Abstract
Histones are characterized by numerous posttranslational modifications that influence gene transcription1,2. However, because of the lack of global distribution data in higher eukaryotic systems3, the extent to which gene-specific combinatorial patterns of histone modifications exist remains to be determined. Here, we report the patterns derived from the analysis of 39 histone modifications in human CD4+ T cells. Our data indicate that a large number of patterns are associated with promoters and enhancers. In particular, we identify a common modification module consisting of 17 modifications detected at 3,286 promoters. These modifications tend to colocalize in the genome and correlate with each other at an individual nucleosome level. Genes associated with this module tend to have higher expression, and addition of more modifications to this module is associated with further increased expression. Our data suggest that these histone modifications may act cooperatively to prepare chromatin for transcriptional activation.
Histones are subject to numerous covalent modifications, including methylation and acetylation, that occur mainly at their N-terminal tails and that can affect transcription of genes1,2,4,5. Extensive studies have established that histone acetylation is primarily associated with gene activation, whereas methylation, depending on its position and state, is associated with either repression or activation5–10. Various models, including the histone code, the signaling network and the charge neutralization model, have been proposed to account for the function of histone modifications11–14. The histone code hypothesis suggests that multiple histone modifications act in a combinatorial fashion to specify distinct chromatin states. However, the extent to which combinatorial patterns of histone modifications exist in the genome is unknown. We have now produced genome-wide maps of 18 histone acetylations (H2AK5ac, H2AK9ac, H2BK5ac, H2BK12ac, H2BK20ac, H2BK120ac, H3K4ac, H3K9ac, H3K14ac, H3K18ac, H3K23ac, H3K27ac, H3K36ac, H4K5ac, H4K8ac, H4K12ac, H4K16ac and H4K91ac) at an individual nucleosome level (see Methods section for data deposition), and analyzed these together with the H2A.Z and 19 histone methylation maps we generated previously15.
We first systematically evaluated the specificities of the acetylation antibodies used in this study (Supplementary Methods, Supplementary Table 1 and Supplementary Fig. 1 online). Competition assays using modified and unmodified peptides indicated that most antibodies showed specificity for the desired acetylation (Supplementary Fig. 1). The H4K5ac and H3K4ac antibodies demonstrated some crossreactivity toward H4K12ac and H3K9ac, respectively, in a condition with excess competitor peptides (Supplementary Fig. 1d,j), and the H4K91ac antibody did not work in protein blotting. Thus, the results for these modifications should be interpreted with caution. Of note, H2AK9ac has not been reported previously, and H3K4ac has only been identified by mass-spectrometry analysis and has not been previously characterized functionally16. Protein blotting indicated that these acetylations indeed exist in human CD4+ T cells (Supplementary Fig. 1j,o). We previously analyzed the genome-wide distribution of H2BK5me1 (ref. 15), and protein blotting data in this study indicated that this methylation exists in human cells and that the H2BK5me1 antibody is specific (Supplementary Fig. 1p).
Next, we determined the genomic distribution patterns of these histone acetylations using the ChIP-Seq technique15, which we previously confirmed yields H3K4me3 distribution patterns similar to those generated by the ChIP-SAGE (GMAT) strategy15,17. To validate the histone acetylation data, we compared the genomic distribution patterns of the K9/K14-diacetylated histone H3 from ChIP-SAGE18 with the separately examined patterns of H3K9ac and H3K14ac in this study (Supplementary Fig. 2 online). We found that the ChIP-Seq acetylation data are reliable and that the previously observed H3K9/K14 diacetylation patterns could be primarily attributed to H3K9 acetylation.
