups during days 2–5 (ANOVA, P = 0.0007, 0.005, 0.01, and 0.0002, respectively) suggest that the mutants were slower learners than their wild-type littermates during this period, but this difference was insignificant by the end of the task. (D,E) Probe tests (PT1 and PT2) were performed (without platform) after training days 7 and 13, respectively. GluN1+/+ mice spent more time in the TQ than GluN1Rneo/+ mice. (F) Representative swim paths from PT1. (TQ) Training quadrant; (AL) adjacent left; (OQ) opposite; (AR) adjacent right; (+) start of swim path; (●) training position of platform.
To test spatial learning, the local cue on the platform was removed and the curtains drawn back to reveal extra maze cues (Fig. 3). Animals were initially trained in place navigation for 7 d (Fig. 3A,B). GluN1+/+ controls significantly reduced their escape latencies during training (ANOVA, F(6,84) = 6.4, P < 0.001). However, GluN1Rneo/+ mice showed no significant decrease in escape latency (F < 1, P > 0.4). The apparent difference in escape latency in Figure 3A on day 1 of training is due to rapid within-day learning by controls. Figure 3B shows the decline in latency across the four trials of day 1 from the comparable starting point in the two groups.
To test whether the mice had acquired any spatial bias for the training quadrant (TQ), mice were given a post-training probe test (Fig. 3D,F). In this probe test (PT1) GluN1+/+ animals showed a clear preference for the TQ (ANOVA, quadrant time in TQ vs. mean of the other three quadrants F(1,24) = 14.2, P < 0.001), which was absent in GluN1Rneo/+ mice (F(1,22) = 1.6, P > 0.2). The mean time spent within the TQ was significantly different between the groups (ANOVA, F(1,23) = 7.2, P = 0.01).
The spatial learning impairment in GluN1Rneo/+ animals was further investigated by a reversal experiment (Fig. 3A,E). The platform position was moved to the opposite pool quadrant (days 8–13). As expected, control GluN1+/+ mice showed an increase in escape latency on day 8 (relative to day 7), reflecting their prior learning of the original platform location, whereas GluN1Rneo/+ did not show an increase, again consistent with the PT1 data indicating no spatial learning in the mutants. While the GluN1+/+ mice showed a significant decline in escape latency (ANOVA, F(5,72) = 10.4, P < 0.0001), GluN1Rneo/+ animals were apparently uninfluenced by the reversal; their subsequent trend toward reduction of escape latencies did not reach significance (F(5,66) = 1.6, P = 0.2).
Probe trials at the end of the experiment (PT2) mirrored the results found in PT1; GluN1+/+ mice showed a significant preference for the new TQ (ANOVA, F(1,24) = 16.0, P < 0.001), which was absent in GluN1Rneo/+ mice (ANOVA, PT2: F(1,22) = 1.4, P = 0.3). Again, there was a significant difference in the mean times spent by either group within the TQ (ANOVA, F(1,23) = 5.6, P = 0.03).
During PT1 and PT2 we observed similar mean swim speeds in both GluN1Rneo/+ and GluN1+/+ mice (26 ± 0.6 cm/sec, 27 ± 0.4 cm/sec), suggesting that the GluN1Rneo/+ animals were unlikely to be impaired by motor dysfunction during the task. In addition, we found that the GluN1Rneo/+ mice retained normal motor coordination when their ability to remain on a rotating rotarod was examined. The mean latency to fall from the accelerating rotarod was almost identical for the two genotypes (Fig. 4A), and the performance between the groups was similar throughout the 3 d of training.
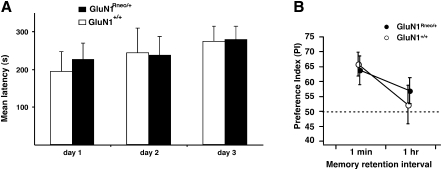
Figure 4.
Normal performance of GluN1Rneo/+ mice in motor-coordination and novel object recognition tasks. (A) Rotarod test. Latencies of GluN1Rneo/+ (n = 12) and wild-type GluN1+/+ littermates (n = 8) over three training days. No difference in performance was observed between the genotypes over the 3 d of training. (B) Novel object recognition. The two genotypes showed equivalent performance when tested shortly (1-min interval) after training. Neither genotype retained a preference for the novel object when tested 1 h after training. GluN1Rneo/+ (n = 8) and wild-type GluN1+/+ littermates (n = 6).
It is possible that the combination in the GluN1Rneo/+ mice of the slightly delayed gain of performance after training with the visible platform and the trend toward a reduced escape latency is caused by these animals retaining some spatial learning capacity and/or that the hidden platform task is somehow confounded by nonspecific factors (Wolfer et al. 1998). The similar results of the reversal experiment make this highly unlikely. The deficits of GluN1Rneo/+ mice revealed in the probe trials support the notion that this targeted mutation is associated with a severe impairment of spatial learning.
GluN1Rneo/+ mice are not impaired in novel object recognition
We also tested these animals in a different learning paradigm, novel object recognition (Fig. 4B), and found that the two genotypes exhibited similar preferences for the novel object 1 min after training. Thus, despite the learning impairment seen in the open field watermaze (Fig. 3), GluN1Rneo/+ mice were able to perform normally in a different learning task. Furthermore, a loss of memory retention in both genotypes was observed when the retention interval was extended to 1 h (Fig. 4B). Limited retention of memory traces in both genotypes was also found in the preference for the goal segment during the probe tests of the annular watermaze task 5 d after training (see below).
