Abstract
Aortic arch artery patterning defects account for approximately 20% of congenital cardiovascular malformations and are observed frequently in velocardiofacial syndrome (VCFS). In the current study, we screened for chromosome rearrangements in patients suspected of VCFS, but who lacked a 22q11 deletion or TBX1 mutation. One individual displayed hemizygous CHD7, which encodes a chromodomain protein. CHD7 haploinsufficiency is the major cause of coloboma, heart defect, atresia choanae, retarded growth and development, genital hypoplasia, and ear anomalies/deafness (CHARGE) syndrome, but this patient lacked the major diagnostic features of coloboma and choanal atresia. Because a subset of CHARGE cases also display 22q11 deletions, we explored the embryological relationship between CHARGE and VCSF using mouse models. The hallmark of Tbx1 haploinsufficiency is hypo/aplasia of the fourth pharyngeal arch artery (PAA) at E10.5. Identical malformations were observed in Chd7 heterozygotes, with resulting aortic arch interruption at later stages. Other than Tbx1, Chd7 is the only gene reported to affect fourth PAA development by haploinsufficiency. Moreover, Tbx1+/–;Chd7+/– double heterozygotes demonstrated a synergistic interaction during fourth PAA, thymus, and ear morphogenesis. We could not rescue PAA morphogenesis by restoring neural crest Chd7 expression. Rather, biallelic expression of Chd7 and Tbx1 in the pharyngeal ectoderm was required for normal PAA development.
Introduction
Velocardiofacial syndrome (VCFS; also referred to as DiGeorge syndrome; incidence, 1 in 4,000 live births) is most commonly associated with an interstitial deletion of 22q11.2. Frequent abnormalities include facial dysmorphism, absent or hypoplastic thymus and parathyroids, velopharyngeal insufficiency, behavioral problems, and congenital heart defects (particularly of the outflow tract and aortic arch; ref. 1). Most of these features have been ascribed to haploinsufficiency of T-box 1 (TBX1), because of mutations in rare patients with no 22q11.2 deletion (2–4) and the excellent phenocopy observed in Tbx1+/LacZ (i.e., Tbx1+/–) mice (5). Tbx1+/– mice display partially penetrant cardiovascular defects such as type B interruption of the aortic arch (IAA-B), tetralogy of Fallot, absent right subclavian artery (ARS; or, more generally, aberrant right subclavian [AbRS]), and ventriculoseptal defects (VSDs), with some thymic hypoplasia (5). At E10.5, on a C57BL/6 background, there is a highly penetrant phenotype of hypoplastic fourth pharyngeal arch artery (PAA; ref. 5). Defective remodeling of the fourth PAA gives malformations such as IAA-B, double aortic arch, and AbRS, although the frequency of these defects is lower than that of the earlier PAA abnormalities, which suggests that some recovery takes place (6). Tbx1–/– mice are embryonic lethal and have a common arterial trunk, aplastic caudal pharyngeal arches, absent thymus, craniofacial malformations, and cleft palate (5, 7, 8). Tissue-specific conditional mutagenesis of Tbx1 reveals a role for the gene in each of the tissues that expresses it: second heart field (9), pharyngeal mesoderm (10), pharyngeal endoderm (11), pharyngeal epithelia (12), and otic epithelium (13).
CHARGE syndrome (incidence, 1 in 10,000 live births) is the nonrandom clustering of coloboma, heart defect, atresia of the choanae, retarded growth and development, genital abnormalities, and ear anomalies/deafness (14). Deletion or mutation of chromodomain helicase DNA-binding protein 7 (CHD7) is the major cause of CHARGE association, accounting for approximately 60% of cases (14). Mutation in SEMA3E has also been reported in 1 patient (15). Phenotypic overlap between CHARGE and VCFS has been remarked upon previously (16–18). Indeed, a number of CHARGE cases (i.e., those fulfilling the diagnostic criteria) with 22q11 deletions have been described (19, 20). A series of N-ethyl-N-nitrosourea (ENU) mutants characterized by dominant circling and hyperactivity provided the first mouse models for CHARGE.
Heterozygous Chd7 ENU mutants, such as whirligig (Whi; refs. 21–23), provide a good model for many of the aspects of CHARGE, in particular eye, inner ear, palatal, choanal, genital, and cardiac defects. Whi heterozygotes demonstrated partial perinatal lethality, which was ascribed to cardiac insufficiency secondary to VSDs. A few embryos appeared to have vascular hemorrhage, which suggests a role for Chd7 in maintaining vascular integrity. The wheels (Whl) locus is presumed to carry a Chd7 mutation on the basis of linkage and phenotype (24). Whl/Whl embryos do not survive past E11.5. At E9.5, homozygotes are growth retarded, but heart tube formation and looping appear normal. In addition, a gene-trapped allele of Chd7 has been previously described that similarly recapitulates the CHARGE phenotype (25). Development of the PAA derivatives and thymus were not commented upon in these prior reports.
Both TBX1 and CHD7 regulate transcription. TBX1 is a member of the T-box DNA binding domain–containing protein family. TBX1 activates transcription in the systems studied, and a number of genes dysregulated in Tbx1 mutants have been described. TBX1 physically interacts with SMAD1, interfering with the ability of SMAD1 to bind SMAD4, and thus has effects on signal transduction (26). CHD7 belongs to a protein family of chromodomain helicase DNA-binding domain proteins. Besides the SNF2-like ATPase/helicase domain, the protein encodes a BRK, a SANT, and a DEXc domain. It is predicted to be involved in ATP-dependent chromatin remodeling (27).
In this study, we conducted comparative genome hybridization on a patient referred because of a phenotype compatible with 22q11 deletion, but with no FISH-detectable deletion or TBX1 mutation. Because this patient was found to be hemizygous for CHD7, our aim was to explore the embryological basis for the overlap of CHARGE and VCFS, particularly with regard to great artery development. We found the genes haploinsufficient in these syndromes — Chd7 and Tbx1, respectively — were in epistasis and that the correct level of their expression was required in pharyngeal ectoderm for proper morphogenesis of the great arteries.
Results
We previously reported an array-based comparative genome hybridization screen of a panel of patients referred for TBX1 sequencing because they had features suggestive of VCFS in the absence of any 22q11 deletion (28). In an extension of this work, we identified a deletion of 8q12, estimated to be 5 Mb by microsatellite marker analysis and FISH, in patient 15 (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/37561DS1). The region encompassed markers D8S1812–D8S544 and BACs RP11-414L7 (61.7 Mb) to RP11-115G12 (65.4 Mb), including CHD7. Moreover, 2 patients with T cell immunodeficiencies typical of VCFS were also seen in our clinic and reported separately as having CHD7 mutations (29). However, patient 15 did not present with 2 of the cardinal diagnostic features of CHARGE, coloboma and choanal atresia. Patient 15 had facial dysmorphism (Supplemental Figure 1) and several problems often seen in VCFS, including learning difficulty with speech delay and major feeding difficulties that required nasogastric nutrition. An echocardiogram revealed IAA-A, a coarctation of the aorta, a large membranous VSD, a 16-mm secundum atrial septal defect, and a bicuspid aortic valve (a mild cardiac outflow tract defect). She also had long, slender fingers and low-set, overfolded ear helices. The IAA seen in VCFS is usually type B, but type A has been observed in association with 22q11 deletion (30). Thus, although the facial features were not typical for VCFS, this phenotypic overlap prompted examination of 22q11, and we questioned whether this overlap in genetic etiology reflected a common embryological pathway affected in the 2 conditions.