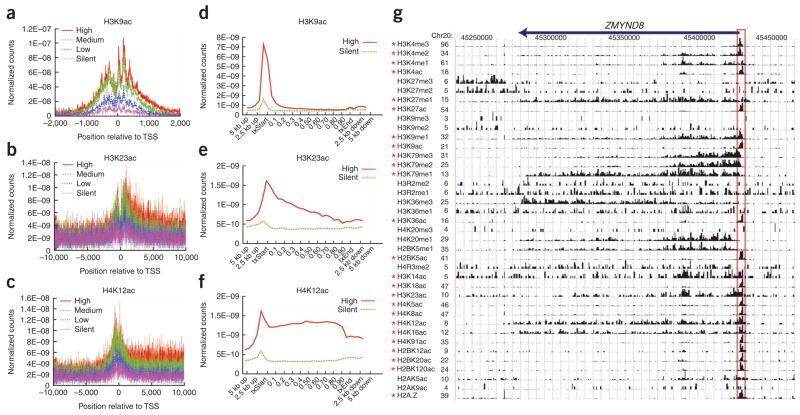
To examine the distribution of the histone acetylations at different functional regions, we generated composite profiles for the region spanning the transcription start sites (TSSs; Fig. 1a–c and Supplementary Fig. 3 online) or the entire gene bodies and extending 5 kb upstream and 5 kb downstream for groups of 1,000 genes according to their expression (Fig. 1d–f and Supplementary Fig. 4 online). We found that all acetylations positively correlated with gene expression, consistent with their involvement in transcriptional activation. However, our data indicate that different acetylations may target different regions of genes. For example, H2AK9ac, H2BK5ac, H3K9ac, H3K18ac, H3K27ac, H3K36ac and H4K91ac are mainly located in the region surrounding the TSS (Fig. 1d and Supplementary Fig. 4), whereas H2BK12ac, H2BK20ac, H2BK120ac, H3K4ac, H4K5ac, H4K8ac, H4K12ac and H4K16ac are elevated in the promoter and transcribed regions of active genes (Fig. 1f and Supplementary Fig. 4). These results are consistent with previous reports that specific histone acetyl transferases (HATs) can associate with different regions of genes. For example, PCAF associates with the elongation-competent RNA Pol II, whereas p300 interacts with the initiation-competent RNA Pol II (ref. 19). Additionally, depletion of GCN5 or PCAF, but not CBP or p300, affects H4K8ac and H3K14ac20. The distribution patterns of these histone acetylations and histone methylations are exemplified by the genomic locus for ZMYND8 (also known as PRKCBP1), which is expressed in CD4+ T cells (Fig. 1g). The promoter region (highlighted in red), which was defined as a 2-kb region surrounding the TSS, is associated with 25 modifications (P <10−7).
Figure 1.
Three distribution patterns of histone acetylations. (a–c) Normalized tag counts of histone acetylation signals surrounding the TSS were indicated for highly active, intermediately active (two levels) and silent genes. Each group represents 1,000 genes with similar expression, as described in Methods. (d–f) Normalized tag counts of histone acetylation signals of 1,000 highly active or silent genes across the gene bodies. The plots extend 5 kb 5′ and 3′ of the genic regions (see Methods). txStart, transcription start site; txEnd, transcription end. (g) Chromatin modification patterns at the ZMYND8 (PRKCBP1) gene locus. Significant modifications in the −1 kb to +1 kb region surrounding the TSS (P <10−7; highlighted in red) are indicated by asterisks on the left.