GluN1Rneo/+ mice show impaired place recognition
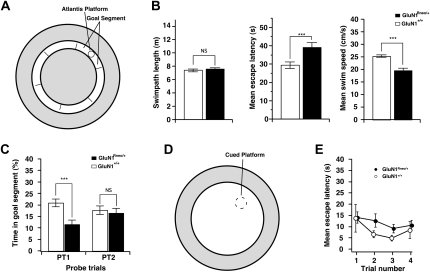
Place recognition was further examined using a task that avoids the need to navigate directly to the correct location. This is an annular watermaze, in which the animals swim in a narrow circular corridor of 16-cm width for a full “lap” of the circle, only after which a hidden escape platform becomes available in the center of one of six target segments (Fig. 5A; Brun et al. 2002; Clark et al. 2005). As the animals learn that this is where escape is possible, they stop investigating the side walls, swim around the circuit to approach this segment, and then slow down their swim speed when they get there. Analysis of acquisition over 5 d showed that path lengths of the wild-type and mutant mice were equivalent at ∼7.5 m (Fig. 5B). However, over the 5 d of training, the mean time to complete this circular path was longer for the mutants (ANOVA, F(1,16) = 24.45, P < 0.001), as their average swim speed was less (ANOVA, F(1,16) = 40.73, P < 0.001). The animals were then given two probe tests (escape platform unavailable) at 24 h after the last day of training and again at 5 d post-training (Fig. 5C). Searching by the GluN1+/+ controls in the goal segment was above chance (one sample t-test, t = 2.45, P < 0.05). The performance of the mutant mice was significantly below the chance level (t = −2.8, P < 0.05), possibly due to a bias to remain in the placement segment, which was always opposite the test segment.
Figure 5.
Impaired place recognition of GluN1Rneo/+ mice. (A) Diagram showing location of the “annulus” swim corridor (16-cm wide, inner ring) within the watermaze. The inaccessible portions of the watermaze (outer ring and inner circle) are shown in gray. It is notionally divided into six segments, one of which is designated the goal segment. The on-demand Atlantis platform comes up after one complete swimming lap of the corridor. (B) Mean swim paths (m), escape latencies (s), and swim speed (cm/sec) for the 5 d of training in the place recognition task. While the swim paths were necessarily the same, mutants took longer to escape and swam more slowly. (C) Percent time spent in the goal segment of the annulus during PT1 (1 d after training) and PT2 (5 d after completion of training). The groups differ on PT1 and only the wild-type mice were above chance. (D) Removal of the inner corridor created a smaller open field watermaze for cue task training (open inner circle). (E) Escape latencies over the four trials of training showed no difference between groups.
There was a highly significant impairment in place recognition (PT1) by the mutants with a much lower proportion of time spent searching the “correct” segment than by controls (ANOVA, F(1,16) = 7.64, P < 0.025), suggesting that they did not learn to search in the goal segment. The group difference dissipated over the 5-d retention interval, such that the groups no longer differed in PT2. An identical pattern was observed with respect to the frequency of swim-path crossings of the correct location by wild-type mice (data not shown). Finally, the inner wall of the annular watermaze was removed and the escape platform marked by a local cue (Fig. 5D,E). Over four trials, there was a trend toward significant trial effect, suggesting some degree of weak learning had occurred in both groups (F(3,48) = 2.45 P = 0.075); however, there was no group by trial interaction (F < 0.1) and no difference was observed between the escape latencies of the mutant and wild-type mice (ANOVA, F(1,16) = 1.05, P > 0.1).
The deficits of GluN1Rneo/+ mice revealed during the training phases of both the reference memory and annular watermaze tasks and in their respective probe tests support the notion that the region-specific alterations of NMDAR function are accompanied by a pronounced impairment of spatial recognition and spatial learning.
Discussion
The objective of this study was to evaluate the consequences of minimal alterations of the NMDAR-mediated coincidence detection system with respect to specific impairment in hippocampal subregions. We have achieved this by investigating a mouse mutant (GluN1Rneo/+) exhibiting a low level of expression of a mutation (GluN1R) within the globally expressed, nonredundant NMDAR subunit, GluN1. Our mutant mouse model bears some similarities to the GluN1 hypomorph described by Mohn and colleagues (Mohn et al. 1999), yet the two models differ in the nature of the expressed GluN1 allele. Mohn and collaborators studied the low-level expression of the wild-type GluN1 allele, whereas we have examined the low-level expression of a mutation within the GluN1 allele (GluN1Rneo/+).
The phenotype of our mouse model is also very different from a heterozygous, or partial loss-of-function mouse model, GluN1+/− (Silva et al. 1997; our own observations; data not shown), in which no obvious learning deficit was detected in a standard genetic background.
