Abstract
The ANL superfamily of adenylating enzymes contains acyl- and aryl-CoA synthetases, firefly luciferase, and the adenylation domains of the modular Non-Ribosomal Peptide Synthetases (NRPSs). Members of this family catalyze two partial reactions, the initial adenylation of a carboxylate to form an acyl-AMP intermediate, followed by a second partial reaction, most commonly, the formation of a thioester. Recent biochemical and structural evidence has been presented that supports the use by this enzyme family of a remarkable catalytic strategy for the two catalytic steps. The enzymes use a 140° domain rotation to present opposing faces of the dynamic C-terminal domain to the active site for the different partial reactions. Support for this Domain Alternation strategy is presented along with an explanation of the advantage of this catalytic strategy on the reaction catalyzed by the ANL enzymes. Finally, the ramifications of this domain rotation in the catalytic cycle of the modular NRPS enzymes are discussed.
The results of numerous investigations on luciferase and other acyl adenylate synthetases indicate that large conformational changes occur in the protein when the specific substrates combine at the active site.
W.D. McElroy, M. DeLuca, J. Travis, (1967)
From a comparison of the allosteric manifestations of the overall reaction and partial reaction (b) it appears that the same conformational state (R), showing a greater affinity for CoA, catalyses both the overall reaction and partial step (b), whereas the T-configuration, favoured in the presence of PPi, seems to catalyse the ATP-forming step (reversal of partial reaction a).
J. Bar-Tana and G. Rose, (1968)
These statements were made over 40 years ago about a family of enzymes that has recently garnered much attention. This family of ligases, which now includes acyl- and aryl-CoA synthetases, the adenylation domains of Non-Ribosomal Peptide Synthetases (NRPSs), and firefly luciferase, catalyzes the activation of a carboxylate substrate with ATP to form an acyl adenylate intermediate that is used in a diverse set of second partial reactions. The study of this family of adenylating enzymes has a long history, dating back to work in the 1950s with acylCoA synthetases. A decade later, biochemical studies of the reactions catalyzed by different members of this family allowed McElroy et al. (1) as well as Bar-Tana and Rose (2) to suggest that large conformational changes play a role in the catalysis of the complete two-step reaction. Bar-Tana and Rose even suggested that the two partial reactions of the acyl-CoA synthetases might be catalyzed by different conformational states. Forty years later, the structural biochemistry of these enzymes has been thoroughly investigated and it is remarkable how prescient these early statements appear to have been. This review describes the biochemical and structural data for a novel catalytic mechanism used by these enzymes and the chemical requirements of the two-step reactions that benefit from this strategy.
The ANL Family of Adenylating Enzymes
The activation of biological carboxylates as thioesters with CoA has been known for over 60 years. The conversion of acetate to acetyl-CoA was first described by Lipmann (3) using a partially purified bacterial enzyme. The mechanism proceeded through an acetyl-phosphate intermediate, which was then converted to acetyl-CoA. A second enzymatic mechanism for the synthesis of acetyl-CoA was identified in the next decade by Berg (4) who purified from baker's yeast a protein he called aceto-CoA kinase that activated acetate not through an acyl-phosphate intermediate but rather through an acyl-adenylate. This study demonstrated the exchange of 32PPPi into ATP in an acetate dependent manner and that the acetyl-AMP intermediate could react with PPi for the formation of ATP or with CoA to form acetyl-CoA and AMP. The formation of acetyl-AMP from ATP and acetate occurred in the absence of CoA, demonstrating the independence of the two partial reactions. Rigorous attempts to separate the active fraction into two distinct enzymes that catalyzed the adenylation and the thioester-forming reactions were unsuccessful, and a single enzyme was proposed to be responsible for the production of acetyl-CoA, AMP, and PPi directly from acetate, ATP, and CoA, proceeding through the acetyl-AMP intermediate. A related medium chain acyl-CoA synthetase was subsequently demonstrated to use a bi-uni-uni-bi ping pong mechanism (2). Such ping-pong kinetics have since been confirmed for many acyl-CoA synthetases, including enzymes with specificity for acetate (5), malonate (6), long chain fatty acids (7), and more complex aryl acids (8, 9).
The possibility that this adenylating enzyme family contained members beyond the acyl- and aryl-CoA synthetases was raised as early as 1967 by McElroy and colleagues who noted the functional similarities of the acyl-CoA synthetases with firefly luciferase, as well as with the reaction of pantoic acid in pantetheine cofactor biosynthesis and the activation of amino acids by amino acyl-tRNA synthetases (1). As sufficient sequence information became available, several reports (10, 11) noted that the acyl-CoA synthetases and luciferase enzymes shared numerous conserved sequence motifs. The amino acyl-tRNA synthetase and pantothenate synthetase enzymes, while functionally similar, share no sequence homology. In the late 1980s, several multidomain proteins that produce bacterial peptide antibiotics or siderophores were also identified that share similar sequence motifs (12-14). This new class of modular enzymes became known as non-ribosomal peptide synthetases (NRPSs) and formed the third subfamily of this adenylating enzyme superfamily.
We and others have referred to this enzyme family as the “adenylate-forming superfamily” of enzymes or the “acyl-AMP forming family of adenylation enzymes”. Neither of these names is particularly satisfying as other acyl-adenylating enzymes exist that do not belong to this family. Using the description of divergent superfamilies described by Gerlt and Babbitt (15), luciferase, acyl-CoA synthetases, and NRPS adenylation domains comprise a Mechanistically Diverse Enzyme Superfamily. The enzymes share ~20% sequence identity, are structurally homologous, and catalyze different overall reactions, while sharing a conserved mechanistic step, the adenylation partial reaction. Other adenylate forming enzymes, such as the amino acyl-tRNA synthetases (16) or the NRPS-independent siderophore (NIS) adenylating enzymes (17), do not belong to this enzyme superfamily and are simply unrelated enzymes that catalyze a similar overall reaction.
In the interests of providing a clear way to describe the enzyme superfamily that is the focus of this review, I propose a new designation of the “ANL superfamily of adenylating enzymes”. This name is derived from the three main subfamilies, namely the Acyl-CoA synthetases, the NRPS adenylation domains, and the Luciferase enzymes.
Functional Diversity and Substrate Specificity in the ANL Adenylating Enzymes
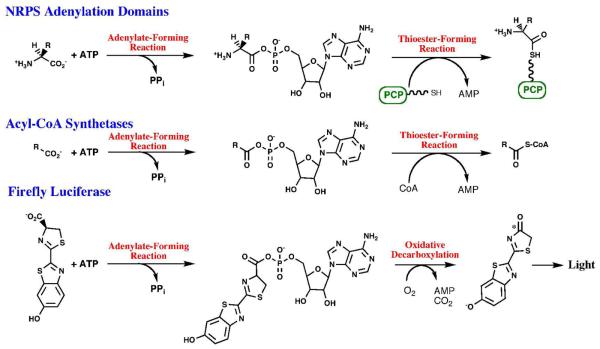
Although distinct in the overall reactions catalyzed, the enzymes within all three subfamilies use a two-step reaction to first activate a carboxylate substrate by reacting with ATP to form the acyladenylate and inorganic PPi (Figure 1). The adenylate, a high energy acid anhydride, provides the activation energy for the second partial reaction. For the acyl-CoA synthetases and NRPS adenylation domains, a pantetheine thiol group attacks the carboxylate carbon, displacing the AMP leaving group, in the second half of the reaction. The hydrolysis of ATP in the first partial reaction therefore creates a higher energy adenylate intermediate that is utilized for the thioester-forming step. In the reaction catalyzed by luciferase, the activated luciferyl-adenylate undergoes an oxidative decarboxylation that results in the formation of an intermediate that subsequently decomposes within the enzyme active site to yield a photon of light. As described below, the unique chemical properties of these reactions have been facilitated by an interesting catalytic mechanism.
Figure 1. Reactions catalyzed by the ANL Superfamily of adenylating enzymes.
Chemical reactions catalyzed by the three subfamilies of the ANL adenylating enzymes. All three reactions include an initial, adenylate-forming reaction to form an acyl-, amino acyl-, or aryl-adenylate with the release of inorganic pyrophosphate. The adenylate intermediate reacts in a second partial reaction to release AMP. For NRPS adenylation domains, the pantetheine cofactor bound to an NRPS peptidyl carrier domain is represented by the linker and SH group.
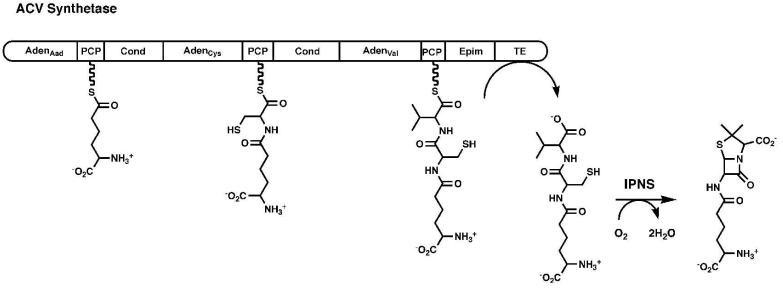
Many excellent reviews have been written describing the fascinating modular architecture of the NRPS enzymes (18-21) and these enzyme assembly lines will be described only briefly here. The multi-domain NRPS enzymes generally contain a single module for the incorporation of each amino acid into the peptide product (Figure 2). During synthesis, the amino acid and peptide intermediates are bound to the pantetheine cofactor of an integrated peptidyl carrier protein domain (22, 23). Within each module is an adenylation domain responsible for activating the amino acid substrate and transferring it to the pantetheine cofactor of the neighboring carrier protein domain. Finally, a condensation domain is required in all modules except the first to catalyze peptide bond formation and transfer the peptide from an upstream to a downstream carrier protein domain, increasing the peptide length by a single residue. This assembly line biosynthesis terminates with a thioesterase domain that cleaves, and often cyclizes, the peptide product. Reflecting the diversity of the peptide products that are produced, some NRPS proteins contain additional internal catalytic domains that are responsible for epimerization or Nmethylation of the constituent amino acids. The ACV Synthetase enzyme (24), for example, is a three-module protein involved in the synthesis of isopenicillin N, a β-lactam antibiotic. The ACV synthetase NRPS produces a tripeptide from α-aminoadipic acid, cysteine, and valine that is then cyclized by the isopenicillin N synthase (25).