We obtained 2 gene-trapped ES lines, XK403 and RRR134, from Baygenomics (31), injected them into blastocysts, and obtained germline transmissions from the resulting chimeric mice. Both lines gave heterozygous mice that were viable and fertile, but displayed the typical circling behavior noted in a series of chemically induced Chd7 mutations. Both homozygotes displayed embryonic lethality by day 11, with the same phenotype previously reported for Chd7-homozygous ENU mutants (22). Expression of the β-gal reporter was identical for the 2 lines and recapitulated the previously published gene expression pattern (22), as determined by in situ hybridization (Supplemental Figure 2). The majority of the work described herein was conducted on the XK403 line, with heterozygotes designated Chd7+/xk (heterozygotes of the RRR134 line are designated Chd7+/rrr). Given that a haploinsufficiency affecting PAA development is the hallmark of Tbx1+/– mice, our analysis concentrated on these structures in Chd7 heterozygotes.
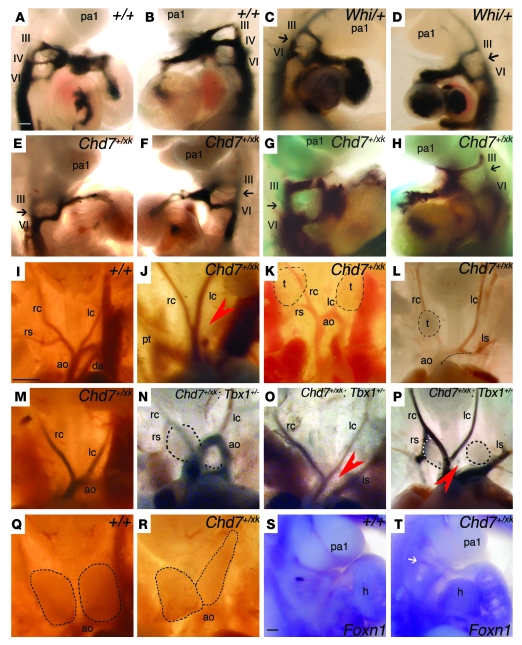
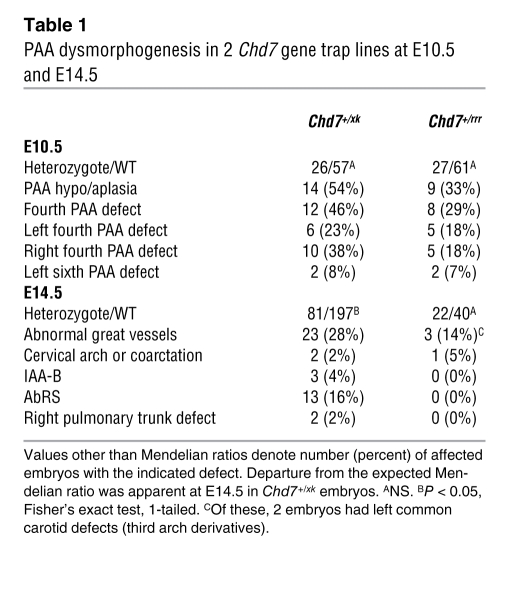
At E10.5, we detected defects of PAA development in 54% of Chd7+/xk embryos, with the fourth PAA particularly affected (Figure 1, A–H, and Table 1), and in 33% of Chd7+/rrr embryos. At E14.5, 4% of Chd7+/xk embryos showed IAA-B, a further 2% exhibited cervical arch or B-segment coarctation, and 16% had AbRS in isolation (Figure 1, I–L, and Table 1), suggestive of some embryonic recovery. The patient studied here had IAA-A; however, we observed no examples of this type of interruption in our mouse models. As far as we are aware, there are no known genetic causes of specifically A-type interruptions. The thymus gland was small, or ectopically placed, in 11% of Chd7+/xk embryos, similar to abnormalities of Tbx1+/– embryos (Figure 1, Q and R). Thymic hypoplasia in Chd7 heterozygotes correlated with downregulation of Foxn1 expression in midgestation embryos (Figure 1, S and T), and this dysmorphogenesis presumably underlies the T cell immunodeficiency reported in CHARGE (29). Thus, Chd7 haploinsufficiency was very similar to Tbx1 haploinsufficiency at the embryological level, especially with regard to structures affected in VCFS. These results were confirmed by analyzing the Whi Chd7 mutant (Figure 1, C and D, and ref. 22). Whereas other homozygous mouse mutants display similar phenotypes, Chd7 and Tbx1 are the only 2 genes reported in which haploinsufficiency is sufficient to produce these specific abnormalities in fourth PAA morphogenesis. At E10.5, expected Mendelian ratios were observed, but a proportion of Chd7+/xk embryos was lost by E14.5 (Table 1).
Figure 1. PAA patterning defects in Chd7+/xk and Chd7+/xk;Tbx1+/– embryos.
Black arrows denote absent fourth PAA; red arrowheads denote absence of arches; dashed lines denote thymic lobes (t). pa1, pharyngeal arch 1; III, third PAA; IV, fourth PAA; VI, sixth PAA; rc, right common carotid; lc, left common carotid; rs, right subclavian; ls, left subclavian; ao, aortic arch; h, heart; pt, pulmonary trunk; da, ductus arteriosus. (A–H) Ink-injected PAAs at E10.5. (A and B) Right and left lateral views of ink-filled WT PAAs after intracardiac ink injection. (C and D) Whi/+ embryos with absence of the fourth PAA (black arrows). (E–H) Chd7+/xk embryos with absent fourth PAA. (I–P) Great vessels in WT (I), Chd7+/xk (J–M), and Chd7+/xk;Tbx1+/– (N–P) embryos at E14.5. (J) IAA-B with a right-sided pulmonary trunk. (K) Cervical aortic arch and hypoplastic, ectopic thymic lobes. (L) ARS with hypoplastic right thymic lobe and left lobe removed to examine the aortic arch. (M) ARS. (N) Right brachiocephalic and cervical right subclavian overlying the right thymic lobe, as well as hypoplastic, irregular, and ectopic thymic lobes. (O) IAA-B and ARS. (P) IAA-B, right brachiocephalic artery overlying right thymic lobe, and cervical right subclavian, as well as hypoplastic, irregular, and ectopic thymic lobes. (Q) WT thymic lobes. (R) Chd7+/xk embryo with irregularly shaped thymic lobes. (S and T) Foxn1 in situ hybridization marking the E10.5 thymic primordium. (S) WT Foxn1 expression in the third pharyngeal pouch. (T) Chd7+/xk embryo. White arrow indicates absent Foxn1 expression. Scale bars: 100 μm (A–H, S, and T); 1 mm (I–R).
Table 1 .
PAA dysmorphogenesis in 2 Chd7 gene trap lines at E10.5 and E14.5
It is likely that the gene trap alleles represent loss of Chd7 function. We believe loss of function to be the case because the heterozygous phenotype is no more severe than that of Whi (missense) or other ENU mutants (22), and a stronger phenotype would be expected in a dominant negative. In addition, the XK403 trap is at the 3′ end of the gene (intron 36 of 37), whereas that of RRR136 is at the 5′ (intron 4; Supplemental Figure 5). It would be highly unusual for such different interruptions to give the same dominant-negative phenotype.
