Abstract
KPC β-lactamases hydrolyze the “last resort” β-lactam antibiotics (carbapenems) used to treat multi-drug resistant infections, and are compromising efforts to combat life-threatening Gram-negative bacterial infections in hospitals worldwide. Consequently, the development of novel inhibitors is essential for restoring the effectiveness of existing antibiotics. The β-lactamase inhibitor protein (BLIP) is a competitive inhibitor of a number of class A β-lactamases. In this study, we characterize the previously unreported interaction between the KPC-2 β-lactamase and BLIP. Biochemical results show that BLIP is an extremely potent inhibitor of KPC enzymes, binding KPC-2 and KPC-3 with subnanomolar affinity. To understand the basis of affinity and specificity in the β-lactamase/BLIP system, the crystallographic structure of the KPC-2/BLIP complex was solved to 1.9 Å resolution. Computational alanine scanning was also conducted to identify putative hot spots in the KPC-2/BLIP interface. Interestingly, the two complexes making up the KPC-2/BLIP asymmetric unit are distinct, and in one structure the BLIP F142 loop is absent, in contrast to homologous structures where it occupies the active site. This finding and other sources of structural plasticity appear to contribute to BLIP’S promiscuity, enabling it to respond to mutations at the β-lactamase interface. Given the continuing emergence of antibiotic resistance, the high-resolution KPC-2/BLIP structure will facilitate its use as a template for the rational design of new inhibitors of this problematic enzyme.
KPC (Klebsiella pneumoniae carbapenemases) β-lactamases confer resistance to extended-spectrum cephalosporins and carbapenems, and have emerged as a significant worldwide threat in the treatment of Gram-negative bacterial infections (1). Along with the frequently encountered homologous TEM and SHV β-lactamases, KPCs are Ambler class A enzymes, but unlike TEM-1 and SHV-1, KPCs are able to hydrolyze “last resort” β-lactam antibiotics, the carbapenems (imipenem, meropenem, doripenem, and ertapenem) used to treat multi-drug resistant infections (2). Although only recently discovered in K. pneumoniae isolates in the United States in 2001, KPC enzymes have spread both globally (United States, China, France, Israel), and to many other Enterobacteriaceae (Enterobacter cloacae, Serratia marcescens, and Escherichia coli) at an alarming rate (1). Furthermore, existing β-lactamase inhibitors have limited efficacy against KPC enzymes: thus, developing an understanding of how to effectively inhibit this enzyme has direct public health consequences. Therefore, by studying the interaction between KPC-2 and a potent protein inhibitor, we hope to glean information useful for the development of novel therapeutics.
BLIP, an 18 kDa protein isolated from the soil bacterium Streptomyces clavuligeris, has been shown to be a potent inhibitor of many class A β-lactamases. BLIP recognizes SHV-1 (K. pneumoniae) with micromolar affinity; TEM-1 (E. coli), SME-1 (S. marcescens), and Bla1 (Bacillus anthracis) with nanomolar affinity; and K1 (Proteus vulgaris) with picomolar affinity (3). To date, several structures of BLIP in complex with TEM-1 and SHV-1 have been determined (4). These structural models show that BLIP interacts with its targets by docking the predominantly polar, concave β-sheet surface onto the enzyme, burying ~2500 Å2 of surface area in a well-hydrated interface. BLIP competitively inhibits its targets with two binding loops that occlude the active site. Key sidechains in each loop mimic interactions observed with the acyl-enzyme intermediate bound to its antibiotic substrate: BLIP F142 occupies a similar position as the benzyl moiety of penicillinG, while BLIP D49 is involved in a hydrogen bond network in the active site, in a similar position as the penicillinG carboxylate.
In addition to its importance for the development of new β-lactamase inhibitors, BLIP interactions present a relevant platform for studying the biochemical determinants of affinity and specificity. Interestingly, the two most closely related of BLIP’S binding partners, TEM-1 and SHV-1, share 67% amino acid identity, yet their affinities for BLIP differ by a thousand-fold (Figure 1). In contrast, TEM-1, SME-1, and Bla1 share only ~30% identity, and show a similar affinity for BLIP. In order to understand the molecular basis of affinity, alanine scanning mutagenesis of BLIP against TEM-1, SHV-1, Bla1, and SME-1 has been reported (5, 6). In addition, the TEM-1/BLIP (and to a lesser extent the SHV-1/BLIP) complex is well characterized experimentally, with mutagenesis, structural, and computational data reported (7).
Figure 1.