Figure 2. Modular Organization of the Non-Ribosomal Peptide Synthetases.
A schematic representation is shown of the ACV synthetase, a three-module protein that is responsible for the synthesis of the linear tripeptide of α-aminoadipic acid (Aad), cysteine, and valine. The linear peptide is subsequently cyclized by the enzyme isopenicillin N synthase. Module 1 contains the adenylation domain and PCP for Aad. Module 2 contains the adenylation and PCP domain for cysteine, as well as the condensation domain that forms the peptide bond between Aad and Cys. The third module contains the adenylation and PCP domain for valine, an epimerization domain that converts l-Val to d-Val, and a condensation domain that transfers the upstream dipeptide to d-Val. The protein terminates with a thioesterase domain that releases the tripeptide.
The choreography that delivers the substrate bound to the NRPS carrier protein domain to the neighboring catalytic domains is undoubtedly complex and is not clearly defined. Recent studies using alanine-scanning mutagenesis have defined the binding surface used by the E. coli EntB PCP domain for interactions with its functional partners, EntF and EntD (26, 27). Additionally, NMR studies have demonstrated that both the PCP domain and the catalytic domains will undergo local conformational rearrangements during their interactions (28-30). In particular, these studies suggest that the PCP domains are dynamic and can adopt multiple conformations and that the catalytic domains are able to stabilize one of the conformations selectively for a functional interaction. Nonetheless, these conformational changes are limited in scale and it appears from a recent multi-domain NRPS structure (31) that more significant conformational rearrangements are required for the proper delivery of amino acyl and peptide substrates to the different catalytic centers.
NRPS adenylation domains can be subdivided into two classes. Most NRPS adenylation domains are integrated into the catalytic module (Figure 2) and activate and load the amino acid on the pantetheine cofactor of an adjacent carrier domain, referred to alternatively as the thiolation domain and the PCP domain (22). Other NRPS adenylation domains however are self-standing and transfer the amino acyl substrate in trans to a separate amino acyl carrier domain. In many cases these isolated adenylation domains load an acyl or aryl capping group that initiates the NRPS peptide (32).
The substrates of the acyl- and aryl-CoA synthetase subfamily of the ANL enzymes are chemically diverse, ranging in size from small acids like acetate and propionate to medium- and long-chain fatty acids and aromatic compounds. The aryl-CoA ligases have been identified in a variety of pathways for the degradation of aromatic compounds, as well as for the biosynthesis of plant metabolites (33).
A subfamily of fatty acyl-AMP ligases (FAALs) has been characterized by Gokhale and colleagues (34, 35). These enzymes share functional features with both the acyl-CoA synthetases and the NRPS adenylation domains. The acyl substrate of these enzymes are fatty acids that are used in the formation of complex lipopeptides, some of which play a role in virulence. Unlike the acyl-CoA synthetases, however, the enzymes of this family transfer the activated fatty acid from the adenylate directly to the pantetheine cofactor of an acyl carrier protein of an NRPS or polyketide synthase cluster (34). In this latter regard, these enzymes share more in common with the NRPS adenylation domains than with the fatty-acyl CoA synthetases. In particular, these enzymes are reminiscent of the self-standing aryl activating of many NRPS siderophore clusters (36) that activate salicylate or 2,3-dihydroxybenzoate and transfer the aromatic acid to an aryl carrier protein domain (32).
As more members of the adenylate-forming family of enzymes were identified, several groups proposed a number of conserved sequence motifs to be important for catalytic activity. The first identified sequence was a serine-, threonine-, and glycine-rich motif (10). This region was deemed a signature sequence for the ANL enzyme family and was designated Motif I. Two additional regions (Motif II and Motif III) were also identified based on sequence conservation (37). A more detailed comparison of the conserved regions exclusively within the NRPS adenylation domains identified ten conserved regions, named A1-A10 (38). The determination of crystal structures of members of this enzyme family allowed a preliminary understanding of the roles of these conserved motifs in the catalytic residues (Table 1). Interestingly, the conservation of several regions that were located at some distance from the active site of the first structures can now be rationalized on the basis of the domain rearrangements described below.
Table 1.
Conserved sequence motifs
| Corea | Motifb | Consensus Sequencec | Position d |
Role |
|---|---|---|---|---|
| A1 | θ(S/T)ϕx(E/Q)θ | Leu30 | Structural Role. Caps a helix at N- terminus of the protein. |
|
| A2 | (R/K/F)θGθ | Arg76 | Structural Role. Torsion angles enforce a glycine at position 78 |
|
| A3 | I | θθx(S/T)(S/T/G)G(S/T)TGxP K |
Ile158 | Phosphate-binding loop, orienting the β,γ- phosphates. |
| A4 | ϕ | His207 | Active site aromatic residue that alternates conformation in the two enzyme states and caps a helix that forms part of the acyl-binding pocket |
|
| A5 | II | ϕ(G/W)x(A/T)E | Tyr304 | Initial aromatic residue stacks agains adenine ring of ATP, Gly or Trp forms wall of active site, Glu binds the Mg2+ ion. |
| A6 | GEx10-14GY | Gly351 | Structural Role. Forms a portion of the distorted β-sheet of the N-terminal domain |
|
| A7 | III | (S/T)GD | Ser383 | The Asp of this motif binds the ribose hydroxyls of ATP and is 100% conserved |
| A8 | Rx(D/K)x6G | Arg400 | The Arg interacts with ribose hydroxyls. The Asp (or rarely Lys) is the hinge residue. The Gly abuts the pantetheine tunnel in the thioester-forming conformation. |
|
| A9 | No conservation within larger family |
|||
| A10 | Px4GKθx(R/K) | Pro486 | The first lysine (Lys492) is present at the active site in the adenylate-forming conformation. |
Core sequence as defined by Marahiel et al (38) for NRPS adenylation domains
Motif defined by Chang et al (37) for aromatic adenylate forming ligases
Single amino acid code, θ is used for medium aliphatic amino acids (A,V,L,I,M). ϕ is used for aromatic amino acids (F,Y,H,W).
Position is provided for the first amino acid residue of the motif in the representative enzyme 4-Chlorobenzoyl-CoA Ligase.
Structural Studies of ANL Family of Adenylating Enzymes
As of July 2009, there are 47 crystal structures deposited in the Protein Data Bank of members of the ANL adenylating enzyme family (Table 2). These structures represent 16 different proteins and have been crystallized in a variety of liganded states, providing a detailed view of the catalytic strategy used by this enzyme family.
Table 2.
Acyl-AMP forming adenylating enzymes that have been structurally characterized
| Protein | Organism | Conformation | PDBs and ligands or mutant structure |
|---|---|---|---|
| Luciferase | P. pyralis | Intermediate | 1LCI, unliganded; 1BA3, bromoform. |
| PheA | B. brevis | Adenylation | 1AMU, AMP + Phe. |
| DhbE | B. subtilis | Adenylation |
1MDF, unliganded; 1MD9, AMP + DHB; 1MD8, DHB-AMP adenylate. |
| Acs | S. enterica | Thioester |
1PG4, adenosine-5′-propylphosphate + CoA; 1PG3, same ligands, acetylated on Lys609; 2P2F, acetate, AMP, COA; 2P20, R584A; 2P2B, V386A; 2P2J, K609A; 2P2M, R194A; 2P2Q, R584E. |
| Acs | S. cerevisiae | Adenylation | 1RY2, AMP |
| CBL | A. sp AL3007 | Adenylation |
1T5S, unliganded; 1T5D, 4CB; 2QVX, 2QVY, I303G; 2QVZ, 2QV0, I303A; 3CW8, 4CB-AMP; 3DLP, D402P. |
| Thioester | 3CW9, 4CPh-CoA + AMP. | ||
| Fatty Acs |
T. thermophilus |
Aden/Intermed | 1ULT, unliganded. |
| Thioester | 1V25, AMPPNP; 1V26, Myristyl-AMP. | ||
| Luciferase | L. cruciata | Adenylation |
2D1Q, AMP; 2D1R, oxyluciferin + AMP; 2D1S, N- (dehydroluciferyl)sulfamoyl adenosine; 2D1T, T217I. |
| Benzoyl-CL |
B. xenovornans |
Adenylation | 2V7B, benzoic acid |
| Med. Acs | H. sapiens | Adenylation |
3C5E, ATP; 2V7E, AMP; 3DAY, AMPPNP; 3EYN, CoA |
| Thioester | 3B7W, unliganded; 3EQ6, butyryl-CoA + AMP. | ||
| SrfA-C | B. subtilis | ~Adenylation | 2VSQ, Leu. Multi-domain NRPS |
| DltA | B. cereus | Adenylation |
3DHV, alanyl-AMP; 3FCC, Mg•ATP; 3FCE, Ca•ATP |
| DltA | B. subtilis | Thioester | 3E7W, AMP; 3E7X, AMP. |
| AAE | M. acetivorans | Thioester | 3ETC, unliganded |
| FAAL28 |
M. tuberculosis |
N-terminal domain only |
3E53, unliganded |
| Fatty Acs | A. fulgidus | Intermediate | 3G7S, unliganded |
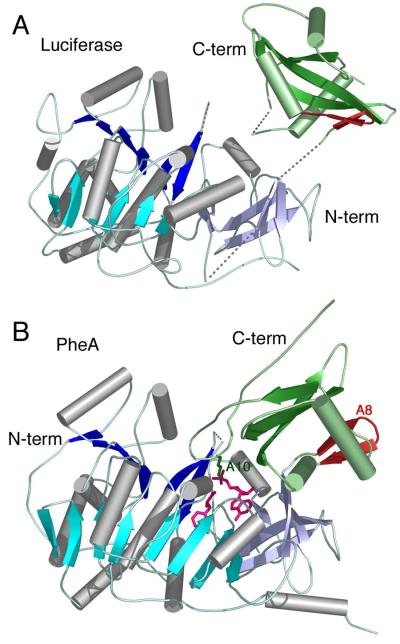
The first crystal structure of a member of the adenylate-forming family of enzymes was of firefly luciferase from P. pyralis (39). This structure identified an overall two-domain architecture with a larger N-terminal domain composed of the first 430 residues and a smaller C-terminal domain of the final 120 residues (Figure 3A). The larger domain contained an ababa domain structure with two large eight stranded β-sheets that surround two α-helices. The N-terminal domain ends with a distorted β-sheet. Following a short disordered loop in the luciferase structure, the C-terminal domain begins with an antiparallel β-sheet that contained two strands, followed by a central 3-stranded β-sheet that was surrounded by helices. The residues connecting the two domains form the A8 motif (Table 1) and are collectively referred to as the A8 loop. Luciferase was crystallized in the absence of ligands. The conserved sequence motifs (40) were used to propose a location of the enzyme active site. In particular, the Gly- and Ser-rich motif I was located at the interface between the N- and C-terminal domains. Conti et al. (39) note that the cleft is likely “too big to accommodate the substrates” and predict closure of the interface upon substrate binding.