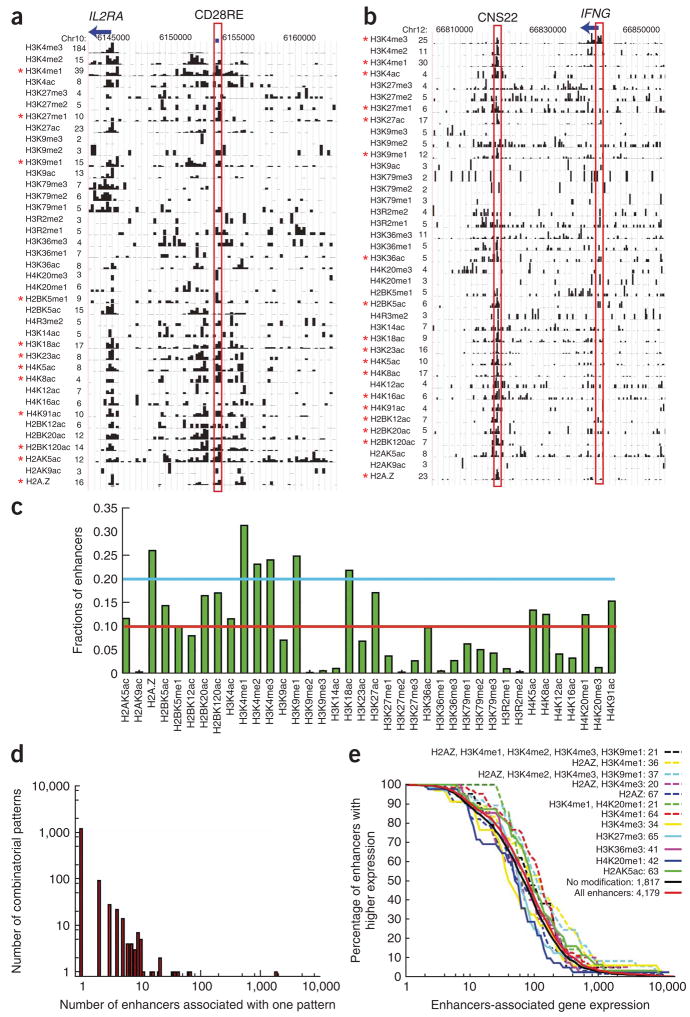
To identify the patterns of histone modifications in an unbiased way, we examined each of the 12,541 gene promoters for association with each of the 18 acetylations, 19 methylations and H2A.Z. Of the possible patterns, only a small fraction exists at promoters. Of 4,339 detected patterns, 1,174 are associated with multiple genes and 3,165 with only one gene each (Fig. 2a). The 13 most prevalent patterns are each associated with more than 62 genes. We next examined the expression of genes in these patterns, using the mean expression of all genes as a reference (Fig. 2b). It seems that we can roughly classify these top patterns into three classes (I, II and III in Fig. 2b) according to expression. Four of six patterns in class I contain H3K27me3 and correlate with low expression. These patterns also contain H3K4me1/2/3, H3K9me1 and H2A.Z but no acetylations. The patterns containing only H3K4me3 or no modification also belong to this class. Class II contains H3K36me3 or a modification backbone consisting of 17 modifications (as discussed below), or the backbone plus H4K16ac, which correlates with intermediate gene expression. Class III shows the highest expression, and it includes H2BK5me1, H4K16ac, H4K20me1 and H3K79me1/2/3 in addition to the modification backbone (Fig. 2b). Our Gene Ontology analysis suggests that genes involved in cellular physiology and metabolism are enriched in the active class III patterns, consistent with their house-keeping roles (data not shown). In contrast, many genes involved in development, cell–cell signaling and synaptic transmission are enriched in the inactive class I patterns, consistent with their not being required for mature T-cell function.
Figure 2.
Patterns of histone modifications associated with promoters. (a) Patterns of histone modifications at promoters. The y axis indicates the number of patterns of 39 histone modifications (see Methods), and the x axis indicates the number of promoters associated with each pattern. (b) Correlation of gene expression with the thirteen most prevalent modification patterns. B, the 17-modification backbone; All, all genes. The number of promoters within each pattern is also indicated. The gene expression is determined using DNA microarrays. See Figure 4a for the composition of ‘backbone’. (c) Correlation of each modification with gene expression. The changes in gene expression (log2) are calculated by comparing the average expression of the subsets of genes with or without a particular modification.
To correlate each modification with gene expression, we compared the average gene expression with or without each modification (Fig. 2c). H3K27me3 was among a group of repressive marks also including H3K27me2, H3K9me2, H3K9me3 and H4K20me3, whereas most other modifications correlated with activation. Although the modification patterns do not uniquely determine the extent of expression, the H3K79me3 and H2BK5ac modifications showed weak correlation with expression within a modification pattern (Supplementary Fig. 5 online).
Enhancers of transcription have been identified using modification patterns15,18,21,22. We previously found that all three H3K4 methylation states and H2A.Z were detected at enhancers15. However, others identified enhancers associated with high H3K4me1 but very low H3K4me3 (refs. 21,23). These results suggest that there are distinctive modification patterns at different enhancers. Indeed, the CD28 response element (CD28RE) enhancer24 of the IL2RA gene is associated with 12 modifications (P <10−7), whereas 18 modifications are detected at the CNS22 enhancer25 of the IFNG gene (Fig. 3a,b).