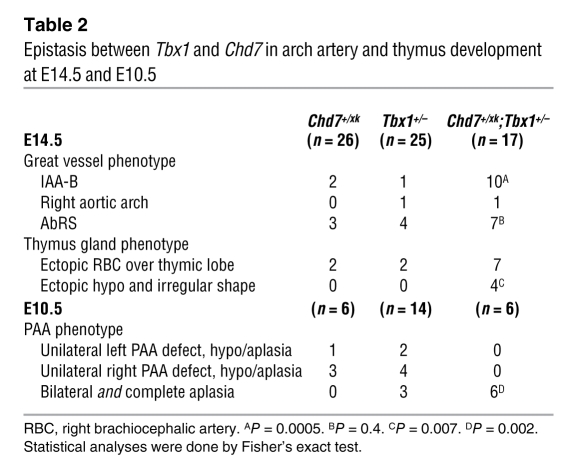
We reasoned that, if Tbx1 and Chd7 were within the same or a convergent developmental pathway, doubly heterozygous mice might show epistasis, a synergistic deficiency resulting in more frequent or severe effects (e.g., with novel defects and/or increased incidence of bilateral defects) than expected. The expected additive effect of both mutations was taken to be a simple sum of the frequency of abnormalities seen in the respective single mutants. The cardiovascular defect most specific for VCFS is IAA-B. It is estimated that 50% (32) to 80% (33) of patients presenting with IAA-B have a 22q11.2 deletion. To test for epistasis, we examined the frequency of IAA-B in Chd7+/xk;Tbx1+/– mice at E14.5 and found a significant increase in the incidence of this malformation (Table 2 and Figure 1, M–P). At E10.5, all 6 Chd7+/xk;Tbx1+/– mice studied had a complete, bilateral fourth PAA aplasia, compared with 1 of 9 single heterozygotes. We also found evidence for epistasis with regard to thymus development (Table 2) and ear development. We next tested the postnatal viability of single and double heterozygotes and found a significant reduction of living double heterozygotes compared with single heterozygotes (Supplemental Table 1).
Table 2 .
Epistasis between Tbx1 and Chd7 in arch artery and thymus development at E14.5 and E10.5
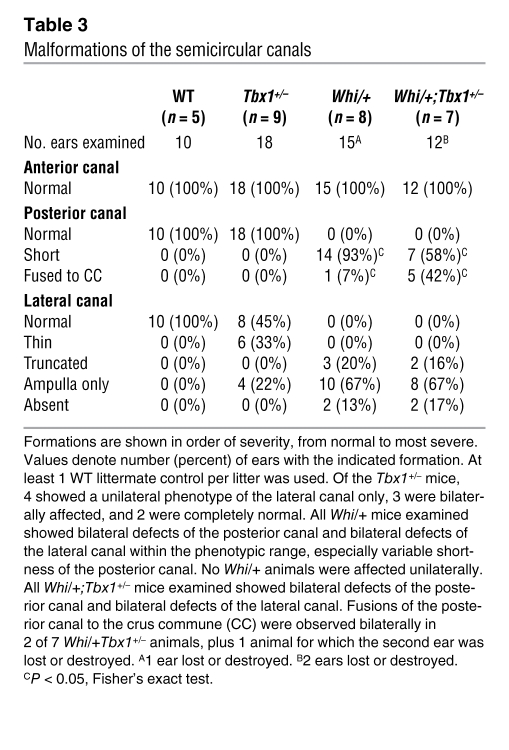
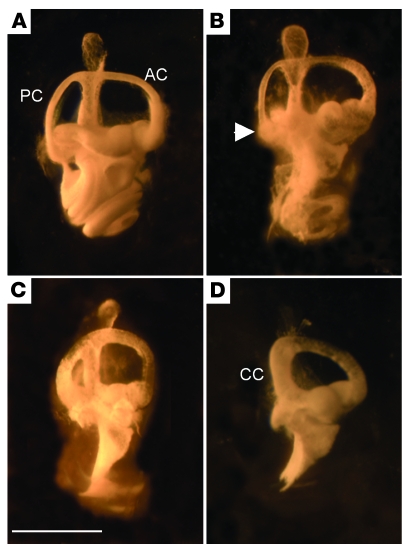
Inner-ear malformations, albeit of different types, have been previously noted in Tbx1-null and Chd7-heterozygous mice (13, 23). We examined paint-filled inner ears from E16.5 embryos to assess the extent of any malformations of the semicircular canals, which have been previously detailed in Chd7+/– mutant mice (21, 23, 24). Tbx1+/– mice had normal anterior and posterior canals, but showed a lateral canal defect in 56% of the ears (Table 3 and Figure 2). The whirligig heterozygote (Whi/+) phenotype was fully penetrant, but showed variability with respect to the severity of lateral canal truncations and posterior canal shortening (Table 3 and ref. 23). In Whi/+;Tbx1+/– mice, the underlying whirligig phenotype was enhanced. An interaction was revealed by more severe shortening of the posterior canal, leading to more Whi/+;Tbx1+/– than Whi/+ mice with posterior canals fused to the crus commune along their length (42% versus 7%; P < 0.05, Fisher’s exact test). The lateral canal defects were similar in the Whi/+;Tbx1+/– and Whi/+ mice. The anterior canal was not affected in any of the genotypes studied.
Table 3 .
Malformations of the semicircular canals
Figure 2. Malformations of the semicircular canals in Whi/+, Tbx1+/–, and Whi/+;Tbx1+/– mice.
(A) Paint-filled semicircular canals of the right ear of an E16.5 WT mouse showing normal canal morphology. (B) Tbx1+/– mouse displaying normal anterior (AC) and posterior canals (PC), but truncation of the lateral canal (arrowhead). (C) Whi/+ mouse showing normal morphology of the anterior canal, truncation of the lateral canal, and some shortening of the posterior canal. (D) Whi/+;Tbx1+/– mouse showing normal anterior canal, fusion of the posterior canal to the crus commune (CC), and absence of the lateral canal. Scale bar: 1 mm.
Tbx1 is thought to have both cell-autonomous and non–cell-autonomous effects on cardiovascular development.
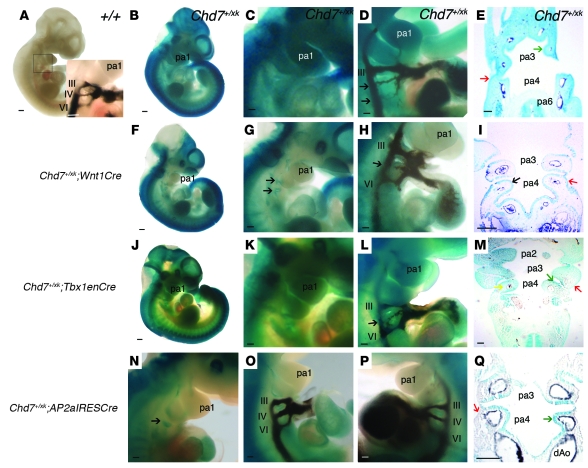
We therefore aimed to refine the tissue requirements for these genes during development. It has been postulated that CHARGE is secondary to a defective neural crest cell contribution during development (34–37). Unlike Tbx1, Chd7 is expressed in the CNS, neuroepithelium, and neural crest (38). In Chd7+/xk embryos, we observed some structural defects of cranial nerves and subtly altered neural crest migration to the caudal pharyngeal region (Figure 3). To test the role of Chd7 in neural crest with regard to PAA formation, we used a Wnt1Cre transgenic line to rescue Chd7 expression in this lineage (39). This is possible because the splice acceptor within the gene trap is flanked by loxP sites; thus, active Cre extinguishes β-gal expression as endogenous Chd7 is restored. Wnt1Cre gave efficient recombination in neural crest–derived tissues, as evidenced by the cleared LacZ staining in heterozygous animals (Figure 4, A–I). However, we continued to observe fourth PAA hypo/aplasia at the expected frequency (Figure 4H and Table 4). The clearance of β-gal expression from the neural crest–derived mesenchyme highlighted that the Chd7-driven reporter remained in the ectoderm and endoderm of E10.5 embryos (Figure 4I). We therefore explored the ability of a variety of Cre drivers to rescue the fourth PAA phenotype.