Sequence alignment between TEM-1, SHV-1, and KPC-2 showing conserved residues (gray), conserved interface residues (purple), and nonconserved interface residues (orange). KPC-2 shares 39% (41%) sequence identity to TEM-1 (SHV-1) overall, and 39% identity when the comparison is restricted to interface residues.
Here we describe the previously uncharacterized interaction between BLIP and KPC-2. The results indicate that BLIP inhibits KPC-2 and KPC-3 β-lactamases with subnanomolar affinity. Diffraction data collected from two crystal forms resulted in two structures of the KPC-2(G175S)/BLIP complex: a 1.9 Å resolution co-structure (PDB ID 3E2L), and a 2.1 Å resolution co-structure (PDB ID 3E2K)1. Overall, the KPC-2/BLIP interaction is similar to the TEM-1/BLIP and SHV-1/BLIP interactions, with the key difference that the BLIP F142 binding loop is absent from the interface in one complex in the asymmetric unit (AU). This displacement seems to be driven by subtly different orientations of BLIP with respect to KPC, as observed between the two complexes contained in the AU. Further examples of BLIP’S plasticity, a feature enabling its broad recognition, along with the biophysical implications of such flexibility are discussed.
Experimental Procedures
Cloning, Protein Expression, and Purification
BLIP was purified as described elsewhere (8). The pBR322 plasmid producing blaKPC-2 was obtained as a kind gift (F. Tenover, CDC, Atlanta GA), and subcloned into a pET-24a(+) based vector behind an OmpA signal sequence; other KPC sequences were obtained by site-directed mutagenesis. KPC proteins were expressed in E. coli BL21(DE3) grown at 30°C by induction with isopropyl-B-D-thiogalactopyranoside. Cells were harvested by centrifugation, and the periplasmic fraction was isolated by osmotic shock. The resulting solution was passed over a phenylboronate column (MoBiTec), and the β-lactamase was eluted with borate (0.5 M borate, pH 7.0, containing 0.5 M NaCl), followed by overnight dialysis against PBS. Using the same buffer, size exclusion chromatography (using a HiLoad 26/60 Superdex 75 column, GE Healthcare) then served as both an additional purification and buffer exchange step. After purification, KPC β-lactamase containing fractions were concentrated, flash frozen, and stored at −80°C. Protein purity was assessed by observation of a single species by SDS polyacrylamide gel electrophoresis. For the initial protein preparation, the mass was verified by MALDI-TOF mass spectrometry to ensure proper processing of the signal sequence. The mass was that expected of the mature protein, with no boronyl adducts. This initial preparation was used for experimental inhibition assays and initial crystallization screens; additional protein purifications were prepared and used for structural studies without verification by mass spectrometry or activity assays.
Crystallization, Data Collection, and Structure Solution
Initial crystallization screens Hampton1&2, Hampton Peg/Ion (Hampton Research), Wizard I–III (Emerald BioSystems) were set using an Oryx8 crystallization robot in sitting drop format. KPC-2 and BLIP were mixed in 1:1 ratios and concentrated to 3.0 mg/mL in 10 mM NaCl, 10 mM BisTris, pH 7.25, dialyzed overnight against the same buffer. Hanging drop trays were set by combining 1μL well solution with 1μL protein solution to pursue initial crystallization hits. After refining conditions, two different forms of diffraction quality crystals were produced by seeding into 20% PEG 8000, 6% ethylene glycol, 100 mM citrate, pH 5.0 (crystal 1), or 20% PEG 8000, 4% ethylene glycol, 100 mM citrate, pH 4.5 (crystal 2). Either 20% ethylene glycol (crystal 1), or 20% xylitol (crystal 2) was added as a cryoprotectant, and crystals were looped and flash frozen in liquid nitrogen.
Datasets were collected at beamline 8.3.1 at the Advanced Light Source (LBNL, Berkeley CA). Diffraction data were scaled and integrated using HKL2000 (9); phases were found by performing sequential searches with PHASER for KPC-2 (PDB ID 20V5) and BLIP monomer taken from the TEM-1/BLIP co-structure (PDB ID 1JTG) (10). Two datasets were processed: one to 1.9Å (spacegroup C2, PDB ID 3E2L) and the other to 2.1 Å (spacegroup P212121, PBD ID 3E2K); both contained two complexes in the AU. Iterations between manual rebuilding in COOT and refinement with PHENIX generated the final structural models (11,12); TLS groups were chosen according to TLSMD (13). Details of the crystallographic data processing and refinement are in given in Table 1. Structure alignments were calculated with LSQMAN (14); all molecular figures were prepared using PyMol2.
Table 1.