Figure 3. Crystal Structures of A. Firefly Luciferase and B. PheA adenylation domain.
Structures of the first two members of the ANL superfamily of enzymes to be characterized by crystallography. The proteins are aligned on the basis of the N-terminal domains. The structure of luciferase (1LCI) displayed an open conformation with few interactions between the N- and C-terminal domains. Several disordered loops are indicated with dashed lines. PheA was co-crystallized in the presence of ATP and phenylalanine (1AMU), and revealed a molecule of AMP and Phe in the active site (pink). The A8 loop, that follows the hinge for domain alternation is shown in red while the A10 lysine that is conserved throughout the entire family is shown in green. In both panels, the N-terminal domain is represented with β-sheets of different shades of blue, while the C-terminal domain is shown in green.
As noted above, early experiments predicted a large conformational change for acyl-CoA synthetases and luciferase (1) on the basis of tritium exchange and thermal inactivation in the presence and absence of ligands. A conformational change was also invoked to explain the stabilization of phenylacetyl-CoA ligase (41). The crystal structure of firefly luciferase thus provided a structural framework to envision this large conformational change. The single structure, however, provided only an initial look at the protein and many additional structural studies were necessary to understand the full conformational mechanism.
The structure of an initiating NRPS adenylation domain, the phenylalanine activating domain of gramicidin synthetase S (GrsA), was determined the following year (42). Importantly, this structure was determined in the presence of the amino acyl substrate phenylalanine and a molecule of AMP (Figure 3B) confirming the predicted location of the active site. The C-terminal domain was rotated by ~90° compared to the orientation seen in the luciferase structure. A universally conserved lysine from the A10 region formed hydrogen bonds to the ribose ring oxygen, the 5′-bridging oxygen, and a carboxylate oxygen of phenylalanine. This suggested a possible catalytic role for this lysine, which was indeed supported by prior biochemical studies of tyrocidin synthetase (40).
These initial structures provided the foundation for a number of studies that investigated the roles of catalytic or substrate specificity residues. In particular, the interaction of the C-terminal lysine from the A10 region (Table 1) was studied by mutagenesis in both luciferase (43) and PrpE, a propionyl-CoA synthetase (44). No activity could be detected for the complete reaction of the K592E mutant of PrpE for example and activity was reduced by over four orders of magnitude for the reverse of the adenylation reaction; the rate of production of propionyl-CoA from the propionyl-AMP intermediate, however, was reduced by only a factor of 2. These studies demonstrated a role of this residue specifically in the adenylation partial reaction.
Subsequent to the structural characterization of PheA, a number of additional structures of enzymes in this family were determined that demonstrated a similar tertiary structure and a similar conformational orientation between the N- and C-terminal domains. These structures included DhbE, the self standing adenylation domain from the bacillibactin NRPS cluster (45), yeast acetyl-CoA synthetase (46), two enzymes that catalyze aryl-CoA synthesis that are involved in the metabolic breakdown of 4-chlorobenzoate (47) and benzoic acid (48), luciferase from L. cruciola (49), and the enzyme DltA from B. cereus, which is involved in the activation of alanine for subsequent alanylation of teichoic acid in cell wall biosynthesis of gram positive bacteria (50, 51). These structures were all determined in the absence of ligands, or in the presence of the acyl substrate, the adenylate, or AMP. Notably, none of these structures contained a bound CoA or thiol acceptor for the second partial reaction (Table 2).
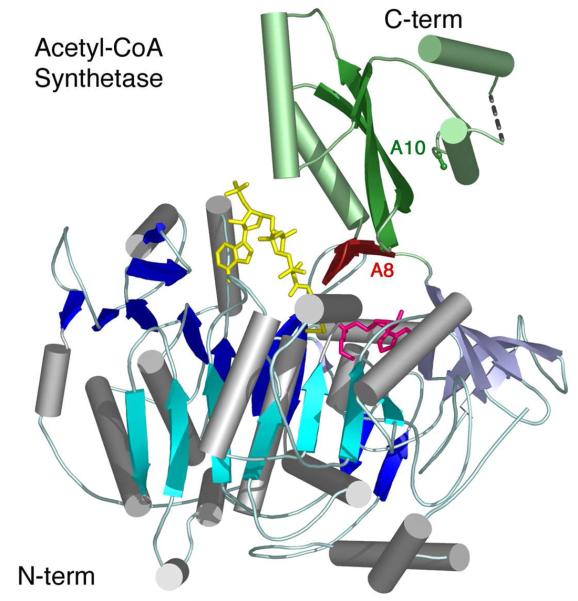
Insights into CoA binding were derived from the crystallization of a bacterial acetyl-CoA synthetase (Acs) bound to adenosine-5′-propylphosphate, a non-hydrolyzable mimic of the adenylate intermediate, and CoA (52). This structure, which appeared to show the enzyme poised to catalyze the thioesterification reaction, located the nucleotide of CoA at the surface of the protein with the pantetheine portion of CoA passing through a pantetheine tunnel that runs between the N- and C-terminal domains to enter the mostly buried adenylate binding site.
The most intriguing feature of this structure was that the C-terminal domain of Acs adopted a dramatically different conformation compared to that seen in the prior structures of PheA and DhbE (Figure 3B). The C-terminal domain of Acs packed against the N-terminal domain forming a more lobular enzyme (Figure 4). Multiple interactions were observed between the two domains, and between the C-terminal domain and the reaction intermediates. In this conformation, the loop containing the A10 lysine residue is 25Å from the active site. In contrast, the A8 β-sheet that initiates the C-terminal domain was rotated into the active site. The luciferase C-terminal domain, which was also observed in a conformation than observed in PheA, did not make any interactions with the N-terminal domain and did not seem to be a functionally relevant conformation.
Figure 4. Crystallographic Structure of Acetyl-CoA synthetase.
The structure is shown of bacterial acetyl-CoA synthetase (1PG4). The enzyme is oriented as other family members are in Figure 3 and includes the ligands adenosine-5′-propyl phosphate (pink) and CoA (yellow). The CoA nucleotide binds on the surface of the N-terminal domain while the pantetheine passes into the enzyme active site via the pantetheine tunnel. The A8 loop that follows the hinge is shown in red. The Cα and Cβ positions of Lys609, the A10 lysine, are shown in green. The N- and C-terminal domains are colored as in Figure 3.
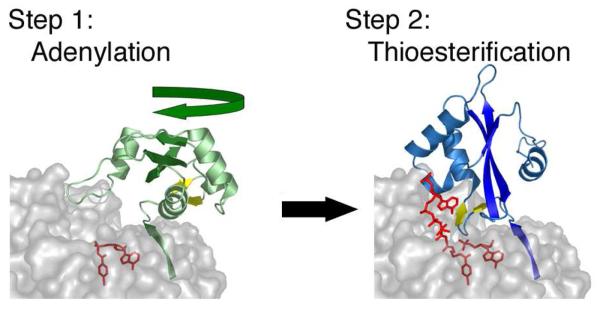
The biochemical data implicating the A10 lysine from the C-terminal domain in catalyzing the adenylate-forming reaction specifically (43, 44) and the structural observation of this new conformational state of Acs bound to CoA provided preliminary support for a novel catalytic strategy (52). In this proposed catalytic strategy, the members of this adenylate-forming family would adopt the PheA-like structure to catalyze the adenylation partial reaction. Upon formation of the adenylate and the release of pyrophosphate, the C-terminal domain would rotate to the orientation observed in the bacterial Acs to form a second conformation that would be used to catalyze the thioester-forming reaction. We adopted the term Domain Alternation, which had been used to describe a large-scale domain rearrangement in methionine synthase (53), to describe this mechanism.