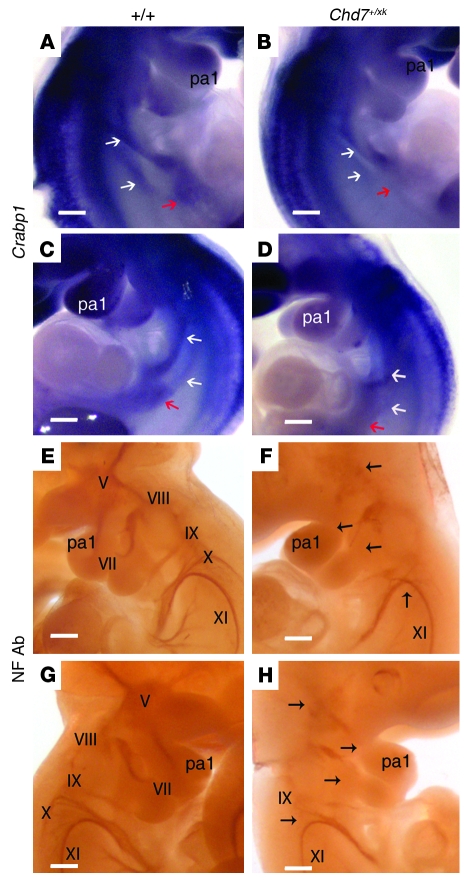
Figure 3. Neural crest and cranial nerve abnormalities in Chd7+/xk embryos.
(A–D) In situ hybridization with a Crabp1 probe, a marker of neural crest cells, at E10.75. White arrows denote 2 streams of neural crest cells populating the fourth and sixth pharyngeal arches. Red arrows show a population of cells in the region of the foregut. (A and C) WT Crabp1 expression at E10.5 in the pharyngeal region. (B and D) Right and left lateral view of Chd7+/xk embryos. The caudal stream of neural crest cells was reduced or absent. (E–H) Immunohistochemistry to detect neurofilaments at E10.5, with complete staining of cranial nerves V (trigeminal), VII (facial), VIII (vestibulocochlear), IX (glossopharyngeal), X (vagus), and XI (accessory). (E and G) In WT embryos, the trigeminal nerve was properly condensed and made full projections into the first branchial arch. (F and H) Chd7+/xk embryos showed reduced staining of all the nerves (expected locations indicated by arrows). The trigeminal nerve was not condensed, and the projections into the branchial arches were not complete. The vagus nerve was reduced and sometimes fused with the glossopharyngeal nerve. Scale bars: 200 μm.
Figure 4. Effects of Cre-mediated rescue of Chd7 mutation on the arch artery phenotype at E10.5.
(A) WT embryo. Inset shows the normal PAA configuration. (B–E) Chd7+/xk embryos. (B) β-geo expression driven by the Chd7 promoter. (C) Magnified view of the pharyngeal region, showing ubiquitous expression. (D) Ink-injected Chd7+/xk embryo, revealing hypoplastic right fourth and sixth PAAs. (E) Reporter expression in all pharyngeal tissues (stronger in endoderm and ectoderm; arrows). Dark staining in arteries is India ink. (F–I) Chd7+/xk;Wnt1Cre embryos. (F) Loss of reporter expression in neural crest–derived structures. (G) Magnified view of the pharyngeal region, with trap retained in pharyngeal endoderm (arrows). (H) Hypoplastic fourth PAA (arrow). (I) Trap expression was lost in the pharyngeal mesenchyme, but remained in epithelia (arrows). (J–M) Chd7+/xk;Tbx1enCre embryos. Whole-mount embryo (J) and magnified views (K) of the pharyngeal region showed reduced X-gal staining. (L) Hypoplastic right fourth PAA (arrow). (M) Coronal section across pharyngeal region of a Chd7+/xk;Tbx1enCre embryo. Staining was reduced in core mesoderm, but was retained in ectoderm (red arrow). (N–Q) Chd7+/xk;AP2aIRESCre embryos. (N) Pharyngeal region, demonstrating loss of reporter staining in pharyngeal mesenchyme, but with staining present in pharyngeal endoderm (arrow). (O and P) Normal PAAs. (Q) Pharyngeal arch section showing loss of reporter activity in ectoderm and neural crest cell–derived mesenchyme, but maintained activity in pharyngeal endoderm (green arrow). dAo, dorsal aorta. Scale bars: 20 mm (A, B, F, and J); 100 μm (C–E, G–I, and K–Q).
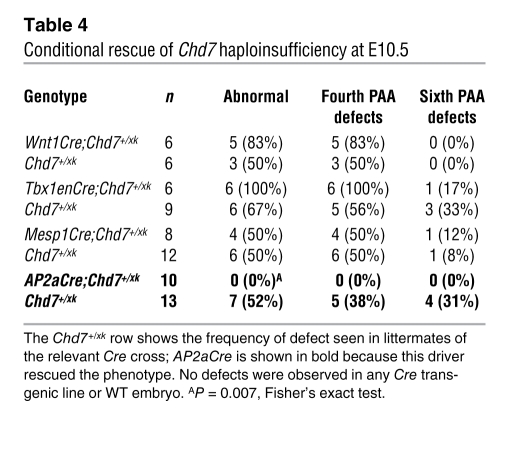
Table 4 .
Conditional rescue of Chd7 haploinsufficiency at E10.5
Given the epistasis with Tbx1, we expected Cre driven by the Tbx1 promoter to rescue the defect. We used a previously reported Tbx1 enhancer–driven Cre (Tbx1enCre; ref. 40) in order to avoid the complication of having a Tbx1Cre knockin (i.e., haploinsufficient) phenotype, but observed no significant rescue (Figure 4, J–M, and Table 4). However, examination of ROSA-R26R reporter expression in crosses with Tbx1enCre mice demonstrated poor recombination in the pharyngeal ectoderm and mosaic expression in the pharyngeal endoderm, but robust mesodermal expression that was broader than expression in WT Tbx1 mice (Supplemental Figure 3). These data suggest that in pharyngeal epithelia, but not in mesoderm, expression of Chd7 is required for PAA morphogenesis. Furthermore, no rescue was observed with a Mesp1Cre cross (Table 4), which directs recombination in mesodermal lineages (41).
Together, these data suggested that, as for Tbx1 (12), Chd7 is required in pharyngeal epithelium for fourth PAA growth and remodeling. To test this, we crossed the Chd7 gene trap mice to AP2aIRESCre mice. AP2aIRESCre drives recombination in the pharyngeal ectoderm and neural crest (42), which, in conjunction with the failure of Wnt1Cre to provide any phenotypic rescue, tests requirement of Chd7 in the pharyngeal ectoderm for normal fourth arch development. Full rescue of the arch artery defects was observed with AP2aIRESCre (Table 4 and Figure 4, N–Q). This finding demonstrated an ectodermal requirement for Chd7 in PAA development; however, because AP2aIRESCre will rescue Chd7 expression in both ectoderm and crest, we cannot rule out a subsidiary contributory role for neural crest expression of Chd7.