Crystallographic data collection, refinement and stereochemistry parameters.
| 3E2L | 3E2K | |
|---|---|---|
| Data Collection: | ||
| Resolution (Å) | 200.00-1.87 | 50.00-2.10 |
| Wavelength (Å) | 1.1159 | 1.1159 |
| Space Group | C2 | P212121 |
| Unit Cell Dimensions (a,b,c) Å | 162.2, 66.4, 82.2 | 41.2, 76.2, 241.4 |
| Unit Cell Angles (α,β,γ)° | 90.0, 101.3, 90.0 | 90.0, 90.0, 90.0 |
| I/σ (last shell) | 12.6 (2.0) | 13.8 (3.2) |
| Rsym (last shell) (%) | 8.4 (46.3) | 12.7(68.9) |
| Completeness (last shell) | 98.2 (82.0) | 99.5 (95.0) |
| No. of reflections | 233,579 | 329,383 |
| unique | 66,559 | 46,130 |
| Refinement: | ||
| Resolution (Å) | 51.79-1.87 | 40.78-2.09 |
| No. of reflections | 66.559 | 46.130 |
| working | 63,183 | 43,801 |
| free (% total) | 3,376 (5.1) | 2,329 (5.1) |
| Rwork (last shell) (%) | 16.9 (25.3) | 19.0 (20.9) |
| Rfree (last shell) (%) | 21.3 (32.6) | 23.4 (24.5) |
| Structure and Stereochemistry | ||
| No. of atoms | 7,011 | 6,523 |
| protein | 6,309 | 6,319 |
| water | 702 | 204 |
| RMSD bond lengths (Å) | 0.008 | 0.005 |
| RMSD bond angles (°) | 1.124 | 0.951 |
Experimental Inhibition Assays
Inhibition constants between β-lactamase and BLIP were determined by competition with the β-lactam substrate nitrocefin (Becton Dickinson Biosciences, Cockeysville, MD). Assays were performed in quadruplicate in a 96-well plate format at 26°C. Assay were performed with 1.0 nM enzyme and BLIP concentrations from 63 pM to 4 nM, with 200 μM nitrocefin, 0.1 mg/mL BSA, and in PBS (pH 7.4). The liquid handling capabilities of the FlexstationIII initiated reactions by the addition of substrate, and absorbance at 486 nm was monitored. Data was normalized to the observed activity without the inhibitor (fractional activity). IC50 values were obtained by plotting fractional activity versus total inhibitor concentration, and fitting to Morrison’s equation (Eq 1) with OriginPro.
| Eq (1) |
The inhibition constant, Ki, was calculated from the IC50 value by applying the Cheng-Prusoff correction (Eq 2) (15).
| Eq (2) |
The Km of the enzyme for the β-lactam substrate, nitrocefin, was experimentally determined in a separate experiment, by fitting to the Henri-Michaelis-Menton equation. The Km and kcat values for KPC-2 determined here are 32 ± 9 μM and 30 ± 3 sec−1 respectively, which agree well with literature values (16).
Computational Alanine Scanning
The EGAD library was employed to calculate the effects of alanine mutation on dissociation energy (7). The energy function includes a linearized vdW potential, coulombic electrostatics term, torsional potential, solvent accessible surface area term, and generalized Born model to describe solvation. Bound and unbound backbone structures were fixed, sidechains at interfacial and neighboring positions were allowed to change conformation in response to mutation.
Unbound structures were generated by translating chains in the bound conformations to a distance of 50 Å apart. The energy of both bound and unbound states were minimized using Monte Carlo optimization followed by a heuristic quench step. Binding energies were calculated as the energy difference between bound and unbound states; the energy change upon mutation was calculated by subtracting the mutant and wild type states.
Calculations were performed for both complexes in the AU of the higher resolution KPC-2/BLIP structure. Resulting energies were similar, and data are shown for the interface between chain A and chain C.