The crystal structures of several other members of this enzyme family have since been determined in this conformation. These structures include the long chain fatty acyl-CoA synthetase from T. thermophilus (54), DltA from B. subtilis (55), and an acyl-adenylating enzyme from the methanogenic bacteria Methanosarcina acetivorans (56). Interestingly, while the Acs structure contained CoA, these latter structures did not contain bound coenzyme A, although in the case of the fatty acyl-CoA synthetase, CoA was included in the crystallization conditions yet was not bound in the crystal structure.
In the last year, two examples of a single enzyme being crystallized in both conformations have been reported. The 4-chlorobenzoyl-CoA ligase (4CBL) from Alcaligenes sp. AL3007 was the subject of extensive structural and kinetic evaluation, as will be discussed below. As part of this study, the enzyme was trapped in both the adenylate-forming conformation bound to the adenylate intermediate as well as to a complex of AMP and the product analog 4-chlorophenacyl-CoA (57). More recently, the structure of the human medium chain Acyl-CoA synthetase has also been determined in both conformational states (58). These alternate structures demonstrate that the domain rotation is a dynamic feature of the ANL enzymes and does not simply reflect differences in tertiary organization between different superfamily members.
Not surprisingly, crystallization of these conformationally flexible enzymes has been challenging and several of the deposited structures exhibit significant disorder in the C-terminal domain. Indeed, a recent structure of an FAAL (34, 59) required the removal of the C-terminal domain altogether to achieve crystallization. As with all structural studies, the careful selection of appropriate inhibitors can support the crystallization of enzymes trapped in relevant conformations. The use of alkyl phosphate esters (52) or adenosyl sulfamate analogs (49) as mimics of the adenylate intermediate, or a substituted phenacyl-CoA thioether (57) as a mimic of the CoA thioester product has enabled the determination of some of the highest resolution structures of members of this conformationally dynamic enzyme family.
The Active Site of the Acyl-AMP Forming Adenylating Enzymes
Acs (52, 60), 4CBL (57), DltA (50, 51, 55), and a human medium chain acyl-CoA synthetase (58) have now been studied structurally with numerous ligand complexes or structures of site-directed mutants. These structures give insights into the catalysis of the individual reactions and illustrate residues responsible for substrate binding. The active sites of these three proteins will be described as representative members of the family.
The carboxylate substrate binds in a pocket located within the N-terminal domain (Figures 3B, 4). The residues that form the binding pocket are quite diverse, reflecting the differences in substrate specificity between different enzyme family members. In fact, we have recently noted (56) that the core of the N-terminal domain of five structurally characterized acyl-CoA synthetases contains only 14 conserved residues (out of 250). Interestingly, a conserved glycine is observed throughout the superfamily except in acetyl- and propionyl-CA synthetase where this glycine is replaced by a tryptophan residue that truncates the acyl binding pocket for the smaller substrates (47).
The amino acyl binding pocket of NRPS adenylation domains is better characterized than the acyl-CoA synthetase pockets. The substrate α-amino group is positioned by a conserved aspartic acid residue that directs the amino acid side chain into a pocket that is chemically complementary to the specific substrate. This feature of the adenylation domain binding pocket has allowed the clustering (61-63) and prediction (64) of specificity of NRPS adenylation domains.
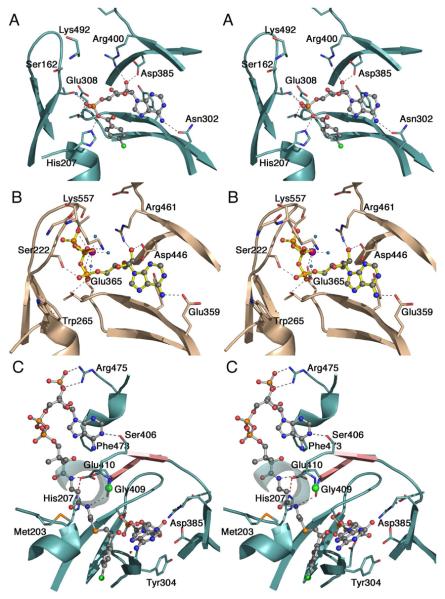
The ATP binding site contains several highly conserved motifs that are present in all family members and perform similar roles in positioning the nucleotide (Figure 5A). The aspartic acid residue of the A7/motif III region, Asp385 of 4CBL, is universally conserved and interacts with one or both ribose hydroxyls. Fifteen residues downstream is the completely conserved Arg residue of the A8 motif, Arg400 in 4CBL, that also interacts with the ribose hydroxyls. An aromatic residue from the A5 motif, Tyr304 in 4CBL, stacks against the adenine base. Two recent structures, DltA and the human medium chain acyl-CoA synthetase (51, 58), illustrate the interactions that occur between protein and the triphosphate moiety. The motif I residues that surround the β- and γ-phosphates of the human medium chain acyl-CoA synthetase (Figure 5B) are well-conserved suggesting that the ATP binding position will be similar in all family members.
Figure 5. Active site and binding interactions of the ANL enzymes.
A. Aryl-AMP binding site of 4CBL determined in the adenylate-forming conformation (3CW8). Conserved residues that interact with the adenylate are shown. Also interacting with the α-phosphate are Thr161, from the P-loop, and Thr307. Tyr304, behind the adenine ring is unlabeled. The side chain of the A10 lysine, Lys492, was disordered beyond Cβ. B. ATP binding site of medium chain Acyl-CoA synthetase in the adenylate-forming conformation (3C5E). The interaction of the Gly- and Ser/Thr-rich P-loop with the triphosphate of ATP is clearly demonstrated. The Mg2+ ion, which bridges the b- and g-phosphates, is shown in purple. Pairs of homologous residues between mAcs and 4CBL are (mAcs listed first with 4CBL residue in parentheses): Ser222 (Thr161), Thr223 (Ser162), Trp265 (His207), Glu359 (Asn302), Tyr361 (Tyr304), Thr364 (Thr307), Glu365 (Glu308), Asp446 (Asp385), and Arg461 (Arg400). C. Binding interactions between CoA and 4CBL observed in the thioester-forming conformation (3CW9). The CoA nucleotide binds on the surface of the 4CBL enzyme, with the nucleotide ring stacking against Phe473. The pantetheine passes to the interior of the protein where the CoA thiol can attack the adenylate intermediate. Here, the thioester bond is modeled by the thioether linkage of 4-chlorophenacyl-CoA. The two strands that form the A8 loop are shown in pink and the Cα position of the universally conserved glycine at position 409 is shown with a green sphere. The β-sheet like interaction between Gly408-Gly409 and the β-alanine moiety of the pantetheine group is indicated with dashed lines. The interaction between His207 and Glu410 that stabilizes the A4 aromatic group (His207) in the new side chain position is shown with a red dashed line. The first turn of the helix at 251-265 is depicted transparently to allow the pantetheine chain to be seen.
The structure of DltA bound to ATP was recently determined and compared to previously characterized structures in the adenylate-forming conformation (51). The authors note that even within this conformation, differences of as much as 40° exist in the orientation of the C-terminal domain. The different orientation causes the invariant A10 lysine residue to interact with different ligand atoms, including the bridging oxygen between the ribose and phosphate and a carboxylate oxygen in PheA (42) and DhbE (45), or a β-phosphate oxygen in the medium chain acyl-CoA synthetase (58) or DltA (51). The authors raise the intriguing possibility that the ANL enzymes adopt a pre-adenylation state bound to ATP and a post-adenylation state upon completion of the adenylation partial reaction. The A10 lysine residue is proposed to track the accumulation of negative charge on the initial attack complex, the transition state for adenylation formation, and finally to the pyrophosphate product prior to product release.
The CoA binding site can be divided into two regions, a nucleotide binding site that is located on the surface of the protein (Figure 5C), and the previously mentioned pantetheine tunnel that runs between the two domains. The tunnel contains a β-sheet like interaction that occurs between the conserved glycine on the A8 loop and the β-alanine group of the pantetheine.
In the three protein structures bound to CoA in a productive conformation, the CoA nucleotide moiety is located in different positions (52, 57, 58). The adenine group of the CoA in the structures of the two acyl-CoA synthetases interacts most closely with the N-terminal domain, while the nucleotide moiety of the CoA ligand in 4CBL binds primarily to the C-terminal domain. In 4CBL, it is sandwiched between two aromatic side chains that are well conserved within other 4CBL enzymes (9) but not in the larger subfamily of acyl-CoA synthetases.
Catalysis of the thioester-forming reaction likely requires the deprotonation of the thiol of CoA to increase the nucleophilicity for attack on the carboxylate in the adenylate intermediate. Surprisingly, no conserved residues were identified in the area surrounding the CoA thiol. Instead, the enzymes may use the helix dipole of the helix that starts with the A4 aromatic residue to provide a positive dipole that could reduce the pKa of the thiol (9).
Kinetic Evidence for the Domain Alternation Hypothesis
The observations of the distinct structures support the domain alternation hypothesis however kinetic studies have been equally important for understanding the catalytic mechanism. In particular, these studies support the involvement of residues from the opposite faces of the C-terminal domain in the distinct partial reactions and identified the catalytic advantage that is gained by the domain alternation.
Following the original experiments on luciferase and PrpE (43, 44), studies have since been performed with all three subfamilies of the acyl-AMP forming adenylating enzymes that support the hypothesis that the two C-terminal conformations are used for the different partial reactions. Mutagenesis studies with luciferase (65), Acs (60), and the EntE self-standing adenylation domain (66), all demonstrate that residues on the A8 loop are important specifically for the thioester-forming partial reaction. As the A8 loop is rotated into the active site only in thioester-forming conformation, this supported the use of the conformation observed with Acs for the thioester-forming reaction.