These data suggest that Chd7 and Tbx1 may share transcriptional targets, either directly or indirectly. Pharyngeal ectodermal signaling regulates mesenchymal cells populating and migrating through the third and fourth pharyngeal arches, and cells respond to a number of signaling cues that include FGFs. Tbx1 is known to be in epistasis with Fgf8, a growth factor required in the ectoderm for PAA morphogenesis and expressed in overlapping domains with Tbx1 at E10.5 (43–45). In Chd7+/xk embryos, we found no alteration in expression of Fgf8, nor in that of genes whose expression level correlates with Fgf signaling, such as Etv5 and Etv4, as assessed by in situ hybridization and real-time quantitative PCR (RTQPCR), respectively (Supplemental Table 2 and data not shown). Moreover, Fgf8 expression in the pharyngeal ectoderm is unaffected by loss of Tbx1 (12), overexpression of Fgf8 in Tbx1-null mice does not rescue the PAA phenotype (46), and Tbx1EnCre-induced Fgf8 conditional mutants do not demonstrate IAA (40). Finally, we found no epistasis between Chd7 and Fgf8 (Supplemental Table 3).
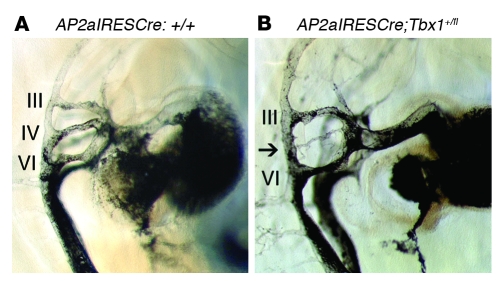
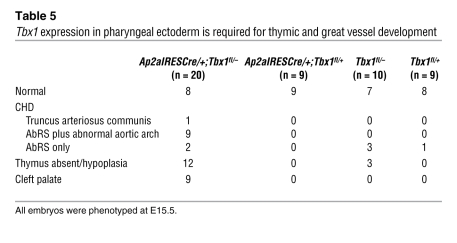
As mentioned above, previously published data point to a requirement for Tbx1 in the pharyngeal epithelium on the basis of conditional Tbx1 ablation with Fgf15Cre (12). To explore this further, we deleted Tbx1 using AP2aIRESCre and Wnt1Cre. Wnt1Cre deletion of Tbx1 caused no abnormality in conditional heterozygous and conditional null mutants (data not shown), consistent with the reported lack of Tbx1 expression in neural crest cells (12). We concluded that any PAA abnormalities found in AP2aIRESCre-deleted Tbx1 conditional mutants would be the result of loss of ectodermal Tbx1. Intracardiac ink injection of mouse embryos at E10.5 revealed fourth PAA hypoplasia in 76% of AP2aIRESCre-deleted conditional heterozygotes (n = 17; Figure 5), thereby recapitulating the fourth PAA defects previously reported in Tbx1+/– mice (5) and in Fgf15Cre-deleted conditional mutants (10). Corresponding great vessel defects were seen at E15.5 (Table 5). Surprisingly, AP2aIRESCre conditional null mutants had a broad spectrum of 22q11DS-like malformations, including aortic arch defects, thymic hypoplasia, and cleft palate. Thymic defects were an unexpected finding that, when considered with previously published data (10, 11), suggest that all 3 germ layers contribute to thymus development. Therefore, although Tbx1 is expressed transiently in pharyngeal ectoderm (10), it plays a critical role in PAA development. A single heart defect, namely VSD, was observed in E15.5 ectodermal Tbx1 mutants, in an embryo with truncus arteriosus communis (Table 5). At E18.5, 1 of 9 Ap2aIRESCre/+;Tbx1fl/– embryos had VSD (data not shown). Overall, our results demonstrated that both Tbx1 and Chd7 are required in the pharyngeal ectoderm for normal PAA development.
Figure 5. Conditional ablation of Tbx1 in pharyngeal ectoderm results in fourth PAA aplasia.
(A) Typical fourth PAA morphology in a Tbx1fl/fl control embryo at E10.5, revealed by intracardiac ink injection. (B) Hypoplasia of the fourth PAA (arrow) in a conditionally heterozygous AP2aIRESCre;Tbx1+/fl embryo at E10.5. Scale bar: 100 μm.
Table 5 .
Tbx1 expression in pharyngeal ectoderm is required for thymic and great vessel development
Discussion
The aorta, pulmonary, subclavian, and carotid arteries comprise the great vessels. They develop via remodeling of a bilaterally symmetrical system of paired PAAs connecting the aortic sac with the dorsal aortae. Disturbance of this process results in vascular malformations that may be associated with heart defects. The genetic networks controlling the formation and remodeling processes have been dissected using human genetics and animal models. Here we showed that, in mouse models of 2 important syndromes associated with great vessel defects, Chd7 and Tbx1 were in epistasis. Although epistasis between different genes mutated in the same human birth defect syndrome has been reported in a number of instances (47–49), interaction between genes haploinsufficient in distinct syndromes is rarer (50). In addition, both these genes were separately required to be expressed at biallelic levels in the embryonic pharyngeal ectoderm, emphasizing the importance of this epithelial tissue in PAA morphogenesis. Pharyngeal ectoderm has an important role in signaling to the neural crest (51, 52). Although FGF8 is known to be vital in this regard, it does not appear to be a major factor downstream of Chd7. The pharyngeal ectoderm makes no direct cellular contribution to the PAAs; thus, the effects of Tbx1 and Chd7 will be non–cell-autonomous. The neural crest is the most likely target tissue of Tbx1 and Chd7, based on proximity and the effects of Chd on neural crest migration seen in Chd7 heterozygotes.
Although small numbers of embryos were examined at E10.5, it is interesting that the proportion of PAA defects seen in Chd7+/xk embryos dropped from 4 of 6 at E10.5 to 5 of 26 at E14.5. This has similarities to the previously reported recovery, or catch-up, in PAA morphogenesis observed in Tbx1 heterozygotes (6). Recovery appeared attenuated in double-mutant embryos at E14.5, with 1 of 17 Chd7+/xk;Tbx1+/– embryos having normal great vessels at this stage. The basis for the recovery phenomenon is unknown. It is possible that reduction of signaling from pharyngeal ectoderm to neural crest is an early event, but that the same signal from other tissues at later stages of crest contribution can effect recovery. In this regard, the absence of observable great vessel defects in Ap2aIRESCre/+;Tbx1fl/+ embryos at E15.5 is interesting in that it represents a full rescue, possibly because of the higher levels of later signaling from tissues biallelic for Tbx1.
We also observed epistasis with regard to thymus development and an overall increase in embryonic and perinatal mortality in double heterozygotes, which suggests that additional abnormalities remain to be detected in these mice. Previous work has demonstrated ingress of inflammatory cells and otitis media in Tbx1+/– mice (53), but our data are the first to our knowledge to show a structural abnormality — a partially penetrant lateral canal hypoplasia — in heterozygotes. This further emphasizes the phenotypic overlap between the 2 haploinsufficiencies. Tbx1 and Chd7 demonstrated epistasis with regard to semicircular canal dysmorphogenesis. In particular, bilateral fusion of the posterior canal to the crus commune was observed in 2 of 7 embryos.