Moreover, our mouse model also differs from the region-specific GluN1 subunit knockouts described by McHugh et al. (2007) and Niewoehner et al. (2007). These studies have examined “loss-of-function” mice with DG region-specific elimination of GluN1 expression in the hippocampus. In contrast, we describe a mouse model with a “gain-of-function” allele of the GluN1 locus (i.e., an allele conferring an altered function, rather than loss-of-function). Common to these mouse models is the impairment of LTP in the DG region with preservation of LTP in area CA1. The behavioral phenotypes, however, are markedly different. The behavioral phenotype in our model could, in principle, be due to as yet undetected electrophysiological extrahippocampal effects of our mutation, which may contribute to the delayed learning of the visible platform task; alternatively, and perhaps in addition, it is possible that the gain-of-function allele imposes a more severe impairment on the network properties of the DG. The latter interpretation is supported by the marked changes in granule cell excitability observed in our mutant mice (Fig. 2C,E), a phenotype not seen in the DG-specific GluN1 knockout (McHugh et al. 2007).
It is not surprising that these very different alleles produce different phenotypes, even if the expression of the electrophysiological phenotype is restricted to a subregion within the hippocampus. While in both cases the electrophysiological phenotype is essentially restricted to the DG, the nature of the phenotype is substantially different. In our study we have observed that behavioral and electrophysiological phenotypes can be expressed when subtle changes occur in nonredundant proteins such as the GluN1 subunit, and that these functional changes can be restricted regionally despite global expression of the GluN1 mutation. In summary, the GluN1Rneo/+ hypomorph presented here is distinct from previously described GluN1 subunit mouse mutants and highlights the phenotypic and localized dysfunctions that become evident when subtle changes in NMDAR function occur.
GluN1R-containing NMDARs are impaired in their coincidence-detection properties (Burnashev et al. 1992; Mori et al. 1992; Single et al. 2000; Rudhard et al. 2003) and only a minority of NMDARs (10%) are affected in GluN1Rneo/+ mice. The heterogeneous population of NMDARs in GluN1Rneo/+ has a reduced macroscopic sensitivity to Mg2+ ions (a gain-of-function of the GluN1R-containing receptors) (Rudhard et al. 2003) and retains Ca2+ signaling (through wild-type receptors). This interpretation is fully compatible with the genetic classification of the GluN1Rneo allele, and its gene product, i.e., GluN1R subunits, as a gain-of-function mutation (Burnashev et al. 1992; Mori et al. 1992; Single et al. 2000; Rudhard et al. 2003).
Thus, NMDAR-dependent signaling in our GluN1Rneo/+ mice is activated under less-stringent conditions than in wild-type mice, in line with the data by Pawlak et al. (2005), who observed a substantial NMDAR-mediated EPSP component in cells expressing a high proportion (i.e., 50%) of GluN1R subunits in the presence of physiological concentrations of Mg2+. As we have shown, the minor alteration of the NMDAR-mediated coincidence detection system in GluN1Rneo/+ mice resulted in severe impairment of spatial learning and spatial navigation and functional deficits in the DG.
Previous studies using lesions and physiological disruption of the DG have revealed abnormal spatial learning in rats (Sutherland et al. 1983; Moser et al. 1998), but left some uncertainty as to whether the manipulations had affected additional regions. We now show that spatial learning is impaired, while short- and long-term plasticity in area CA1 in vivo appears to be unaffected. Our data suggest that the spatial learning deficits observed in GluN1Rneo/+ mice can be caused by functional disruption in DG properties in the absence of CA1 dysfunction, in contrast to other genetic mouse models (Mayford and Kandel 1999; Matynia et al. 2002; McHugh et al. 2007; Niewoehner et al. 2007). Conversely, selective CA1 dysfunction can also cause impairments in spatial learning and place cells (McHugh et al. 1996; Tsien et al. 1996). It may seem surprising that such a striking behavioral phenotype accompanies such an apparently subtle change in NMDAR coincidence detection, given that the distributed-associative nature of hippocampal circuitry is one that should enable normal function to continue in the face of partial damage (Marr 1971; Rolls and Treves 1998). However, “graceful degradation” in the face of partial damage is a property observed in neural networks when each of the processing elements are themselves normal (McNaughton and Morris 1987). Here, in contrast, individual granule cells will have a complement of normal receptors at which NMDAR coincidence detection is functioning correctly (90%) and a subset of other receptors at which it does not (10%). Thus, unlike a partial structural lesion, all neurons within the DG will be potentially affected by this genetic alteration. Similarly, all will be affected by the changes in feedback and feed-forward inhibition.
Our first use in mice of Moser's annular watermaze task was motivated in part by a Brun et al (2002) observation that rats with CA3 lesions that interrupt the hippocampal trisynaptic circuit have normal CA1 place fields and can learn to recognize a spatial location under conditions where directed navigation is unnecessary. Such animals are, however, impaired in learning a standard watermaze task that appears to require the integrity of the trisynaptic circuit, including the DG. We therefore thought it likely that GluN1Rneo/+ mice, with an apparently specific dentate impairment within the hippocampal formation, might be able to learn an annular watermaze. Our findings do not bear out this supposition. Either the GluN1Rneo/+ mice have other functional abnormalities that affect spatial recognition memory (e.g., in the neocortex) or processing in the trisynaptic hippocampal circuit when there is aberrant activity-dependent plasticity in each of the granule cells of DG that has more deleterious effects on hippocampal function than a frank lesion.
The absence of LTP in the DG and its preservation in CA1 is, a priori, difficult to explain. The requirement for NMDARs in the initiation of LTP in both regions is well established (Bliss and Collingridge 1993). LTP in area CA1 of GluN1Rneo/+ mice suggests a functional NMDAR system in that region, consistent with a majority of wild-type NMDARs in these mice.