Figure 3.
Patterns of histone modifications at enhancers. (a) The histone modification pattern at the CD28RE enhancer (highlighted in red) of the IL2RA gene. Significant modifications are indicated by asterisks on the left. (b) Histone modification patterns at the IFNG gene and its downstream enhancer, CNS22, are shown. Significant modifications at CNS22 are indicated by asterisks on the left. (c) The fractions of enhancers associated with each of the 38 modifications. (d) Patterns of histone modifications at 4,179 DNase hypersensitive sites. The y axis indicates the number of patterns, and x axis indicates the number of hypersensitive sites associated with each pattern. (e) Correlation analysis of gene expression with the ten largest modification patterns by assigning an enhancer to the TSS of the nearest known gene. All, all DNase I hypersensitive sites. The number of hypersensitive sites associated with each pattern is indicated.
We next examined histone modification patterns at 4,179 intergenic DNase I hypersensitive sites26, excluding those in promoter regions and miRNA genes, as a set of putative enhancers. As compared to the whole genome, these putative enhancer regions showed an elevated sum of all modifications (Supplementary Fig. 6a online). Each of H2A.Z, H3K4me1, H3K4me2, H3K4me3, H3K9me1 and H3K18ac associated with more than 20% of enhancers (Fig. 3c). Several modifications were associated with very few enhancers, including both repressive marks (H3K9me2, H3K9me3 and H3K27me2) and activating marks (H2AK9ac and H3K14ac) (Fig. 3c). Among the total of 1,389 patterns, 195 are associated with multiple hypersensitive sites and 1,194 associated with a distinct hypersensitive site (Fig. 3d). The top 12 patterns include those associated with only one modification or combinations of H3K4me1, H3K4me2, H3K4me3, H3K9me1, H3K36me3, H4K20me1 and H2A.Z (Fig. 3e). No modifications were detected at 1,817 hypersensitive sites, perhaps indicating nucleosome-free regions or insufficient sequencing. These data suggest that there are multiple patterns associated with enhancers. However, we did not find significant correlations between modification patterns at enhancers and gene expression (Fig. 3e and Methods).
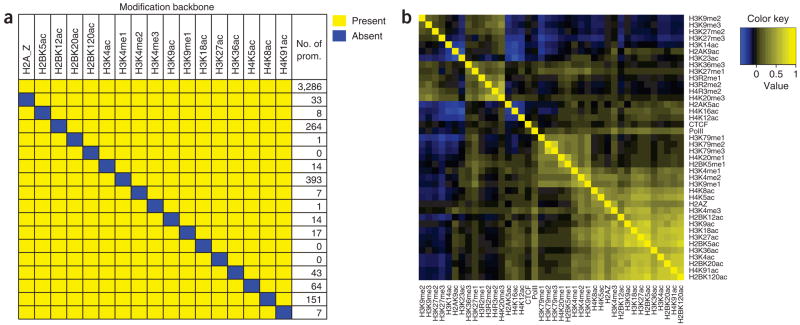
These results suggest that multiple histone modifications can associate with critical regulatory elements of transcription. To identify modifications that may function together to modify chromatin, we searched for robust modification features at promoter regions. This analysis revealed a modification ‘backbone’ consisting of 17 modifications (H2A.Z, H2BK5ac, H2BK12ac, H2BK20ac, H2BK120ac, H3K4ac, H3K4me1, H3K4me2, H3K4me3, H3K9ac, H3K9me1, H3K18ac, H3K27ac, H3K36ac, H4K5ac, H4K8ac and H4K91ac) that were present in 821 different patterns and associated with a total of 3,286 genes. To examine whether the modifications in this group tend to coexist, we counted the promoters missing one of these modifications. Notably, only a small number of promoters (0–393) were identified as missing one of these 17 modifications, as compared to 3,286 promoters that had all 17 modifications (Fig. 4a). This backbone of modifications is robust against perturbations (Supplementary Fig. 6b). Furthermore, none of the promoters possessing the backbone had significant levels of the repressive mark H3K27me3. We verified the existence of the ‘backbone’ modification by principal component analysis, which revealed that 16 of the 17 modifications are highly clustered in the first principal component (Supplementary Fig. 7 online) and provided further evidence that the modifications in the backbone can be classified in the same group. Genes with this backbone seem to have higher expression than the genes with no modifications (Fig. 2b, class II).