One obvious explanation for the multiple examples of Tbx1;Chd7 interaction would be a direct regulatory relationship between Tbx1 and Chd7. As a putative chromatin remodeler is expressed throughout development, the most likely scenario would be Chd7 modulating Tbx1 expression. However, testing at a variety of stages by in situ hybridization and RTQPCR analysis of each gene in embryos heterozygously mutated at the other locus did not demonstrate such a relationship between E8.5 and E10.5 (Supplemental Table 2 and Supplemental Figure 4). It is quite possible that changes in expression are too subtle to be detected in heterozygotes and that the severe growth delay seen in Chd7-null embryos obviates meaningful intralitter comparisons. We are currently deriving mice with a conditional allele of Chd7; it will be interesting to analyze the expression of Tbx1 in embryos in which Chd7 has been selectively ablated in ectodermal lineages. Conditional mutants will be invaluable in exploring additional tissue-specific roles for Chd7 — in tissues such as the neural crest and brain — and may therefore shed further light on the developmental pathology of CHARGE.
Despite Tbx1;Fgf8 epistasis, expression analysis of Fgf8 and Fgf signaling readouts in Chd7 mutants gave no support to the hypothesis that Fgf8 is a common target. Moreover, we failed to detect evidence of epistasis between Chd7 and Fgf8. Together, these data suggest that an Fgf8-independent ectoderm–to–neural crest signaling pathway is operative in great vessel development and controlled by Tbx1 and Chd7. Candidate downstream signaling molecules include Pdgf (54), semaphorins (55), slits and ephrins (56), and Vegf (57).
TBX1 is generally thought to be a transcriptional activator (3, 9, 58, 59). CHD7 has previously been shown to selectively interact with methylated H3K4 and H3K9, but not with acetylated histones, implying a role in transcriptional repression (60). For instance, CHD7 can bind phosphorylated SETDB1 and nemo-like kinase and repress PPARγ transactivation (60). Chromatin immunoprecipitation reveals that CHD7 binding correlates with a subset of H3K4Me sites, often distal to transcriptional start sites and within DNAse hypersensitive sites; binding to these sites can enhance transcription (60). Tbx1 was not a major binding site in DLD1 or ES cells (27). Although this might imply that Tbx1 and Chd7 do not act together at the same promoters, the ideal tissue and stage of development have not been examined. Alternatively, their downstream pathways may converge at a later point.
In summary, our data provide an embryological basis for the phenotypic overlap between the 22q11 deletion and CHARGE syndromes. The major respective genes involved, Tbx1 and Chd7, were both required in pharyngeal ectoderm for fourth PAA growth and remodeling and were in epistasis in this pathway. As the ectoderm makes no direct contribution to the PAAs, Tbx1 and Chd7 presumably regulate signaling from ectoderm to mesenchymal cells populating the arches, in a pathway in parallel to that involving Fgf8. Further dissection of this genetic and signaling network will be required for a fuller understanding of aortic arch malformations. Similarly, further work is required to investigate potential epistasis during development of second heart field structures and noncardiovascular structures. It is possible that gene interactions between loss-of-function alleles, or partial loss-of-function alleles, similar to that observed between Tbx1 and Chd7, underlies or modifies congenital heart defects in humans. At the clinical level, examining 22q11 in CHARGE cases without CHD7 mutation, and CHD7 in VCFS-like cases without 22q11 deletion or TBX1 mutation, may be a useful adjunct to current practice.
Methods
Study approval.
All human studies were undertaken with approval of the relevant ethical and licensing authorities; in the United Kingdom, this was the UCL-ICH/Great Ormond St. Hospital Research Ethics Committee (08/H0713/82). All patients provided informed consent for the molecular analysis conducted during this work, and separate consent was obtained for the patient photograph in Supplemental Figure 1 (OGO32; Ethical Committee of University Hospitals, Catholic University of Leuven). Animal experiments were conducted with licensing approval from the UK Home Office, after having first been approved by University College London’s animal ethics committee.
Comparative genome hybridization and FISH.
The protocols used have been described previously (28).
Mouse mutants, breeding, and genotyping.
The following mouse lines were obtained from A. Baldini (Telethon Institute of Genetics and Medicine, Napoli, Italy): AP2aIRESCre (42), Tbx1+/LacZ (5), Tbx1+/fl (12), and Tbx1mcm/+ (9). J. Epstein (University of Pennsylvania, Philadelphia, Pennsylvania, USA) provided Tbx1enCre (40). A. Moon (University of Utah, Salt Lake City, Utah, USA) provided AP2aIRESCre. M.A. Basson (King’s College London, London, United Kingdom) provided Fgf8Δ2,3/+ (61). Mesp1Cre (RBRC011450; ref. 41) was obtained from the RIKEN Bioresource Center. Wnt1Cre (39) was from P. Riley (Institute of Child Health, London, United Kingdom), and Whi (22) was from K.P. Steel (Sanger Centre, Cambridge, United Kingdom). Mice were maintained on a C57BL/6 or CD1 background. The mouse ES cell lines XK403 and RRR136 were obtained from Baygenomics, used to establish mouse lines using standard procedures, and bred onto a C57BL/6 background; Tbx1 and Chd7 mutants were backcrossed to this background for at least 5 generations. A schematic of the gene trap is given in Supplemental Figure 5. Embryos were harvested from timed matings and fixed overnight at room temperature in 4% formaldehyde. Mice were genotyped by PCR on yolk sacs or ear biopsies. Cre-expressing mice were determined using the following primers: forward, TGGAAAATGCTTCTGTCCGTTTGC; reverse, AACGAACCTGGTCGAAATCAGTG. Cre-expressing mice gave a product of approximately 300 bp, and wild-type mice no product, when visualized by gel electrophoresis. RRR mice were genotyped by the following primers: β-Geo forward, GTTTCAATATTGGCTTCATC; β-Geo reverse, CGCCGACGGCACGCTGATTG. Mutants gave a product of approximately 300 bp, and WT mice had no product. XK403 mice were genotyped using a 3-primer PCR: XKFx36, CAGGAGAAGAAAGGGTTCCTG; XKRx37, GGCAGGTCCTTCATTGGA; XKRgt, GTTTTCCCAGTCACGACGTT. Heterozygous mice produced 2 products, of about 1,500 and about 236 bp; wild-type mice produced the 236-bp product only.
RTQPCR.
cDNA was prepared from whole embryos using standard methods. Whole embryos were homogenized in 600 μl buffer RLT and purified using the RNeasy procedure (Qiagen). To avoid contamination with DNA, a DNase digestion step was included in the purification process using RNase free DNase I (Qiagen). cDNA was synthesized from 1 μg total RNA with 3 μl of 100 ng/μl random primers (Promega) and mixed with water to a total volume of 37 μl. This mixture was incubated at 65°C for 5 minutes. We added 10 μl of 5× reverse transcriptase buffer and 2 μl of 10 mM dNTP together with 1 μl 200 U/μl Superscript reverse transcriptase II (Invitrogen) to a total volume of 50 μl and incubated at 37°C for 1 hour and 90°C for 5 minutes to inactivate the reaction. Quantitative PCR was performed with Tbx1 Assay on Demand Mm00448948_m1 (Applied Biosystems) with mouse GAPDH used as an endogenous control (Applied Biosystems). Primers for Chd7 were synthesized by Sigma-Aldrich: forward, GGAGAACCCTGAGTTTGCTG; reverse, CCCTGAAGTAGAGGCGACAG. Tubulin was used as an endogenous control: forward, TCACTGTGCCTGACTTACC; reverse, GGAACATAGCCGTAACTGC. Reactions were performed in quadruplicate on a 7900HT Fast Real Time PCR System (Applied Biosystems). Relative quantitation was calculated by the ΔΔCt method (62).