So why is LTP in the DG absent? We can exclude differential expression of the hypomorphic allele in the two areas (Fig. 1E). However, it is likely that the signaling pathways downstream from NMDARs in the DG are different and more sensitive to minor alterations of Ca2+ signaling than those in CA1. For example, in area CA1, early LTP depends on autonomous calcium-independent αCaMKII signaling (Giese et al. 1998), while this is not the case for the DG (Cooke et al. 2006).
An alternative explanation, that an increase in direct tonic inhibitory modulation by GABAergic synapses might suppress LTP in the DG (Freund and Buzsaki 1996; Errington et al. 1997) is supported by the reduction in the amplitude of the population spike in GluN1Rneo/+ mutants. Against this hypothesis, however, we failed to find abnormalities in the level of the inhibitory markers GAD 67 or the numbers of inhibitory interneurons in the GluN1Rneo/+ mice. The functional alterations that we have found in the DG may well have resulted from a slightly modified, activity-dependent development of the DG network; alternatively, it may represent an acute alteration of NMDAR-mediated signaling, despite the absence of any obvious change of the hippocampal transcriptome or major synaptic proteins.
In addition to the suppression of LTP we have described a second electrophysiological phenotype in the DG of GluN1Rneo/+ mutant mice. The suppression of the population spike by recurrent inhibition at short paired-pulse intervals lasts considerably longer in the mutants, and the massive 10-fold facilitation seen at longer intervals in wild-type animals is much less developed. These properties indicate a functional enhancement of feedback inhibition and a dampening of feed-forward disinhibition relative to wild-type animals (Freund and Buzsaki 1996) in the absence of a change in the number of GAD 67 positive cells or GAD 67 protein levels. One possibility to link the two electrophysiological phenotypes in the DG is that back-propagation of action potentials (APs) into the dendritic tree might play an important role in the initiation of LTP in the DG.
Whatever the precise mechanism that leads to the impairment of short- and long-term plasticity in the DG, it is remarkable that the subtle mutation that we have introduced into the NMDAR system manifests itself in such a distinct and localized manner in different subregions of the hippocampus. This finding highlights the notion that different brain circuits may be governed by specialized assemblies of signaling pathways that contribute to the developmental refinement and electrical control of synaptic connectivity, even when these pathways share core components such as the nonredundant, globally expressed GluN1 subunit of the NMDAR.
Materials and Methods
Mouse strains
The GluN1Rneo/+ and GluN1+/− strains and their genotyping have been described previously (Rudhard et al. 2003). GluN1Rneo/+ × GluN1Rneo/+ breeding produced litters with a Mendelian distribution of genotypes (22 wild-type, 58 GluN1Rneo/+, and 27 GluN1Rneo/Rneo). Mice were bred into a C57BL/6J background and experiments were done with at least four backcrosses. No differences between the generations were observed. All work was undertaken under the auspices of the UK Home Office Project and Personal Licenses held by the authors in their designated laboratories.
Data analysis
Averages are given as mean ± SEM unless otherwise noted. Individual tests used have been indicated where relevant.
mRNA analysis
Plaque assay
Poly(A)+ -RNA was prepared from whole brains by first isolating total RNA using Tri-reagent solution (Sigma) followed by poly(A)+ -RNA preparation (Fast Track 2.0 kit, Invitrogen). RT-PCR and differential oligonucleotide hybridization from dual plaque filters have been described previously (Rudhard et al. 2003).
Single-cell RT-PCR
The cytoplasmic contents of single CA1 pyramidal neurons or DG granule cells (from acute GluN1Rneo/+ adult hippocampal slices) were harvested using constant negative pressure into a patch pipette containing 52 mM Tris-Cl (pH 8.5), 78 mM KCl, 3.1 mM MgCl2, 80 U/mL RNase inhibitor (Roche). The contents of the pipette tip plus the broken-off tip was added into a 0.5 mL RT-PCR tube containing a dissolved (50 μL) Ready-To-Go RT-PCR bead (Amersham Pharmacia Biotech) plus primers (0.25 μg of oligo-dT and first round GluN1 primers [mGluN1-Seq51-s, 5′->3′] GTGATGCTGTACCTGCTGGACCGC, exon 14 and mGluN1-Seq52-a, CCCCTGCCATGTTCTCAAAAGTGA, exon 19). The RT-PCR was incubated for 30 min at 42°C (Biometra Uno II Thermocycler) for first-strand cDNA synthesis, followed by first-round PCR cycling (30 cycles touchdown-PCR [94°C, 30 sec; 68°C –0.5°C every second cycle, 30 sec; 72°C, 90 sec]) and a further 15 cycles ([94°C, 30 sec; 46°C, 30 sec; 72°C, 45 sec]). A 1:10 dilution of this reaction was used as a template for the second-round PCR (primers mGluN1-PCR101-s and mGluN1-PCR104-a [Rudhard et al. 2003]; 30 cycles [94°C, 30 sec; 52°C, 30 sec; 72°C, 60 sec]). This set of nested GluN1 primers binds to exons 14 and 19, respectively (Fig. 1A). The amplicons were then digested and subcloned for GluN1R mRNA quantification. The relative abundance of GluN1+ and GluN1R transcripts was determined as the ratio of the number of plaques hybridizing with one probe to the total number of GluN1 transcript-containing plaques as described (Rudhard et al. 2003). A number of GluN1+ and GluN1R-containing plaques were verified by DNA sequencing, revealing exclusively transcripts that were spliced identically to the wild-type GluN1+ allele.