To analyze more rigorously the effects of mutations on this enzyme family, the 4CBL enzyme was extensively mutated and subjected to kinetic analyses (9) that complement the structural characterization of the two conformations (57). Mutations were constructed in over 20 different residues of the 4CBL protein and were analyzed by steady-state kinetics. Rate constants were determined for the individual partial reactions for a subset of 10 of the mutant enzymes. The mutations were studied for their effects on k1 and k2, the forward rate constants for the adenylation and thioesterification steps of the reaction, respectively. Not surprisingly, mutations in the ATP binding pocket had dramatic effects on k1. Mutations in two residues that are located on the “thioester-forming face” of the C-terminal domain that stack against the adenine ring of CoA (Trp440 and Phe473) showed no effect on the adenylation partial reaction but 200-fold decreases in k2, the rate of the thioester-forming partial reaction.
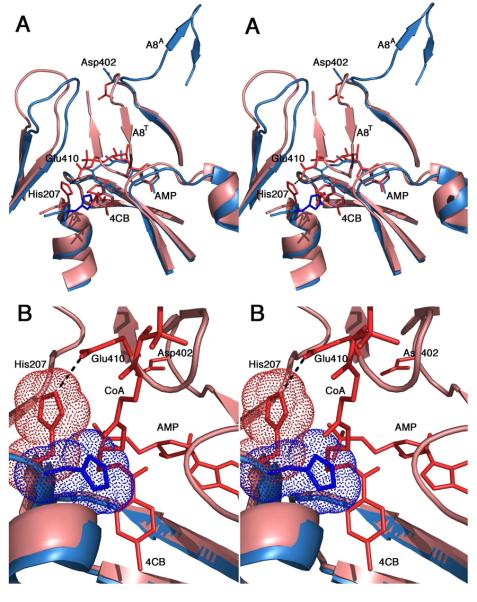
The most interesting results from this study relate to His207 and Glu410. His207 is conserved as an aromatic residue as part of the A4 motif (38). The precise identity of this residue correlates with the subfamily within the ANL family with NRPS adenylation domains most often containing a phenylalanine, small chain acyl-CoA synthetases containing a tryptophan, and larger chain acyl-CoA synthetases and luciferase enzymes containing a histidine. The His207 residue of 4CBL exhibits a side chain torsional rotation in the two observed crystal structures. In the adenylate-forming conformation, the side chain is rotated towards the acyl carboxylate with a side chain χ1 torsion angle of −166°. In the thioester-forming conformation, the χ1 torsion angle is -−60°, with side chain rotated away from the carboxylate (Figure 6). Upon further consideration, this initially minor observation provided one explanation for the catalytic advantage of the domain alternation strategy.
Figure 6. Impact of domain alternation on the orientation of the A4 aromatic residue.
A. Superposition of the crystal structures of 4CBL in both the adenylate-forming (3CW8, blue) and thioester-forming (3CW9, pink) conformations. The view is oriented so that the reader is looking down the pantetheine tunnel into the active site. The A8 loop is shown in both conformations, indicated by the superscript (A8A and A8T for the adenylate- and thioester-forming conformations, respectively). The 4-chlorobenzoyl adenylate (blue) and the 4-chlorophenacyl thioester and AMP (red) are shown. The terminal β-alanine and cysteamine groups of the pantetheine chain are shown. In the thioester-forming conformation, the A8 loop rotates into the active site (A8T), where Glu410 interacts with His207 to rotate the side chain away from the 4CB molecule. B. Closer view of the active site. The orientation is rotated slightly from panel A allowing observation of more of the pantetheine chain. The His207 side chain is shown with van der Waals surface. In the adenylate-forming structure, His207 occludes the pantetheine group from approaching the active site.
In the thioester-forming conformation of 4CBL, His207 forms a hydrogen bond with Glu410, which is located on the A8 loop and follows a universally conserved glycine residue (Gly409 in 4CBL). The His207 mutation resulted in a ~100-fold decrease in both the k1 and k2 rate constants. The Glu410 mutation had no effect on k1 but caused an 800-fold decrease in k2 despite the fact that this residue does not contact any of the reacting ligands. These results were interpreted to demonstrate that the His207 played a role in both partial reactions, while the Glu410 residue played a role in only the thioester-forming step. The interaction between His207 and Glu410 in the second conformation (Figure 5C) was seen to be highly important for stabilizing this state of the enzyme (9).
The A8 motif contains a conserved hinge residue, most commonly an aspartic acid but also commonly present as a lysine. This residue undergoes main chain torsion angle rotations that are responsible for the majority of the difference between the two conformations. Indeed, the main chain dihedral angles of the neighboring residues are not altered between the two studies (67). The dynamics of this hinge residue has been explored structurally and kinetically by mutation of the 4CBL hinge residue, Asp402, to a proline residue (67). The ϕ/ψ angles of the proline residue allow the enzyme to adopt the adenylate-forming conformation; indeed, the crystal structure of the D402P mutant showed the overall fold of the enzyme was nearly identical to the wild-type enzyme in conformation I. The restraints imposed by the proline ring however prevent the enzyme from easily transitioning from conformation I to the thioester-forming conformation. Kinetic measurements showed that while the rate of the adenylation partial reaction was reduced ~3-fold, the rate constant for the thioester-forming reaction was reduced by four orders of magnitude.
Catalytic Role of Domain Alternation
Careful consideration of the structural and kinetic results provides insight into the role of the domain movement in the catalytic cycle. The side chain torsion of the A4 aromatic residue is conserved in the homologous crystal structures. In the adenylate-forming conformation, the A4 motif aromatic residue is located close to the carboxylate carbon where it positions this moiety to attack the α-phosphate and displace PPi. The χ1 side chain torsion angle for the aromatic residue is −176±6° in six members of the family that were crystallized in the adenylate-forming conformation bound with the adenylate intermediate or with ligands that mimic this state. Three exceptions were observed: the yeast Acs (46), human medium chain acyl-CoA synthetase (58), and the SrfA-C multi-domain NRPS (31), all of which lack both AMP and the acyl substrate. Thus it appears that when the acyl-adenylate is present, the aromatic residue from the A4 motif is directed at the carboxylate. In contrast, in the thioester-forming state, the A4 aromatic residue is rotated out of the active site and exhibits an average χ1 torsion angle of −59±14° in six different structures.
Examining the structures of the family members that contain CoA provides an explanation for this necessary rotation. The pantetheine tunnel is obstructed by the A4 aromatic residue in the side chain conformation observed in the adenylate-forming conformation (Figure 6). The rotation of the side chain allows the pantetheine group access to the carboxylate. The domain rotation brings the A8 loop into the active site where it provides an environment that promotes the rotation of the A4 aromatic residue to the −60° side chain orientation. The specific environment that the A8 residues provide depends on the identity of the A4 residue. In proteins that contain an A4 histidine, a glutamic acid side chain is most commonly present on the A8 loop to form a hydrogen bond with the histidine imidazole group. In proteins that contain a phenylalanine or tryptophan A4 residue, the A8 loop more commonly contains a tyrosine or histidine that promotes the hydrophobic environment.
Thus, one role of the domain alternation appears to be to provide a new environment for the aromatic residue of the A4 motif to remove it from the pantetheine tunnel and allow access to the adenylate intermediate. Associated with this movement in the 4CBL enzyme, there are numerous residues on the thioester-specific face of the C-terminal domain that are involved in binding the CoA nucleotide (57). Two hydrophobic residues sandwich the nucleotide adenine ring. Interestingly, the two other protein structures containing CoA, the bacterial Acs (52) and the human medium chain acyl-CoA synthetase (58), both bind to CoA in a manner that is different from 4CBL and from each other. In both of these structures the CoA nucleotide does not interact with the mobile C-terminal domain.
Considering the chemical requirements for the partial reactions performed by this enzyme family, one can understand the role that domain alternation plays in the catalytic mechanism. The adenylation reaction requires the nucleophilic attack of the negatively charged carboxylate from the acyl substrate on the negatively charged α-phosphate of ATP, displacing PPi. To accomplish this reaction, the enzyme must properly orient the acyl substrate. The enzyme thus appears to use the A4 aromatic residue to constrict the substrate binding pocket, properly positioning the carboxylate for nucleophilic attack on the α-phosphate. This required feature of the adenylation active site becomes a hindrance to the second partial reaction. In the thioester-forming reaction, the thiol of the CoA or pantetheine group must be able to approach the carboxylate carbon to displace the AMP leaving group and form the thioester linkage. Domain alternation provides the A4 residue with an appropriate new environment to induce its rotation out of the active site.
The domain rotation also creates the pantetheine tunnel, appropriate to bind the thiol substrate. In all three CoA bound structures, the backbone residues on the A8 loop, the universally conserved glycine and the residue preceding it, hydrogen bond to the amide nitrogen of the β-alanine moiety of the pantetheine. Additionally, as noted above, the C-terminal domain also can contain residues that are involved in binding the CoA nucleotide (57).
The structures of these proteins also provide insights into the timing of the domain rotation with respect to the catalytic cycle. Recent structures of members of the ANL family bound to ATP (51, 58) confirms an earlier proposal (57) that the β- and γ-phosphates would bind in a cavity filled by the A8 loop in the thioester-forming conformation. This prediction of the location of the binding position of the phosphates assumed the inline displacement of the PPi upon nucleophilic attack of the carboxylate on the α-phosphate. The steric clash between the A8 loop and the PPi binding pocket dictates that PPi must be released prior to the domain rotation from the adenylate-forming to the thioester-forming conformation. Additionally, the rotation of the A4 aromatic residue suggests that the pantetheine group cannot bind productively to the enzyme in the adenylate-forming conformation. The aromatic side chain occludes the thiol of the pantetheine from approaching the active site and suggests that the domain rotation to the thioester-forming conformation must precede binding of CoA.