X-gal staining, histology, and RNA in situ hybridization.
To visualize β-gal activity, formaldehyde-fixed embryos were stained using X-gal substrate according to standard procedures. Stained embryos were photographed as whole-mount specimens, dehydrated, embedded in paraffin wax, and cut into 6-μm histological sections for photography. RNA in situ hybridization experiments were performed according to a previously published protocol (63).
Immunohistochemistry.
Formaldehyde-fixed whole embryos were washed with PBS plus 1% Tween 20 (PTW) dehydrated through an alcohol/PTW series, bleached (10% H2O2 in 80% MeOH/20% DMSO), and washed in blocking solution (5% sheep serum with 1 mg/ml BSA). They were then incubated in primary antibody (2H3; Developmental Studies Hybridoma Bank, University of Iowa) diluted 1:200 in blocking solution overnight at 4°C. Embryos were washed again in blocking solution and then incubated with the secondary antibody HRP-conjugated anti-IgG (Sigma-Aldrich) diluted 1:200 in blocking solution overnight at 4°C. Embryos were washed again in PTW. The color reaction was developed using liquid DAB (Sigma-Aldrich).
Ink injections and paint filling.
To visualize the aortic arch artery system at different developmental stages, embryos were injected with India ink via the outflow tract of the heart. Inner ears were dissected from E16.5 embryos, fixed, and cleared in methyl salicylate, and the lumen of each was filled with white paint as described previously (21).
Statistics.
Intergroup comparisons of mouse embryo phenotypes were made using Fisher’s exact test. A P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported by the British Heart Foundation and the European Union (to P.J. Scambler); by the Wellcome Trust (to K. Prescott and C. Shaw-Smith); by the National Heart, Lung, and Blood Institute, NIH, and the Dulbecco Telethon Institute (to E. Illingworth); and by the Medical Research Council (to M.A. Basson). We thank Antonio Baldini for helpful advice and provision of mouse mutants, Anne Moon for AP2aIRESCre, and Jon Epstein for Tbx1enCre.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 119:3301–3310 (2009). doi:10.1172/JCI37561.
References
- 1. Scambler, P.J. 2006. DiGeorge syndrome and velocardiofacial syndrome (VCFS). InEncyclopedia of life sciences . D. Cooper, editor. John Wiley & Sons, Ltd. Chichester, United Kingdom. doi:10.1038/npg.els.0006074. [Google Scholar]
- 2.Yagi H., et al. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/S0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 3.Paylor R., et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: Implications for 22q11 deletion syndrome. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zweier C., Sticht H., Aydin-Yaylagul I., Campbell C.E., Rauch A. Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. Am. J. Hum. Genet. 2007;80:510–517. doi: 10.1086/511993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindsay E.A., et al. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. . Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay E.A., Baldini A. Recovery from arterial growth delay reduces penetrance of cardiovascular defects in mice deleted for the DiGeorge syndrome region. Hum. Mol. Genet. 2001;10:997–1002. doi: 10.1093/hmg/10.9.997. [DOI] [PubMed] [Google Scholar]
- 7.Jerome L.A., Papaioannou V.E. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. . Nat. Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 8.Merscher S., et al. TBX1 is responsible for the cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. . Cell. 2001;104:619–629. doi: 10.1016/S0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 9.Xu H., et al. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development. 2004;131:3217–3227. doi: 10.1242/dev.01174. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., Huynh T., Baldini A. Mesodermal expression of Tbx1 is necessary and sufficient for pharyngeal arch and cardiac outflow tract development. Development. 2006;133:3587–3595. doi: 10.1242/dev.02539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnold J.S., et al. Inactivation of Tbx1 in the pharyngeal endoderm results in 22q11DS malformations. Development. 2006;133:977–987. doi: 10.1242/dev.02264. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z., et al. Tbx1 expression in pharyngeal epithelia is necessary for pharyngeal arch artery development. Development. 2005;132:5307–5315. doi: 10.1242/dev.02086. [DOI] [PubMed] [Google Scholar]
- 13.Xu H., et al. Tbx1 regulates population, proliferation and cell fate determination of otic epithelial cells. Dev. Biol. 2006;302:670–682. doi: 10.1016/j.ydbio.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blake K.D., Prasad C. CHARGE syndrome. Orphanet. J. Rare. Dis. 2006;1:34. doi: 10.1186/1750-1172-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lalani S.R., et al. SEMA3E mutation in a patient with CHARGE syndrome. J. Med. Genet. 2004;41:e94. doi: 10.1136/jmg.2003.017640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iafolla A.K., McKonkie-Rosell A., Kahler S.G.1990CHARGE association with complete or partial DiGeorge sequence in 4 patients [abstract]. Am. J. Hum. Genet. 47622349951 [Google Scholar]
- 17.de Lonlay-Debeney P., et al. Features of DiGeorge syndrome and CHARGE association in five patients. J. Med. Genet. 1997;34:986–989. doi: 10.1136/jmg.34.12.986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Digilio M.C., et al. Radial aplasia and chromosome 22q11 deletion. J. Med. Genet. 1997;34:942–944. doi: 10.1136/jmg.34.11.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emanuel B.S., Budarf M.L., Sellinger B., Goldmuntz E., Driscoll D.A.1992Detection of microdeletions of 22q11.2 with fluorescence in situ hybridisation (FISH): diagnosis of DiGeorge syndrome (DGS), velo-cardio-facial (VCF) syndrome, CHARGE association and conotruncal cardiac malformations [abstract]. Am. J. Hum. Genet. 5111609788 [Google Scholar]
- 20.Devriendt K., Swillen A., Fryns J.P. Deletion in chromosome region 22q11 in a child with CHARGE association. Clin. Genet. 1998;53:408–410. doi: 10.1111/j.1399-0004.1998.tb02755.x. [DOI] [PubMed] [Google Scholar]
- 21.Kiernan A.E., et al. ENU mutagenesis reveals a highly mutable locus on mouse Chromosome 4 that affects ear morphogenesis. Mamm. Genome. 2002;13:142–148. doi: 10.1007/s0033501-2088-9. [DOI] [PubMed] [Google Scholar]
- 22.Bosman E.A., et al. Multiple mutations in mouse Chd7 provide models for CHARGE syndrome. Hum. Mol. Genet. 2005;14:3463–3476. doi: 10.1093/hmg/ddi375. [DOI] [PubMed] [Google Scholar]
- 23.Hawker K., Fuchs H., Angelis M.H., Steel K.P. Two new mouse mutants with vestibular defects that map to the highly mutable locus on chromosome 4. Int. J. Audiol. 2005;44:171–177. doi: 10.1080/14992020500057434. [DOI] [PubMed] [Google Scholar]
- 24.Alavizadeh A., et al. The Wheels mutation in the mouse causes vascular, hindbrain, and inner ear defects. Dev. Biol. 2001;234:244–260. doi: 10.1006/dbio.2001.0241. [DOI] [PubMed] [Google Scholar]