RNA expression profiling
The array experiment was performed by Genome Systems (now Incyte Genomics; mouse GEM 1 microarray) on pooled hippocampal poly(A)+-RNA from three animals of each genotype; for details, see Specht and Schoepfer (2001).
Immunoblotting and antibodies
Total brain homogenate (from P0 pups) and hippocampal homogenates (adult animals) were analyzed by Western blot as described (Specht and Schoepfer 2001; Rudhard et al. 2003). For details about antibodies see Supplemental material.
Histology
Preparation of brain tissue for histology and Nissl staining has been described (Chen et al. 2002). Serial coronal sections from 5–7-mo-old littermates were collected in groups of three. One section from each group was taken for immunohistochemistry and the other for Nissl staining.
Immunohistochemistry
Free-floating sections were washed 3× in PBS (20 mM sodium phosphate, 150 mM NaCl, pH 7.5), and endogenous peroxidases were exhausted with 3% hydrogen peroxide (5 min), followed by three washes in PBS. Sections were blocked in 3% goat serum in TBS (50 mM Tris-Cl, 154 mM NaCl, pH 7.4) for 30 min, incubated overnight with anti-GAD 67 1:2000 in 1% goat serum in TBS at 4°C. Following three washes in TBS, the sections were incubated with goat anti-rabbit HRP-conjugated secondary antibody 1:500, for 1 h at room temperature. After two more washes with TBS, sections were developed with nickel-enhanced DAB intensification reaction for 5 min in the dark. Sections were washed twice in TBS, mounted on glass slides, dried overnight, dehydrated, cleared in Histoclear (National Diagnostics), and coverslipped in DPX (BDH-Merck).
Behavioral analysis
Subjects
Male mice (5–8-mo-old) were used, with the experimenter blind to genotype. GluN1Rneo/+ animals from GluN1+/+ × GluN1Rneo/+ breeding were compared with wild-type littermates. For analysis of GluN1+/− animals, offspring from GluN1+/+ × GluN1+/− breeding were used.
Rotarod
Motor coordination and learning were assessed using an accelerating rotarod for mice (Ugo Basile, No. 7650, 3-cm diameter of rotor/15-cm height/60-cm width). Mice were placed on the rotarod at an initial speed of 5 rpm for 30 sec. If mice fell during this period, they were once more placed on the rotarod. The rotarod was then gradually accelerated to 40 rpm over a period of 4 min, and then maintained for a further 30 sec. A short habituation phase for the mice (three short runs) was carried out prior to the actual training period. During the 3-d training period animals were subjected to three consecutive trials with an intertrial interval (ITI) of 30 min. The average latencies of trials 2 and 3 of each day were taken as a measure of motor coordination.
Novel object recognition
Animals were tested using equipment and similar protocols described by Chen et al. (2000), except that habituation consisted of daily 10-min periods in the empty arena over a 5-d period. Memory recall for the novel object was examined under two memory delay intervals, 1 min and 1 h. For details see Supplemental material.
Watermaze apparatus
We used both a standard open field watermaze (2-m diameter pool; 20-cm diameter platform; platform area/pool area 1/100) and an “annular watermaze” (made by placing two circular stainless steel walls of 1.40-m diameter and 1.08-m diameter placed within the standard pool). In both tasks, we used opaque water at 25°C ± 1°C in a room filled with extramaze cues (Chen et al. 2002). The apparatus included automated swim-path monitoring with an overhead video camera and processed using the LabView-based computer program Watermaze.
Standard open field watermaze
Visible platform training: Mice were trained to a pseudorandomly located platform with a local cue protruding out of the water surface with curtains drawn around the pool to occlude extramaze cues (four trials/day, 6 d, maximum trial duration 90 sec; ITI 10 min; platform moved randomly after each trial). Hidden platform training: Mice were trained to a hidden platform with the extramaze cues visible (four trials/day, 7 d, 30 sec spent on the platform at the end of each trial; ITI 10 min; max trial duration 90 sec). For the reversal phase of this experiment, training was for a further 6 d, training days 8–13, to an escape platform in the opposite quadrant. The back-crossing into C57BL/6J had resulted in some animals carrying the chromosomal deletion Del(6)Snca1Slab, that does not result in any obvious phenotype or affect spatial learning (Specht and Schoepfer 2001; Chen et al. 2002). Probe test: 24 h after the last training trial, mice were placed in the pool for 60 sec with the platform absent. The start position was opposite to whatever TQ had been used for an individual animal. Probe tests (PT1 and PT2) were given after days 7 and 13, respectively. The percentage time spent in each quadrant during the trial constituted the primary measure of task performance.
Annular watermaze
The animals were required to swim from a randomly chosen start segment in a narrow circular corridor of 16-cm width for a full “lap” of the circle (i.e., return to same segment), after which a hidden escape platform became available in the center of the target segment. In this study, an “Atlantis Platform” was used that was initially at the bottom of the pool and then rose, on command, after completion of one lap. Training consisted of four trials/block, two blocks/day, separated by a 1-h interval, within-block ITI at 10 min. Upon finding and climbing onto the platform, the animals were allowed to stay there for 30 sec before being removed to their cages. The probe tests (PT1 and PT2) lasted 60 sec and ended with the Atlantis platform coming up to provide escape. Probe trials always started from the segment opposite to the platform.