Figure 4.
Correlation of histone modifications in the genome. (a) A group of 17 modifications, the ‘backbone’ in Figure 2b, tend to coexist. The numbers of promoters associated with all the 17 modifications or missing any one modification (the blue rectangle) are indicated on the right. (b) Tags are binned into nonoverlapping 200-bp bins along the genome. Using the tallies for each bin, we calculated a pairwise Spearman correlation coefficient for each pair of modifications to create a correlation coefficient matrix. A heat map was then generated from this matrix.
The tendency of the 17 modifications to coexist suggests that some of these modifications may be present at the same nucleosome. We tested this hypothesis by carrying out a pairwise correlation analysis at a single-nucleosome level of 18 histone acetylations, 18 histone methylations, CTCF, Pol II and H2A.Z using 200-bp windows for the entire genome (see Supplementary Table 2 online for the top correlated pairs (r > 0.4; P < 0.01)). A heat map based on the mean correlation coefficients indicates that a cluster of strong positively correlated pairs was observed at the lower-right corner (Fig. 4b). All 14 modifications within this cluster (r > 0.4; P < 0.01) belong to the 17-modification backbone module found at promoters. Another cluster of positively correlated pairs contains modifications enriched in transcribed regions of genes, including H4K20me1, H2BK5me1, H3K9me1, H3K4me1/2 and H3K79me1/2/3. Pol II clustered together with these modifications rather than the promoter-centric modifications. Modifications related to gene silencing, including H3K27me2, H3K27me3, H3K9me2 and H3K9me3, were clustered together (Fig. 4b). Many correlated pairs, including H3K4me1-H3K9me1 (r = 0.51; P < 0.01), H3K9me1-H3K27me1 (r = 0.31; P < 0.01) and H3K9me2-H3K27me2 (r = 0.28; P < 0.01), identified in this study have been detected in one single modified peptide using a mass spectrometry strategy16.
Mutually exclusive acetylation and methylation are detectable at several lysines, including H2BK5, H3K4, H3K9, H3K27 and H3K36. However, except for the H3K9 acetylation and methylation, little is known about the functional and spatial differences of modification variance at other lysines. Our data indicate that all acetylations of these five lysines are enriched in the promoter regions of active genes (Fig. 5), whereas the predominant region of enrichment for the various methylations depends on the specific lysine. H3K4me1 and H3K4me2 were elevated in the vicinity of promoters, whereas mono-methylated H2BK5, H3K9 and H3K27 were elevated throughout the transcribed regions of active genes (Fig. 5). Localization in different regions of genes suggests that these modifications have different functions. For example, H3K9ac was elevated in promoter regions, consistent with a role in transcriptional initiation20, whereas H4K12ac was detected at high levels in transcribed regions, suggesting a role in transcriptional elongation19,27.
Figure 5.
Characteristic spatial distribution of mutually exclusive modifications. (a) The distribution of H2BK5me1 (upper) and H2BK5ac (lower) surrounding the ZNF101 gene. (b) The composite profiles of H2BK5me1 (green) and H2BK5ac (red) surround the TSS regions of 1,000 active genes. (c) The distribution patterns of H3K36me3 (upper), H3K36me1 (middle) and H3K36ac (lower) surrounding the ZNF101 gene. (d) The composite profiles of H3K36ac (red) are compared with the two states of H3K36 methylation (H3K36me1 and H3K36me3) surrounding the TSS regions of 1,000 active genes. (e) The distribution patterns of H3K4ac and the three states of H3K4 methylation surrounding the promoter region of the SMAP gene. (f) The composite profiles of H3K4ac (red) are compared with the three states of H3K4 methylation (H3K4me1, H3K4me2 and H3K4me3) surrounding the TSS regions of 1,000 active genes. (g) The distribution patterns of H3K27ac and the three states of H3K27 methylation surrounding the SMAP gene. (h) The composite profiles of H3K27ac (red) are compared with the three states of H3K27 methylation (H3K27me1, H3K27me2 and H3K27me3) surrounding the TSS regions of 1,000 active genes. (i) The distribution patterns of H3K9ac and the three states of H3K9 methylation surrounding the SMAP gene. (j) The composite files of H3K9ac (red) are compared with the three states of H3K9 methylation (H3K9me1, H3K9me2 and H3K9me3) surrounding the TSS regions of 1,000 active genes.