- 25.Hurd E.A., et al. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm. Genome. 2007;18:94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- 26.Fulcoli F.G., Huynh T., Scambler P.J., Baldini A. Tbx1 regulates the BMP-Smad1 pathway in a transcription independent manner. PLoS ONE. 2009;4:e6049. doi: 10.1371/journal.pone.0006049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Schnetz M.P., et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prescott K., et al. A novel 5q11.2 deletion detected by microarray comparative genomic hybridisation in a child referred as a case of suspected 22q11 deletion syndrome. Hum. Genet. 2005;116:83–90. doi: 10.1007/s00439-004-1195-6. [DOI] [PubMed] [Google Scholar]
- 29.Writzl K., Cale C.M., Pierce C.M., Wilson L.C., Hennekam R.C. Immunological abnormalities in CHARGE syndrome. Eur. J. Med. Genet. 2007;50:338–355. doi: 10.1016/j.ejmg.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Volpe P., et al. 22q11 deletions in fetuses with malformations of the outflow tracts or interruption of the aortic arch: impact of additional ultrasound signs. Prenat. Diagn. 2003;23:752–757. doi: 10.1002/pd.682. [DOI] [PubMed] [Google Scholar]
- 31.Skarnes W.C., et al. A public gene trap resource for mouse functional genomics. Nat. Genet. 2004;36:543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewin M.B., et al. A genetic etiology for interruption of the aortic arch type B. Am. J. Cardiol. 1997;80:493–497. doi: 10.1016/S0002-9149(97)00401-3. [DOI] [PubMed] [Google Scholar]
- 33.Rauch A., et al. Incidence and significance of 22q11.2 hemizygosity in patients with interrupted aortic arch. Am. J. Med. Genet. 1998;78:322–331. doi: 10.1002/(SICI)1096-8628(19980724)78:4<322::AID-AJMG4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 34.Siebert J.R., Graham J.M., MacDonald C. Pathologic features of the CHARGE association: support for involvement of the neural crest. Teratology. 1985;31:331–336. doi: 10.1002/tera.1420310303. [DOI] [PubMed] [Google Scholar]
- 35.Beckwith J.B. Cardiovascular malformations and the neural crest. Pediatr. Radiol. 1989;19:122–123. doi: 10.1007/BF02387901. [DOI] [PubMed] [Google Scholar]
- 36.Squires L.A., Dieffenbach A.Z., Betz B.W. Three malformation complexes related to neural crest development. Brain Dev. 1998;20:183–185. doi: 10.1016/S0387-7604(98)00013-8. [DOI] [PubMed] [Google Scholar]
- 37.Sanlaville D., et al. Phenotypic spectrum of CHARGE syndrome in fetuses with CHD7 truncating mutations correlates with expression during human development. J. Med. Genet. 2006;43:211–217. doi: 10.1136/jmg.2005.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aramaki M., et al. Embryonic expression profile of chicken CHD7, the ortholog of the causative gene for CHARGE syndrome. Birth Defects Res. A Clin. Mol. Teratol. 2007;79:50–57. doi: 10.1002/bdra.20330. [DOI] [PubMed] [Google Scholar]
- 39.Danielian P.S., et al. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998;8:1323–1326. doi: 10.1016/S0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 40.Brown C.B., et al. Cre-mediated excision of Fgf8 in the Tbx1 expression domain reveals a critical role for Fgf8 in cardiovascular development in the mouse. Dev. Biol. 2004;267:190–202. doi: 10.1016/j.ydbio.2003.10.024. [DOI] [PubMed] [Google Scholar]
- 41.Saga Y., et al. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 42.Macatee T.L., et al. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vitelli F., et al. A genetic link between Tbx1 and fibroblast growth factor signaling. Development. 2002;129:4605–4611. doi: 10.1242/dev.129.19.4605. [DOI] [PubMed] [Google Scholar]
- 44.Park E.J., et al. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frank D.U., et al. An Fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitelli F., et al. Fgf8 expression in the Tbx1 domain causes skeletal abnormalities and modifies the aortic arch but not the outflow tract phenotype of Tbx1 mutants. Dev. Biol. 2006;295:559–570. doi: 10.1016/j.ydbio.2006.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Badano J.L., et al. Heterozygous mutations in BBS1, BBS2 and BBS6 have a potential epistatic effect on Bardet-Biedl patients with two mutations at a second BBS locus. Hum. Mol. Genet. 2003;12:1651–1659. doi: 10.1093/hmg/ddg188. [DOI] [PubMed] [Google Scholar]
- 48.Badano J.L., et al. Dissection of epistasis in oligogenic Bardet-Biedl syndrome. Nature. 2006;439:326–330. doi: 10.1038/nature04370. [DOI] [PubMed] [Google Scholar]
- 49.Sato-Jin K., et al. Epistatic connections between microphthalmia-associated transcription factor and endothelin signaling in Waardenburg syndrome and other pigmentary disorders. FASEB J. 2008;22:1155–1168. doi: 10.1096/fj.07-9080com. [DOI] [PubMed] [Google Scholar]
- 50.Tory K., et al. High NPHP1 and NPHP6 mutation rate in patients with Joubert syndrome and nephronophthisis: potential epistatic effect of NPHP6 and AHI1 mutations in patients with NPHP1 mutations. J. Am. Soc. Nephrol. 2007;18:1566–1575. doi: 10.1681/ASN.2006101164. [DOI] [PubMed] [Google Scholar]
- 51.Basch M.L., Bronner-Fraser M. Neural crest inducing signals. Adv. Exp. Med. Biol. 2006;589:24–31. doi: 10.1007/978-0-387-46954-6_2. [DOI] [PubMed] [Google Scholar]
- 52.Kameda Y. Hoxa3 and signaling molecules involved in aortic arch patterning and remodeling. Cell Tissue Res. 2009;336:165–178. doi: 10.1007/s00441-009-0760-7. [DOI] [PubMed] [Google Scholar]
- 53.Liao J., et al. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Hum. Mol. Genet. 2004;13:1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- 54.Ding H., et al. The mouse Pdgfc gene: dynamic expression in embryonic tissues during organogenesis. Mech. Dev. 2000;96:209–213. doi: 10.1016/S0925-4773(00)00425-1. [DOI] [PubMed] [Google Scholar]
- 55.Toyofuku T., et al. Repulsive and attractive semaphorins cooperate to direct the navigation of cardiac neural crest cells. Dev. Biol. 2008;321:251–262. doi: 10.1016/j.ydbio.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 56.Kuriyama S., Mayor R. Molecular analysis of neural crest migration. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:1349–1362. doi: 10.1098/rstb.2007.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stalmans I., et al. VEGF: A modifier of the del22q11 (DiGeorge) syndrome? Nat. Med. 2003;9:173–182. doi: 10.1038/nm819. [DOI] [PubMed] [Google Scholar]
- 58.Ataliotis P., Ivins S., Mohun T.J., Scambler P.J. XTbx1 is a transcriptional activator involved in head and pharyngeal arch development in Xenopus laevis. Dev. Dyn. 2005;232:979–991. doi: 10.1002/dvdy.20276. [DOI] [PubMed] [Google Scholar]
- 59.Nowotschin S., et al. Tbx1 affects asymmetric cardiac morphogenesis by regulating Pitx2 in the secondary heart field. Development. 2006;133:1565–1573. doi: 10.1242/dev.02309. [DOI] [PubMed] [Google Scholar]
- 60.Takada I., et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat. Cell Biol. 2007;9:1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- 61.Meyers E.N., Lewandoski M., Martin G.R. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat. Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 62.Yuan J.S., Wang D., Stewart C.N., Jr. Statistical methods for efficiency adjusted real-time PCR quantification. Biotechnol. J. 2008;3:112–123. doi: 10.1002/biot.200700169. [DOI] [PubMed] [Google Scholar]
- 63.Streit A., et al. Chordin regulates primitive streak development and the stability of induced neural cells, but is not sufficient for neural induction in the chick embryo. Development. 1998;125:507–519. doi: 10.1242/dev.125.3.507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.