Electrophysiology
For in vivo experiments, animals were anesthetized with urethane (1.5 g/kg body weight, intraperitoneal) and held in a stereotaxic apparatus. For experiments in area CA1, the glass recording electrode was placed 2.0-mm lateral and 2.0-mm caudal to Bregma and a metal bipolar stimulating electrode (Rhodes Electromedical) was placed at the same coordinates on the contralateral side. The LTP induction protocols were based on previous experiments and found to be the most effective for anesthetized mice (ML Errington, unpubl.).
For experiments in the DG, the glass recording electrode was inserted 2.1-mm caudal and 1.7-mm lateral to bregma. A metal bipolar stimulating electrode was inserted 3.1-mm lateral to lambda and advanced into the angular bundle to activate fibers of the perforant path. Constant current pulses (50 μs, 70–120 μA) were delivered at 30-sec intervals. Electrodes were adjusted to produce maximal responses in the cell body layer. Significance of potentiation was tested on group average potentiation in 2-min intervals during 40–60 min after induction relative to baseline during 5 min before induction. For LTP experiments and measurements of paired-pulse interaction on the population spike, the stimulus intensity was adjusted to produce an initial negative population spike with an amplitude of 1–2 mV. The tetanus consisted of six series of six trains of six stimuli delivered at 400 Hz with 200 msec between trains and 20 sec between series, at twice the intensity of test stimulation. For paired-pulse experiments on the field EPSP, the stimulus strength was lowered to produce an EPSP amplitude of ∼1 mV for the first response.
For CA1 experiments electrode positions were adjusted to maximize the response in the stratum radiatum. The test stimulus intensity was adjusted to give a field EPSP with a slope of 40%–50% of the maximum obtainable. The conditioning stimulus consisted of two trains of 50 stimuli at 100 Hz, given at the same intensity as the test stimulus, with an ITI of 60 sec.
Acknowledgments
We thank members of the Laboratory for Cognitive Neuroscience for help and advice with the watermaze experiments, Edda Thiels for discussions on the project, and Biological Services at UCL, Edinburgh University, and NIMR for colony maintenance. This work was funded by the Wellcome Trust through a Senior Research Fellowship (R.S.), postdoctoral fellowship (C.M.T.), and studentship (Y.H.R.); by support from the MRC (M.L.E., R.G.M.M., and T.V.P.B.) and from the BBSRC (R.S.). G.F.R. was a recipient of an Emmy Noether fellowship (DFG).
Footnotes
[Supplemental material is available online at http://www.learnmem.org.]
Article is online at http://www.learnmem.org/cgi/doi/10.1101/lm.1316909.
References
- Behe P, Stern P, Wyllie DJ, Nassar M, Schoepfer R, Colquhoun D. Determination of NMDA NR1 subunit copy number in recombinant NMDA receptors. Proc R Soc Lond B Biol Sci. 1995;262:205–213. doi: 10.1098/rspb.1995.0197. [DOI] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL. A synaptic model of memory: Long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Bliss T, Errington M, Fransen E, Godfraind JM, Kauer JA, Kooy RF, Maness PF, Furley AJ. Long-term potentiation in mice lacking the neural cell adhesion molecule L1. Curr Biol. 2000;10:1607–1610. doi: 10.1016/s0960-9822(00)00865-4. [DOI] [PubMed] [Google Scholar]
- Brun VH, Otnass MK, Molden S, Steffenach HA, Witter MP, Moser MB, Moser EI. Place cells and place recognition maintained by direct entorhinal-hippocampal circuitry. Science. 2002;296:2243–2246. doi: 10.1126/science.1071089. [DOI] [PubMed] [Google Scholar]
- Burnashev N, Schoepfer R, Monyer H, Ruppersberg JP, Gunther W, Seeburg PH, Sakmann B. Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science. 1992;257:1415–1419. doi: 10.1126/science.1382314. [DOI] [PubMed] [Google Scholar]
- Castner SA, Williams GV. Tuning the engine of cognition: A focus on NMDA/D1 receptor interactions in prefrontal cortex. Brain Cogn. 2007;63:94–122. doi: 10.1016/j.bandc.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Chen G, Chen KS, Knox J, Inglis J, Bernard A, Martin SJ, Justice A, McConlogue L, Games D, Freedman SB, et al. A learning deficit related to age and β-amyloid plaques in a mouse model of Alzheimer's disease. Nature. 2000;408:975–979. doi: 10.1038/35050103. [DOI] [PubMed] [Google Scholar]
- Chen PE, Specht CG, Morris RG, Schoepfer R. Spatial learning is unimpaired in mice containing a deletion of the α-synuclein locus. Eur J Neurosci. 2002;16:154–158. doi: 10.1046/j.1460-9568.2002.02062.x. [DOI] [PubMed] [Google Scholar]