Adenylation Domains of the Non-Ribosomal Peptide Synthetase
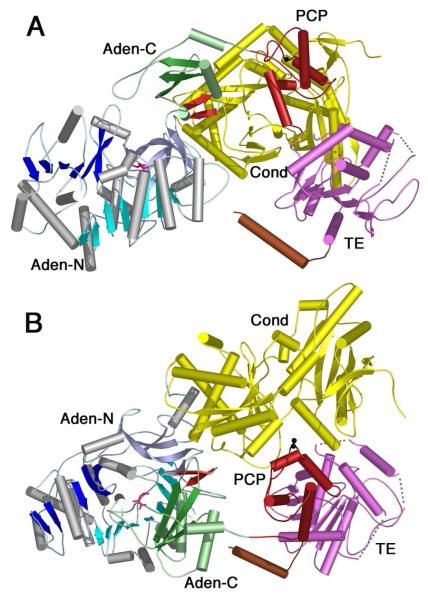
Much structural and biochemical evidence exists for the domain alternation hypothesis in the acyl-CoA synthetases. Limited evidence supports a role in the A8 loop specifically in the thioester-forming reaction of the NRPS adenylation domains (66). Very recently the crystal structure of a four domain NRPS was determined (31). The 1274 residue SrfA-C protein contains a full NRPS module organized as condensation, adenylation, peptidyl carrier, and thioesterase domains. This remarkable structure illustrated that the adenylation domain was positioned in a conformation that was similar, though not identical, to the adenylate-forming conformation (Figure 7A). The C-terminal domain is rotated away from the N-terminal domain by ~40° compared to the PheA structure, resulting in a more open active site. The active site of the adenylation domain contains the amino acyl substrate leucine, but does not contain AMP.
Figure 7. Crystal structure of the SrfA-C termination module from Surfactin NRPS cluster.
The structure contains the domains organized as Condensation-Adenylation-PCP-Thioesterase, from N- to C-terminus (2VSQ). The N-terminal domain is colored as other members of the ANL family with N-terminal domain containing β-sheets of blue and the C-terminal domain shown in green. The A8 loop is shown as the two-stranded β-sheet in red and a molecule of leucine is shown in pink in the adenylate-binding pocket. The Condensation domain (yellow), PCP domain (red) and thioesterase domain (purple) are shown. The C-terminal purification tag formed a helix that is shown in brown. The cofactor binding site, Ser1003, was mutated to an alanine and is shown in black. In panel A, the adenylation domain is oriented as for other members of the ANL family in Figures 3 and 4. The C-terminal domain most closely represents the adenylate-forming conformation, although it is opened by ~40° compared to other enzymes. In panel B, the image is rotated by ~90° around the X-axis to depict the presentation of the cofactor binding site to the condensation domain active site cleft. The pantetheine cofactor is not present in the structure yet would be unable to reach the adenylation domain active site without a large conformational change.
The PCP domain of the SrfA-C protein is positioned to interact productively with the upstream condensation domain (Figure 7B). The serine residue on which the pantetheine would be placed was mutated to an alanine to produce homogeneous apo-protein. Nonetheless, the serine residue is close enough to the condensation domain active site that the structure observed is likely the conformation used in the condensation domain reaction. In contrast, the PCP domain is not positioned where it could donate the pantetheine arm to either the adenylation domain or the thioesterase domain. The authors note that a conformational rearrangement may be required to reposition the PCP to interact with the alternate catalytic sites. The C-terminal domain rotation of the adenylation domain was suggested as an attractive candidate to play a role in the progression of the nascent peptide from the active site of one catalytic domain to the next. Extensive interactions exist between the condensation domain and the N-terminal sub-domain of the adenylation domain, which may serve as a “foundation” for each module. Upon completion of the adenylation partial reaction, the rotation of the C-terminal sub-domain of the adenylation domain could be used to transport the pantetheine cofactor into the adenylation domain active site where it would become amino acylated in the thioesterification partial reaction using the conformation observed in Acs. Release of the loaded PCP domain, the thioester product of the adenylation domain complete reaction, would accompany a rotation of the adenylation C-terminal sub-domain back to the conformation observed in PheA or SrfA-C. This would enable the delivery of the aminoacylated pantetheine cofactor to the upstream condensation domain for peptide bond formation. The conformational mechanism to direct the substrate to the downstream thioesterase domain remains to be determined.
This required domain rearrangement was modeled in a recent structural report of DltA (55). This structure was determined in the thioester-forming conformation bound to AMP. Extensive manual modeling was performed to generate a potential cycle of the DltA-catalyzed reaction (55). The reaction imitated in the unliganded, apo-state in which the C-terminal domain was in the open orientation seen in the original luciferase structure (39). The C-terminal domain of DltA was then modeled into the orientation observed in PheA to mimic the adenylation partial reaction. The orientation of the triphosphates of modeled ATP is consistent with the subsequently determined structure of the human medium chain Acs bound to ATP (58). The structure of DltA in the thioester-forming conformation was also used to model the interaction of DltA with the carrier protein DltC using the pantetheine group from the structure of Acs (52). The other constraint used in modeling the interaction derived from the distance between the C-terminus of the adenylation domain and the N-terminus of the carrier protein, which we had suggested to be a maximum of 20-25Å if the peptide linker were fully extended (66). This value was determined by comparison of the sequences of the linker joining the adenylation and PCP domains in multidomain NRPSs. In the structure of the SrfA-C multi-domain NRPS, the distance between the termini is 14Å.
Crystallographic studies of multi-domain NRPS enzymes have been challenged by the difficulty in creating a conformationally uniform population of protein molecules. The SrfA-C structure does show the overall architecture of a complete termination module of an NRPS (31) however it was necessary to mutate the phosphopantetheine binding site to obtain a uniform apo population of protein. Mutation of the hinge residue of the adenylation domains is one way to reduce the conformational flexibility of these large multi-domain enzymes. The structure of the D402P mutant of 4CBL demonstrates that the proline mutation forces 4CBL to adopt the adenylate forming conformation (67). A similar mutation to the hinge of NRPS adenylation domains may be a useful tool for reducing the conformational flexibility of adenylation domains and may allow the crystallization of larger, multi-domain NRPS proteins.
Conformational Changes in Other Adenylating Enzymes
The domain alternation hypothesis is presented as a strategy that enzymes of the ANL superfamily have adopted that allows them to catalyze the two-step adenylation and thioesterification reactions. As other ligases use an adenylation step to activate an acyl substrate, one might ask if other enzymes use a similar mechanism to stabilize the adenylation partial reaction and then allow the nucleophilic displacement of AMP in a second step.
A detailed description of other adenylate forming ligases, including amino acyl-tRNA synthetases (16, 68), NIS synthetases (17, 69), and ubiquitin ligases (70), is beyond the scope of this review. An analysis of the structures of multiple members of these families at different steps along the reaction coordinate identifies in some cases limited domain movements however the domain alternation strategy described for the ANL family of enzymes seems unique.
Why then have the ANL enzymes adopted the domain alternation catalytic strategy when other unrelated enzymes apparently accomplish similar chemistry without the dramatic conformational change? In these ligases, the energetically difficult step in the reaction is the initial nucleophilic attack of the carboxylate substrate on the α-phosphate of ATP. The ATP is held firmly in place through specific interactions with the triphosphate, the ribose hydroxyls, and the adenine base. The challenge to the ligase then is to direct the carboxylate substrate to the α-phosphate to promote the adenylate formation. It is reasonable to propose that the earliest members of the ANL enzyme superfamily were acyl-CoA synthetases that use chemically uninteresting fatty acids. Because the acyl substrates did not contain additional groups that the enzyme could use to position the carboxylate, the ANL enzymes evolved the A4 aromatic residue to position the carboxylate substrate and use domain alternation to allow access to this atom in the thioester-forming reaction.
The enzyme ligases mentioned above do not require a domain alternation strategy because their carboxylate substrates (amino acids in amino acyl-tRNA synthetases, di- or tricarboxylate compounds like citrate or α-ketoglutarate in the NIS synthetases, or protein molecules for ubiquitin ligases) contain numerous functional groups that can provide specific binding interactions between the adenylating enzyme and the nucleophilic substrate. The presence of these groups obviates the need to tightly surround the carboxylate carbon, leaving this atom accessible for attack in the second partial reaction. Even the glycyl-tRNAGly synthetase enzyme (71), which has a relatively simple amino acyl substrate, uses multiple interactions, including a hydrogen bond to the glycine α-proton, to position the carboxylate substrate precisely. These multiple interactions leave the glycine carboxylate carbon open for attack by the tRNA acceptor chain in the second partial reaction.
Summary
The importance of dynamics and conformational changes to proteins is now well established and allows enzymes to shield reactive intermediates and to induce the alignment of substrates and reactive catalytic groups from the protein. Several examples of large scale domain rotations have been described that are distinct from the simple closing of catalytic loops that are often on the order of 10-20°. Generally, conformational changes that involve domain rotations larger than 50° are used to transport a substrate or intermediate between active sites. What makes the domain alternation of the ANL enzyme family unique is that this strategy allows the enzyme to present two different faces of a single protein domain to a single active site that catalyzes both reactions.
The extensive structural and functional investigations of the ANL enzyme family over the last decade have not only identified this catalytic strategy but have also explained the catalytic advantage that is derived from the use of the domain alternation. This is yet another example of the ways that enzymes continue to fascinate us with the ability to use unpredictable mechanisms to catalyze challenging chemical reactions. That this understanding explains observations first made over 40 years ago is particularly satisfying and points once again to the value of the complementary approaches of detailed kinetic and structural investigation.