It has been proposed that histone modifications can cross-talk28 and that multiple modifications with similar functions can reinforce the robustness of a chromatin state14. In this report, we identify a large number of histone modification patterns at promoters and potential enhancers. In particular, we identify a backbone module consisting of 17 modifications that is found at 25% of human promoters. These modifications tend to colocalize at gene promoters, and most of them are correlated with each other at an individual nucleosome level. Although the genes associated with these modifications tend to have higher expression, the histone modifications themselves do not uniquely determine expression but may function cooperatively to prepare chromatin for transcriptional activation.
As with the findings of other genome-wide ChIP analyses, our results rely on antibody specificity. Many of the antibodies that we used are well characterized, but some need to be improved further. We also cannot exclude the possibility of antibody occlusion between adjacent modifications because of steric interference. Finally, because the ChIP step requires a population of hundreds of thousands or even millions of cells, the pattern obtained for each modification is an average ‘snapshot’, containing contributions from heterogeneous modification states of different cells.
METHODS
T-cell purification, antibodies, ChIP and ChIP-Seq
Human CD4+ T cell purification and ChIP-Seq analysis for histone acetylations were done as previously described15. The ChIP-Seq analysis for histone methylations was reported previously15 except for that of H3K79me1, H3K79me2 and H3K79me3, which was done using an improved ChIP procedure29. The raw sequence tags for histone acetylations and H3K79 methylations in human CD4+ T cells have been deposited in the Short Read Archive (see Accession codes section below). We have also generated 5.5 million additional sequence reads for H3K27me3 in human CD4+ T cells and deposited them in the Short Read Archive. The histone acetylation maps and BED files can be found online (see URLs section below).
The validation of antibodies is described in Supplementary Methods, Supplementary Table 1 and Supplementary Figure 1. The expression information for CD4+ T cells was obtained from the GNF SymAtlas gene expression atlas30. GNF expression probes were mapped to UCSC Known Genes to obtain expression information in CD4+ T cells for 12,726 known genes. These genes were ranked by expression and broken up into 12 sets of 1,000 genes, starting with the most expressed set. The normalized tag counts were obtained by normalizing the tag counts in 5-bp windows (Fig. 1a–c) or in 2-kb windows (Fig. 1d–f) by the total base pairs.
Identification of patterns and modules of histone modifications
The coverage was examined by scaling analysis (Supplementary Methods). We defined a promoter region as a region of 2 kb around TSS with 1 kb upstream and 1 kb downstream. For each modification and promoter region, we counted the tags located within the promoter region for that modification. The modification on a promoter under consideration was deemed significant when the tag count was higher than a threshold, which was determined by a P value taken from a background model of Poisson distribution parameterized by the genome-wide tag density. In order to ensure high confidence in each of our designations of a significant promoter, we chose a very stringent threshold requiring a P value of 10−7. The P value of 10−7 was a result of using a P value of 10−3 on a single call along with the Bonferroni correction for simultaneous multiple calls. This method was applied to every promoter for each modification. Hence, each promoter was given a binary string to represent the presence or absence of each modification. A similar method was applied to the analysis of combinatorial patterns at a set of putative enhancers, which were defined as a DNase hypersensitive region with extensions of 200 bp both upstream and downstream. To examine if correlations between histone modifications at enhancers and gene expression exist, we assigned an enhancer to the TSS of the nearest known gene. However, we cannot exclude the possibility that the enhancer may not act as a major determinant of the expression of the gene under the assay condition, that the assigned gene is not the true target of the enhancer, or both.