- Clark RE, Broadbent NJ, Squire LR. Hippocampus and remote spatial memory in rats. Hippocampus. 2005;15:260–272. doi: 10.1002/hipo.20056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Olsen R, Peters JA, Spedding M. Ligand gated ion channels. Neuropharmacology. 2009;56:1. doi: 10.1016/j.neuropharm.2008.08.036. [DOI] [PubMed] [Google Scholar]
- Cooke SF, Wu J, Plattner F, Errington M, Rowan M, Peters M, Hirano A, Bradshaw KD, Anwyl R, Bliss TV, et al. Autophosphorylation of αCaMKII is not a general requirement for NMDA receptor-dependent LTP in the adult mouse. J Physiol. 2006;574:805–818. doi: 10.1113/jphysiol.2006.111559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington ML, Bliss TV, Morris RJ, Laroche S, Davis S. Long-term potentiation in awake mutant mice. Nature. 1997;387:666–667. doi: 10.1038/42625. [DOI] [PubMed] [Google Scholar]
- Forrest D, Yuzaki M, Soares HD, Ng L, Luk DC, Sheng M, Stewart CL, Morgan JI, Connor JA, Curran T. Targeted disruption of NMDA receptor 1 gene abolishes NMDA response and results in neonatal death. Neuron. 1994;13:325–338. doi: 10.1016/0896-6273(94)90350-6. [DOI] [PubMed] [Google Scholar]
- Freund TF, Buzsaki G. Interneurons of the hippocampus. Hippocampus. 1996;6:347–470. doi: 10.1002/(SICI)1098-1063(1996)6:4<347::AID-HIPO1>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Giese KP, Fedorov NB, Filipkowski RK, Silva AJ. Autophosphorylation at Thr286 of the α-calcium-calmodulin kinase II in LTP and learning. Science. 1998;279:870–873. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- Gruart A, Munoz MD, Delgado-Garcia JM. Involvement of the CA3-CA1 synapse in the acquisition of associative learning in behaving mice. J Neurosci. 2006;26:1077–1087. doi: 10.1523/JNEUROSCI.2834-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebb DO. The organization of behavior. Wiley; NY: 1949. [Google Scholar]
- Jedlicka P, Schwarzacher SW, Winkels R, Kienzler F, Frotscher M, Bramham CR, Schultz C, Bas Orth C, Deller T. Impairment of in vivo θ-burst long-term potentiation and network excitability in the dentate gyrus of synaptopodin-deficient mice lacking the spine apparatus and the cisternal organelle. Hippocampus. 2009;19:130–140. doi: 10.1002/hipo.20489. [DOI] [PubMed] [Google Scholar]
- Jia Z, Agopyan N, Miu P, Xiong Z, Henderson J, Gerlai R, Taverna FA, Velumian A, MacDonald J, Carlen P, et al. Enhanced LTP in mice deficient in the AMPA receptor GluR2. Neuron. 1996;17:945–956. doi: 10.1016/s0896-6273(00)80225-1. [DOI] [PubMed] [Google Scholar]
- Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- Kristiansen LV, Huerta I, Beneyto M, Meador-Woodruff JH. NMDA receptors and schizophrenia. Curr Opin Pharmacol. 2007;7:48–55. doi: 10.1016/j.coph.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Li Y, Erzurumlu RS, Chen C, Jhaveri S, Tonegawa S. Whisker-related neuronal patterns fail to develop in the trigeminal brainstem nuclei of NMDAR1 knockout mice. Cell. 1994;76:427–437. doi: 10.1016/0092-8674(94)90108-2. [DOI] [PubMed] [Google Scholar]
- Marr D. Simple memory: A theory for archicortex. Philos Trans R Soc Lond B Biol Sci. 1971;262:23–81. doi: 10.1098/rstb.1971.0078. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Grimwood PD, Morris RG. Synaptic plasticity and memory: An evaluation of the hypothesis. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- Matynia A, Kushner SA, Silva AJ. Genetic approaches to molecular and cellular cognition: A focus on LTP and learning and memory. Annu Rev Genet. 2002;36:687–720. doi: 10.1146/annurev.genet.36.062802.091007. [DOI] [PubMed] [Google Scholar]
- Mayford M, Kandel ER. Genetic approaches to memory storage. Trends Genet. 1999;15:463–470. doi: 10.1016/s0168-9525(99)01846-6. [DOI] [PubMed] [Google Scholar]
- McHugh TJ, Blum KI, Tsien JZ, Tonegawa S, Wilson MA. Impaired hippocampal representation of space in CA1-specific NMDAR1 knockout mice. Cell. 1996;87:1339–1349. doi: 10.1016/s0092-8674(00)81828-0. [DOI] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, Tonegawa S. Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science. 2007;317:94–99. doi: 10.1126/science.1140263. [DOI] [PubMed] [Google Scholar]
- McNaughton BL, Morris RGM. Hippocampal synaptic enhancement and information-storage within a distributed memory system. Trends Neurosci. 1987;10:408–415. [Google Scholar]
- Meng Y, Zhang Y, Jia Z. Synaptic transmission and plasticity in the absence of AMPA glutamate receptor GluR2 and GluR3. Neuron. 2003;39:163–176. doi: 10.1016/s0896-6273(03)00368-4. [DOI] [PubMed] [Google Scholar]
- Mohn AR, Gainetdinov RR, Caron MG, Koller BH. Mice with reduced NMDA receptor expression display behaviors related to schizophrenia. Cell. 1999;98:427–436. doi: 10.1016/s0092-8674(00)81972-8. [DOI] [PubMed] [Google Scholar]