Finally, the study of multiple members of this enzyme family has provided tremendous insights into the catalytic cycle of the modular NRPS proteins. These assembly line proteins require the transfer of substrates between different catalytic domains and the recent structural advances demonstrate that the pantetheine cofactor is not sufficiently long to act solely as a swinging arm to transport the substrates. Instead, it appears that coordinated conformational movements are required to carry out this elaborate dynamic cycle. The domain alternation strategy of the NRPS adenylation domains is likely a necessary component of this modular protein family that ensures proper delivery of the bound intermediates to the catalytic domains.
Acknowledgments
I am very grateful to Drs. Debra Dunaway-Mariano and Bruce Branchini for helpful comments and discussion throughout the course of our study of the ANL family of adenylating enzymes. I would also like to thank the members of my laboratory, Albert Reger, Eric Drake, Manish Shah, Jesse Sundlov, and Carter Mitchell, for their work on this project and their contributions made to the understanding of these enzymes.
Glossary Terms
- ANL Superfamily of Adenylating Enzymes
An enzyme superfamily containing acyl- and aryl-CoA synthetases, the adenylation domains of NRPS modular proteins, and firefly luciferase.
- Modular Enzyme
Enzymes that contain multiple catalytic domains joined in a single polypeptide to catalyze sequential biosynthetic steps.
- Non-Ribosomal Peptide Synthetase
A modular enzyme that is responsible for the synthesis of peptide antibiotics and peptide siderophores. The amino acid and peptide intermediates are covalently bound to a peptide carrier domain through a pantetheine cofactor during synthesis by the individual catalytic domains.
- Domain Alternation
A catalytic strategy where an enzyme uses large scale domain rotations to catalyze different steps of a multistep reaction. The conformational changes are distinct from simple opening and closing of an active site loop.
- Peptidyl Carrier Protein
A four-helix dynamic protein domain that contains a pantetheine cofactor bound to a conserved serine residue, on which amino acid and peptide intermediates are bound during NRPS synthesis.
Abbreviations
- Acs
Acetyl-CoA synthetase
- ArCP
aryl-carrier protein
- 4CB
4-chlorobenzoate
- 4CBL
4-chlorobenzoyl-CoA Ligase
- DHB
2,3-dihydroxybenzoic acid
- FAAL
fatty acyl-AMP ligase
- NIS
NRPS-independent siderophore
- NRPS
Non-ribosomal peptide synthetase
- PCP
peptidyl-carrier protein
- PheA
phenylalanine-adenylating domain of GrsA
- PPi
inorganic pyrophosphate.
Footnotes
Work from my laboratory was supported in part by NIH grant GM-068440.
References
- 1.McElroy WD, DeLuca M, Travis J. Molecular uniformity in biological catalyses. The enzymes concerned with firefly luciferin, amino acid, and fatty acid utilization are compared. Science. 1967;157:150–160. doi: 10.1126/science.157.3785.150. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Tana J, Rose G. Studies on medium-chain fatty acyl-coenzyme a synthetase. Enzyme fraction I: mechanism of reaction and allosteric properties. Biochem J. 1968;109:275–282. doi: 10.1042/bj1090275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipmann F. Enzymatic synthesis of acetyl phosphate. J Biol Chem. 1944;155:55–70. [Google Scholar]
- 4.Berg P. Acyl adenylates; an enzymatic mechanism of acetate activation. J Biol Chem. 1956;222:991–1013. [PubMed] [Google Scholar]
- 5.Farrar WW, Plowman KM. Kinetics of acetyl-CA synthetase–1. Mode of addition of substrates. International J. Biochem. 1975;6:537–542. [Google Scholar]
- 6.Kim YS, Kang SW. Steady-state kinetics of malonyl-CoA synthetase from Bradyrhizobium japonicum and evidence for malonyl-AMP formation in the reaction. Biochem J. 1994;297(Pt 2):327–333. doi: 10.1042/bj2970327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Melton EM, Quackenbush S, DiRusso CC, Black PN. Mechanistic studies of the long chain acyl-CoA synthetase Faa1p from Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:1246–1253. doi: 10.1016/j.bbalip.2007.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian Y, Suk DH, Cai F, Crich D, Mesecar AD. Bacillus anthracis o-Succinylbenzoyl-CoA Synthetase: Reaction Kinetics and a Novel Inhibitor Mimicking Its Reaction Intermediate. Biochemistry. 2008 doi: 10.1021/bi801311d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu R, Cao J, Lu X, Reger AS, Gulick AM, Dunaway-Mariano D. Mechanism of 4-chlorobenzoate:coenzyme a ligase catalysis. Biochemistry. 2008;47:8026–8039. doi: 10.1021/bi800698m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babbitt PC, Kenyon GL, Martin BM, Charest H, Slyvestre M, Scholten JD, Chang KH, Liang PH, Dunaway-Mariano D. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry. 1992;31:5594–5604. doi: 10.1021/bi00139a024. [DOI] [PubMed] [Google Scholar]
- 11.Turgay K, Krause M, Marahiel MA. Four homologous domains in the primary structure of GrsB are related to domains in a superfamily of adenylate-forming enzymes. Mol Microbiol. 1992;6:529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 12.Diez B, Gutierrez S, Barredo JL, van Solingen P, van der Voort LH, Martin JF. The cluster of penicillin biosynthetic genes. Identification and characterization of the pcbAB gene encoding the alpha-aminoadipyl-cysteinyl-valine synthetase and linkage to the pcbC and penDE genes. J Biol Chem. 1990;265:16358–16365. [PubMed] [Google Scholar]
- 13.Hori K, Yamamoto Y, Minetoki T, Kurotsu T, Kanda M, Miura S, Okamura K, Furuyama J, Saito Y. Molecular cloning and nucleotide sequence of the gramicidin S synthetase 1 gene. J Biochem (Tokyo) 1989;106:639–645. doi: 10.1093/oxfordjournals.jbchem.a122909. [DOI] [PubMed] [Google Scholar]
- 14.Rusnak F, Faraci WS, Walsh CT. Subcloning, expression, and purification of the enterobactin biosynthetic enzyme 2,3-dihydroxybenzoate-AMP ligase: demonstration of enzyme-bound (2,3-dihydroxybenzoyl)adenylate product. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 15.Gerlt JA, Babbitt PC. Divergent evolution of enzymatic function: mechanistically diverse superfamilies and functionally distinct suprafamilies. Annu Rev Biochem. 2001;70:209–246. doi: 10.1146/annurev.biochem.70.1.209. [DOI] [PubMed] [Google Scholar]
- 16.Francklyn CS. DNA polymerases and aminoacyl-tRNA synthetases: shared mechanisms for ensuring the fidelity of gene expression. Biochemistry. 2008;47:11695–11703. doi: 10.1021/bi801500z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Challis GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem. 2005;6:601–611. doi: 10.1002/cbic.200400283. [DOI] [PubMed] [Google Scholar]
- 18.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 19.Kopp F, Marahiel MA. Where chemistry meets biology: the chemoenzymatic synthesis of nonribosomal peptides and polyketides. Curr Opin Biotechnol. 2007;18:513–520. doi: 10.1016/j.copbio.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Walsh CT. The chemical versatility of natural-product assembly lines. Acc Chem Res. 2008;41:4–10. doi: 10.1021/ar7000414. [DOI] [PubMed] [Google Scholar]
- 21.Weissman KJ, Muller R. Protein-protein interactions in multienzyme megasynthetases. Chembiochem. 2008;9:826–848. doi: 10.1002/cbic.200700751. [DOI] [PubMed] [Google Scholar]
- 22.Lai JR, Koglin A, Walsh CT. Carrier protein structure and recognition in polyketide and nonribosomal peptide biosynthesis. Biochemistry. 2006;45:14869–14879. doi: 10.1021/bi061979p. [DOI] [PubMed] [Google Scholar]
- 23.Mercer AC, Burkart MD. The ubiquitous carrier protein--a window to metabolite biosynthesis. Nat Prod Rep. 2007;24:750–773. doi: 10.1039/b603921a. [DOI] [PubMed] [Google Scholar]
- 24.MacCabe AP, van Liempt H, Palissa H, Unkles SE, Riach MB, Pfeifer E, von Dohren H, Kinghorn JR. Delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase from Aspergillus nidulans. Molecular characterization of the acvA gene encoding the first enzyme of the penicillin biosynthetic pathway. J Biol Chem. 1991;266:12646–12654. [PubMed] [Google Scholar]
- 25.Schofield CJ, Baldwin JE, Byford MF, Clifton I, Hajdu J, Hensgens C, Roach P. Proteins of the penicillin biosynthesis pathway. Curr Opin Struct Biol. 1997;7:857–864. doi: 10.1016/s0959-440x(97)80158-3. [DOI] [PubMed] [Google Scholar]
- 26.Lai JR, Fischbach MA, Liu DR, Walsh CT. Localized protein interaction surfaces on the EntB carrier protein revealed by combinatorial mutagenesis and selection. J Am Chem Soc. 2006;128:11002–11003. doi: 10.1021/ja063238h. [DOI] [PubMed] [Google Scholar]
- 27.Lai JR, Fischbach MA, Liu DR, Walsh CT. A protein interaction surface in nonribosomal peptide synthesis mapped by combinatorial mutagenesis and selection. Proc Natl Acad Sci U S A. 2006;103:5314–5319. doi: 10.1073/pnas.0601038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G. Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koglin A, Lohr F, Bernhard F, Rogov VV, Frueh DP, Strieter ER, Mofid MR, Guntert P, Wagner G, Walsh CT, Marahiel MA, Dotsch V. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature. 2008;454:907–911. doi: 10.1038/nature07161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koglin A, Mofid MR, Lohr F, Schafer B, Rogov VV, Blum MM, Mittag T, Marahiel MA, Bernhard F, Dotsch V. Conformational switches modulate protein interactions in peptide antibiotic synthetases. Science. 2006;312:273–276. doi: 10.1126/science.1122928. [DOI] [PubMed] [Google Scholar]