Results
BLIP’S conserved binding mode
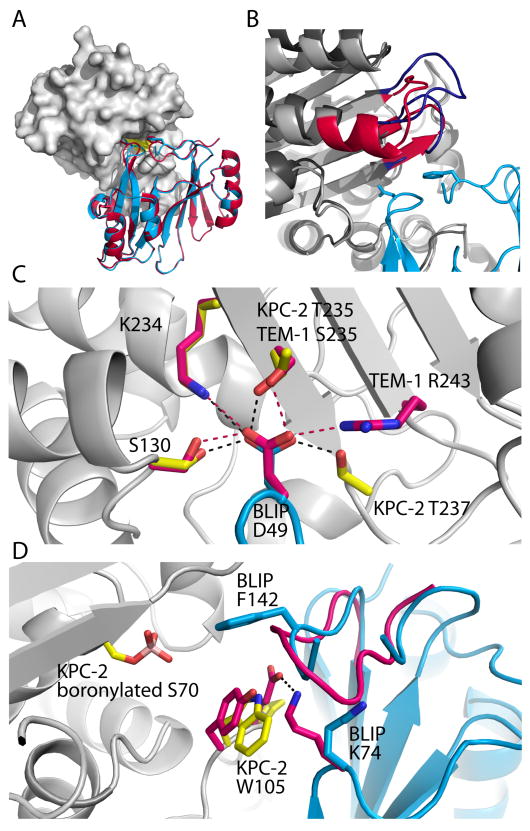
BLIP shows high affinity for KPC enzymes: the Ki values for KPC-2, KPC-2 (G175S), and KPC-3 are 84 ± 3, 69 ± 6, and 250 ± 20 pM respectively. The 1.9 Å structure of the KPC-2/BLIP complex is globally similar to the TEM-1/BLIP and SHV-1/BLIP interfaces (Figure 2A). Structural alignments between the KPC-2/BLIP and TEM-1/BLIP or SHV-1/BLIP complexes yield overall Cα RMSDs of 1.3 Å in both cases (388 Cα atoms aligned for the TEM comparison, and 395 Cα atoms aligned for the SHV comparison). The KPC-2 and TEM-1 backbone structures differ only slightly in the vicinity of the BLIP interface. TEM-1 residues 270–274 are at the N-terminus of the last α-helix, while the corresponding KPC-2 residues are in an extended loop conformation (Figure 2B). The extended KPC-2 loop residues lose solvent accessible surface area upon binding BLIP, but do not make specific intersubunit interactions. Formation of the KPC-2/BLIP complex buries 2852 Å2 of surface area (interface 1, between chains A and C) or 2507 Å2 (interface 2, between chains B and D) in heavily hydrated interfaces. Both KPC-2/BLIP and TEM-1/BLIP interfaces have similar numbers of polar and vdW interactions - there are 10 hydrogen bonds and 1 salt bridge at the KPC-2/BLIP interface, which is comparable to the 9 hydrogen bonds and 3 salt bridges at the TEM-1/BLIP interface (Supporting Information).
Figure 2.
Architecture of the KPC-2/BLIP interface. A) BLIP interacts with KPC-2 (gray) similar to its interaction with TEM-1; BLIP D49 and F142 loops occupy the active site (yellow). BLIP from the TEM-1 co-structure (red - PDB ID 1JTG) is aligned with BLIP in the KPC-2/BLIP structure (cyan - PDB ID 3E2L). B) KPC-2 contains a few extra residues that lose solvent accessible surface area upon binding BLIP, compared to TEM-1. KPC-2 residues 266–276 form an extended loop (blue), whereas the comparable TEM-1 residues (red) continue in an α-helix. C) The hydrogen bonds (black dashes) formed by BLIP D49 (cyan) to KPC-2 sidechains (yellow) and the corresponding hydrogen bonds (red dashes) and participants in the TEM-1/BLIP complex (bright pink). D) The inhibitor proteins are aligned from the KPC-2/BLIP (cyan) and TEM-1/BLIP (bright pink) co-structures. Inspection of the KPC-2 sidechains (yellow) and BLIP (cyan) compared to the TEM-1/BLIP conformers (bright pink) shows that in the TEM-1 interface, BLIP K74 participates in a salt bridge (black dash) across the interface with TEM-1 E104, which is lacking in the KPC-2/BLIP interface. The boronylated S70 is also shown.
Reminiscent to its interaction with TEM-1 and SHV-1, BLIP D49 forms a hydrogen bonding network in the active site of KPC-2 (Figure 2C). The BLIP D49 carboxylate hydrogen bonds to KPC-2 side chains S130, K234, T235, and T237, and also forms a salt bridge to KPC-2 residue K234. This network is closely imitated in the TEM-1/BLIP and SHV-1/BLIP interfaces, where BLIP D49 interacts with similar residues: S130, K234, S235, and R243. In contrast to the SHV-1 and TEM-1 structures, however, one of the two complexes in the asymmetric unit (AU) lacks observable electron density for the BLIP F142 binding loop (BLIP residues 139–144). The BLIP F142 loop is also displaced from the SHV-1(D104K)/BLIP structure (discussed later). The 1.9 Å and 2.1 Å resolution KPC-2/BLIP structures were nearly identical, with the exception that the BLIP F142 loop could not be built in either complexes contained in the AU of the 2.1 Å resolution structure. (Except where noted, the discussion below is relevant to the 1.9 Å resolution structure).