- Mori H, Masaki H, Yamakura T, Mishina M. Identification by mutagenesis of a Mg2+-block site of the NMDA receptor channel. Nature. 1992;358:673–675. doi: 10.1038/358673a0. [DOI] [PubMed] [Google Scholar]
- Morris RG, Garrud P, Rawlins JN, O'Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- Moser EI, Krobert KA, Moser MB, Morris RG. Impaired spatial learning after saturation of long-term potentiation. Science. 1998;281:2038–2042. doi: 10.1126/science.281.5385.2038. [DOI] [PubMed] [Google Scholar]
- Nakazawa K, Quirk MC, Chitwood RA, Watanabe M, Yeckel MF, Sun LD, Kato A, Carr CA, Johnston D, Wilson MA, et al. Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science. 2002;297:211–218. doi: 10.1126/science.1071795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner B, Single FN, Hvalby O, Jensen V, Meyer zum Alten Borgloh S, Seeburg PH, Rawlins JN, Sprengel R, Bannerman DM. Impaired spatial working memory but spared spatial reference memory following functional loss of NMDA receptors in the dentate gyrus. Eur J Neurosci. 2007;25:837–846. doi: 10.1111/j.1460-9568.2007.05312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlak V, Jensen V, Schupp BJ, Kvello A, Hvalby O, Seeburg PH, Kohr G. Frequency-dependent impairment of hippocampal LTP from NMDA receptors with reduced calcium permeability. Eur J Neurosci. 2005;22:476–484. doi: 10.1111/j.1460-9568.2005.04226.x. [DOI] [PubMed] [Google Scholar]
- Rolls ET, Treves A. Neural networks and brain function. Oxford University Press; Oxford, UK: 1998. [Google Scholar]
- Rudhard Y, Kneussel M, Nassar MA, Rast GF, Annala AJ, Chen PE, Tigaret CM, Dean I, Roes J, Gibb AJ, et al. Absence of whisker-related pattern formation in mice with NMDA receptors lacking coincidence detection properties and calcium signaling. J Neurosci. 2003;23:2323–2332. doi: 10.1523/JNEUROSCI.23-06-02323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk F, Morris RG. Dissociation between components of spatial memory in rats after recovery from the effects of retrohippocampal lesions. Exp Brain Res. 1985;58:11–28. doi: 10.1007/BF00238949. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Paylor R, Wehner JM, Tonegawa S. Impaired spatial learning in α-calcium-calmodulin kinase II mutant mice. Science. 1992a;257:206–211. doi: 10.1126/science.1321493. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Stevens CF, Tonegawa S, Wang Y. Deficient hippocampal long-term potentiation in α-calcium-calmodulin kinase II mutant mice. Science. 1992b;257:201–206. doi: 10.1126/science.1378648. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Frankland PW, Marowitz Z, Friedman E, Laszlo GS, Cioffi D, Jacks T, Bourtchuladze R. A mouse model for the learning and memory deficits associated with neurofibromatosis type I. Nat Genet. 1997;15:281–284. doi: 10.1038/ng0397-281. [DOI] [PubMed] [Google Scholar]
- Single FN, Rozov A, Burnashev N, Zimmermann F, Hanley DF, Forrest D, Curran T, Jensen V, Hvalby O, Sprengel R, et al. Dysfunctions in mice by NMDA receptor point mutations NR1(N598Q) and NR1(N598R) J Neurosci. 2000;20:2558–2566. doi: 10.1523/JNEUROSCI.20-07-02558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specht CG, Schoepfer R. Deletion of the α-synuclein locus in a subpopulation of C57BL/6J inbred mice. BMC Neurosci. 2001;2:11. doi: 10.1186/1471-2202-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland RJ, Whishaw IQ, Kolb B. A behavioural analysis of spatial localization following electrolytic, kainate- or colchicine-induced damage to the hippocampal formation in the rat. Behav Brain Res. 1983;7:133–153. doi: 10.1016/0166-4328(83)90188-2. [DOI] [PubMed] [Google Scholar]
- Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor-dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- Whitlock JR, Heynen AJ, Shuler MG, Bear MF. Learning induces long-term potentiation in the hippocampus. Science. 2006;313:1093–1097. doi: 10.1126/science.1128134. [DOI] [PubMed] [Google Scholar]
- Wilson DA, Linster C. Neurobiology of a simple memory. J Neurophysiol. 2008;100:2–7. doi: 10.1152/jn.90479.2008. [DOI] [PubMed] [Google Scholar]
- Wolfer DP, Stagljar-Bozicevic M, Errington ML, Lipp HP. Spatial memory and learning in transgenic mice: Fact or artifact? News Physiol Sci. 1998;13:118–123. doi: 10.1152/physiologyonline.1998.13.3.118. [DOI] [PubMed] [Google Scholar]
- Zamanillo D, Sprengel R, Hvalby O, Jensen V, Burnashev N, Rozov A, Kaiser KM, Koster HJ, Borchardt T, Worley P, et al. Importance of AMPA receptors for hippocampal synaptic plasticity but not for spatial learning. Science. 1999;284:1805–1811. doi: 10.1126/science.284.5421.1805. [DOI] [PubMed] [Google Scholar]