We identified the modification backbone by finding the common features of the top patterns (that is, those associated with the largest number of genes) and by using the ‘single-mutation’ analysis. In ‘single-mutation’ analysis, we considered the size of the gene set for patterns that differ from the constructed backbone by only one modification. We carried out principal component analysis using the tag counts for nonoverlapping 200-bp windows in the 2-kb region surrounding the TSS.
Pairwise correlation analysis
In order to assign tags to the centers of the nucleosomes to which they correspond, we shifted the sequence tag locations +65 bp for hits on the positive strand and −65 bp for hits on the negative strand. Using these locations, we placed tags into nonoverlapping 200-bp bins along the genome, with the bin for each tag determined by the middle of the tag.
To determine the correlation between a pair of modifications, we divided the genome into 100 windows of these 200-bp bins and determined the correlation for each window. We calculated the mean correlation and the s.d. of the correlation from all of the windows of bins using a Pearson correlation coefficient. Resampling was then used to determine a significance P value for each correlation coefficient. This empirical calculation was necessary because the count data does not directly fit any distribution that could be used to produce an analytic P value. To illustrate the P-value calculation, we take the pair of modifications A and B as an example. For the modifications, the counts in 200-bp bins produced colinear arrays of integers that correspond to locations on the genome. These arrays were then shuffled so that for each location in the array, the bin for modification A was taken from a different location than the bin for modification B. We took this shuffled genome to represent the null background from which we determined the significance. We then divided the shuffled genome into 100 windows and determined the correlation for each window. The comparison of the mean correlation to the correlations from the shuffled genome was used to determine significance. We carried out a t-test to compare the actual correlation and the correlations between the shuffled genomes. From the t-test results, we calculated a P value using a Gaussian assumption for the distribution of t-test scores. A heat map was then produced from the mean correlation coefficients. For this analysis, we excluded any bins in the genome that lacked histone modifications, because the addition of paired 0s skews the calculation of a Pearson correlation coefficient.
URLs
Histone acetylation maps and BED files, http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcellacetylation.html.
Accession codes
NCBI Short Read Archive: raw sequence tags for histone acetylations and H3K79 methylations in human CD4+ T cells have been deposited with accession code SRA000287.
Acknowledgments
We thank W. Leonard for comments. This work was supported by the Intramural Research Program of the US National Institutes of Health, National Heart, Lung, and Blood Institute (K.Z.) and by an NIH grant HG001696 (M.Q.Z.). J.A.R. is supported by an NIH training grant to New York University and a New York University McCracken Fellowship.
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
Z.W. and K.Z. designed the study; Z.W. performed the experiments; A.B., S.C., K.C. and T.-Y.R. contributed to the study; C.Z., J.A.R, D.E.S., W.P. and M.Q.Z. analyzed the data; Z.W., J.A.R., W.P., M.Q.Z. and K.Z. wrote the paper.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 5.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 6.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Bulger M. Hyperacetylated chromatin domains: lessons from heterochromatin. J Biol Chem. 2005;280:21689–21692. doi: 10.1074/jbc.R500004200. [DOI] [PubMed] [Google Scholar]
- 8.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 9.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 10.Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 11.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 12.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 13.Wade PA, Pruss D, Wolffe AP. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 14.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 15.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Garcia BA, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 17.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho H, et al. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 21.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 22.Roh TY, Wei G, Farrell CM, Zhao K. Genome-wide prediction of conserved and nonconserved enhancers by histone acetylation patterns. Genome Res. 2007;17:74–81. doi: 10.1101/gr.5767907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HP, Kim BG, Letterio J, Leonard WJ. Smad-dependent cooperative regulation of interleukin 2 receptor alpha chain gene expression by T cell receptor and transforming growth factor-beta. J Biol Chem. 2005;280:34042–34047. doi: 10.1074/jbc.M505833200. [DOI] [PubMed] [Google Scholar]
- 25.Hatton RD, et al. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity. 2006;25:717–729. doi: 10.1016/j.immuni.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 26.Crawford GE, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carey M, Li B, Workman JL. RSC exploits histone acetylation to abrogate the nucleosomal block to RNA polymerase II elongation. Mol Cell. 2006;24:481–487. doi: 10.1016/j.molcel.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr Opin Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 29.Steger DJ. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]