- 31.Tanovic A, Samel SA, Essen LO, Marahiel MA. Crystal Structure of the Termination Module of a Nonribosomal Peptide Synthetase. Science. 2008 doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 32.Quadri LE. Assembly of aryl-capped siderophores by modular peptide synthetases and polyketide synthases. Mol Microbiol. 2000;37:1–12. doi: 10.1046/j.1365-2958.2000.01941.x. [DOI] [PubMed] [Google Scholar]
- 33.Ferrer JL, Austin MB, Stewart C, Jr., Noel JP. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids. Plant Physiol Biochem. 2008;46:356–370. doi: 10.1016/j.plaphy.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arora P, Goyal A, Natarajan VT, Rajakumara E, Verma P, Gupta R, Yousuf M, Trivedi OA, Mohanty D, Tyagi A, Sankaranarayanan R, Gokhale RS. Mechanistic and functional insights into fatty acid activation in Mycobacterium tuberculosis. Nat Chem Biol. 2009;5:166–173. doi: 10.1038/nchembio.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trivedi OA, Arora P, Sridharan V, Tickoo R, Mohanty D, Gokhale RS. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- 36.Gehring AM, Mori I, Walsh CT. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 37.Chang KH, Xiang H, Dunaway-Mariano D. Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry. 1997;36:15650–15659. doi: 10.1021/bi971262p. [DOI] [PubMed] [Google Scholar]
- 38.Marahiel MA, Stachelhaus T, Mootz HD. Modular Peptide Synthetases Involved in Nonribosomal Peptide Synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 39.Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 40.Gocht M, Marahiel MA. Analysis of core sequences in the D-Phe activating domain of the multifunctional peptide synthetase TycA by site-directed mutagenesis. J Bacteriol. 1994;176:2654–2662. doi: 10.1128/jb.176.9.2654-2662.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Blanco H, Reglero A, Rodriguez-Aparicio LB, Luengo JM. Purification and biochemical characterization of phenylacetyl-CoA ligase from Pseudomonas putida. A specific enzyme for the catabolism of phenylacetic acid. J Biol Chem. 1990;265:7084–7090. [PubMed] [Google Scholar]
- 42.Conti E, Stachelhaus T, Marahiel MA, Brick P. Structural basis for the activation of phenylalanine in the non- ribosomal biosynthesis of gramicidin S. Embo J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Branchini BR, Murtiashaw MH, Magyar RA, Anderson SM. The role of lysine 529, a conserved residue of the acyl-adenylate- forming enzyme superfamily, in firefly luciferase. Biochemistry. 2000;39:5433–5440. doi: 10.1021/bi9928804. [DOI] [PubMed] [Google Scholar]
- 44.Horswill AR, Escalante-Semerena JC. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonella enterica: residue Lys592 is required for propionyl-AMP synthesis. Biochemistry. 2002;41:2379–2387. doi: 10.1021/bi015647q. [DOI] [PubMed] [Google Scholar]
- 45.May JJ, Kessler N, Marahiel MA, Stubbs MT. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc Natl Acad Sci U S A. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jogl G, Tong L. Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP. Biochemistry. 2004;43:1425–1431. doi: 10.1021/bi035911a. [DOI] [PubMed] [Google Scholar]
- 47.Gulick AM, Lu X, Dunaway-Mariano D. Crystal structure of 4-chlorobenzoate:CoA ligase/synthetase in the unliganded and aryl substrate-bound states. Biochemistry. 2004;43:8670–8679. doi: 10.1021/bi049384m. [DOI] [PubMed] [Google Scholar]
- 48.Bains J, Boulanger MJ. Biochemical and structural characterization of the paralogous benzoate CoA ligases from Burkholderia xenovorans LB400: defining the entry point into the novel benzoate oxidation (box) pathway. J Mol Biol. 2007;373:965–977. doi: 10.1016/j.jmb.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 49.Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006;440:372–376. doi: 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- 50.Du L, He Y, Luo Y. Crystal structure and enantiomer selection by D-alanyl carrier protein ligase DltA from Bacillus cereus. Biochemistry. 2008;47:11473–11480. doi: 10.1021/bi801363b. [DOI] [PubMed] [Google Scholar]
- 51.Osman KT, Du L, He Y, Luo Y. Crystal structure of Bacillus cereus D-alanyl carrier protein ligase (DltA) in complex with ATP. J Mol Biol. 2009;388:345–355. doi: 10.1016/j.jmb.2009.03.040. [DOI] [PubMed] [Google Scholar]
- 52.Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. The 1.75 A crystal structure of acetyl-CoA synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry. 2003;42:2866–2873. doi: 10.1021/bi0271603. [DOI] [PubMed] [Google Scholar]
- 53.Bandarian V, Pattridge KA, Lennon BW, Huddler DP, Matthews RG, Ludwig ML. Domain alternation switches B(12)-dependent methionine synthase to the activation conformation. Nat Struct Biol. 2002;9:53–56. doi: 10.1038/nsb738. [DOI] [PubMed] [Google Scholar]
- 54.Hisanaga Y, Ago H, Nakagawa N, Hamada K, Ida K, Yamamoto M, Hori T, Arii Y, Sugahara M, Kuramitsu S, Yokoyama S, Miyano M. Structural basis of the substrate-specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. J Biol Chem. 2004;279:31717–31726. doi: 10.1074/jbc.M400100200. [DOI] [PubMed] [Google Scholar]
- 55.Yonus H, Neumann P, Zimmermann S, May JJ, Marahiel MA, Stubbs MT. Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains. J Biol Chem. 2008;283:32484–32491. doi: 10.1074/jbc.M800557200. [DOI] [PubMed] [Google Scholar]
- 56.Shah MB, Ingram-Smith C, Cooper LL, Qu J, Meng Y, Smith KS, Gulick AM. The 2.1Å crystal structure of an acyl-CoA synthetase from Methanosarcina acetivorans reveals an alternate acyl binding pocket for small branched acyl substrates. Proteins. 2009 doi: 10.1002/prot.22482. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reger AS, Wu R, Dunaway-Mariano D, Gulick AM. Structural characterization of a 140° domain movement in the two-step reaction catalyzed by 4-chlorobenzoate:CoA ligase. Biochemistry. 2008;47:8016–8025. doi: 10.1021/bi800696y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kochan G, Pilka ES, von Delft F, Oppermann U, Yue WW. Structural snapshots for the conformation-dependent catalysis by human medium-chain Acyl-coenzyme A synthetase ACSM2A. J Mol Biol. 2009;388:997–1008. doi: 10.1016/j.jmb.2009.03.064. [DOI] [PubMed] [Google Scholar]
- 59.Goyal A, Yousuf M, Rajakumara E, Arora P, Gokhale RS, Sankaranarayanan R. Crystallization and preliminary X-ray crystallographic studies of the N-terminal domain of FadD28, a fatty-acyl AMP ligase from Mycobacterium tuberculosis. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:350–352. doi: 10.1107/S1744309106005938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reger AS, Carney JM, Gulick AM. Biochemical and Crystallographic Analysis of Substrate Binding and Conformational Changes in Acetyl-CoA Synthetase. Biochemistry. 2007;46:6536–6546. doi: 10.1021/bi6026506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 62.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 64.Lautru S, Deeth RJ, Bailey LM, Challis GL. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 65.Branchini BR, Southworth TL, Murtiashaw MH, Wilkinson SR, Khattak NF, Rosenberg JC, Zimmer M. Mutagenesis evidence that the partial reactions of firefly bioluminescence are catalyzed by different conformations of the luciferase C-terminal domain. Biochemistry. 2005;44:1385–1393. doi: 10.1021/bi047903f. [DOI] [PubMed] [Google Scholar]
- 66.Drake EJ, Nicolai DA, Gulick AM. Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the EntE adenylation domain. Chem Biol. 2006;13:409–419. doi: 10.1016/j.chembiol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 67.Wu R, Reger AS, Lu X, Gulick AM, Dunaway-Mariano D. The Mechanism of Domain Alternation in the Acyl-Adenylate Forming Ligase Superfamily Member 4-Chlorobenzoate: Coenzyme A Ligase. Biochemistry. 2009;48:4115–4125. doi: 10.1021/bi9002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sekine S, Nureki O, Dubois DY, Bernier S, Chenevert R, Lapointe J, Vassylyev DG, Yokoyama S. ATP binding by glutamyl-tRNA synthetase is switched to the productive mode by tRNA binding. EMBO J. 2003;22:676–688. doi: 10.1093/emboj/cdg053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmelz S, Kadi N, McMahan SA, Song L, Oves-Costales D, Oke M, Liu H, Johnson KA, Carter L, Botting CH, White MF, Challis GL, Naismith JH. Structure and function of AcsD a new class of adenylating enzyme that catalyzes enantioselective citrate desymmetrization in pathogenicity-conferring siderophore biosynthesis. Nat Chem Bio. 2009;5:174–182. doi: 10.1038/nchembio.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee I, Schindelin H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell. 2008;134:268–278. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 71.Arnez JG, Dock-Bregeon AC, Moras D. Glycyl-tRNA synthetase uses a negatively charged pit for specific recognition and activation of glycine. J Mol Biol. 1999;286:1449–1459. doi: 10.1006/jmbi.1999.2562. [DOI] [PubMed] [Google Scholar]