Two distinct KPC-2/BLIP complexes are contained within the AU. Most striking is the lack of involvement of the BLIP F142 loop in the second interface, and more subtly, BLIP displays a different orientation relative to the enzyme. Aligning the enzymes within the AU gives a Cα RMSD of 0.3 Å (259 aligned atoms), while the bound inhibitors have a Cα RMSD of 1.9 Å (157 aligned atoms); a similar alignment of inhibitors gives a Cα RMSD of 0.3 Å, with a Cα RMSD of 2.2 Å for the cognate enzymes. Conformational differences observed in crystallographically determined structures must be examined carefully, with consideration that differences may result from crystal packing artifacts. However, close inspection of crystal packing in the presented KPC-2/BLIP structures failed to suggest causative interactions.
The noted lack of involvement of the BLIP F142 loop in the second interface (within the AU) seems to be due to the subtle rotation and displacement of BLIP with respect of KPC-2. Alignment between the inhibitors along with their respective KPC-2 partners, shows that van der Waals (vdW) clashes would exist between F142 of the BLIP F142 loop and the alternate KPC-2 active site. The alternate orientation between binding partners, though subtle, may be sufficient to prevent the F142 sidechain from occupying its canonical role in the enzyme active site. Despite the overall offset of BLIP between the complexes, the D49 sidechain positioning is conserved, consistent with the geometrical constraints of BLIP D49’s hydrogen bonding requirements in the enzyme active site.
Active site configurations
Subtle active site features have been proposed to be important for carbapenemase activity (KPC-2, SME-1, NMC-A, GES-1), such as the disulfide bond between conserved Cys69 and Cys238 residues, and a shallower, more accessible, active site (17–21). In KPC-2, the entrance to the active site is flanked on opposite sides by residue W105 (conserved as a Trp or His in carbapenemases, and Tyr in non-carbapenemases) and residue R220. Besides its importance in the active site of carbapenemases, KPC-2 W105 is adjacent to both BLIP binding loops. KPC-2 W105 occupies different rotamers in the bound and unbound states; without a change in rotamer, KPC-2 W105 is positioned to form a vdW clash with BLIP Y50 and Y51. In the BLIP interface, KPC-2 W105 adopts a rotamer compatible with the canonical conformation of the BLIP D49 loop. However, KPC-2 W105 does affect the conformation of BLIP K74, which is a hot spot in the TEM-1 interface. Comparison of K74 between the TEM-1/BLIP and KPC-2/BLIP structures reveals that it adopts drastically different conformers in the two structures (Figure 2D). In the TEM-1/BLIP interface, BLIP K74 is involved in a salt bridge across the interface with TEM-1 E104, contributing 1–2 kcal/mol to affinity (as evidenced by the TEM-1 E104A mutation and the TEM-1 E104A/BLIP K74A double mutant) (22). Conversely, the corresponding KPC-2 residue, P104, is not capable of forming a salt bridge interaction with BLIP K74. Furthermore, if BLIP K74 adopted the TEM-like sidechain conformation when binding KPC-2, it would form a vdW clash with KPC-2 W105. To avoid this clash, BLIP K74 is oriented away from KPC-2, but towards the BLIP F142 loop, affecting its conformation. BLIP K74 is positioned to form a vdW clash with BLIP S139 from the TEM-like conformation, however BLIP S139 lacked electron density during refinement (Figure 2D).
While the monomer structure of KPC-2 exhibited a bound bicine molecule in the active site (18), the 1.9 Å KPC-2/BLIP structure reported here includes an adduct to the catalytic serine, S70 (Figure 2D). The boronylated serine is likely a remnant from the affinity purification protocol (see Experimental), and is present in both complexes in the AU of the higher resolution structure, and absent from the lower resolution structure. The modified serine residue makes vdW contact across the interface with BLIP D49, while in the lower resolution structure, this area is occupied by water molecules. Altogether, the boronylation does not appear to affect the interface structure: the lower resolution structure lacked this modification, and BLIP is similarly placed with respect to KPC-2.
Computational Prediction of Hot Spots
It has been noted that only a subset of residues contribute substantially towards binding affinity; positions that result in a large change in binding energy (>1.5 kcal/mol) upon mutation to alanine are termed “hot spots” (23). Both computational and experimental approaches have been utilized for the identification of hot spots. Towards this aim, the EGAD library, a C++ implementation of the EGAD protein design algorithm, was applied to the KPC-2/BLIP interface. In a recent study, EGAD was shown to capture quantitative energetics, and to be useful in predicting experimentally known hot spots for a range of protein complexes (24, 25).
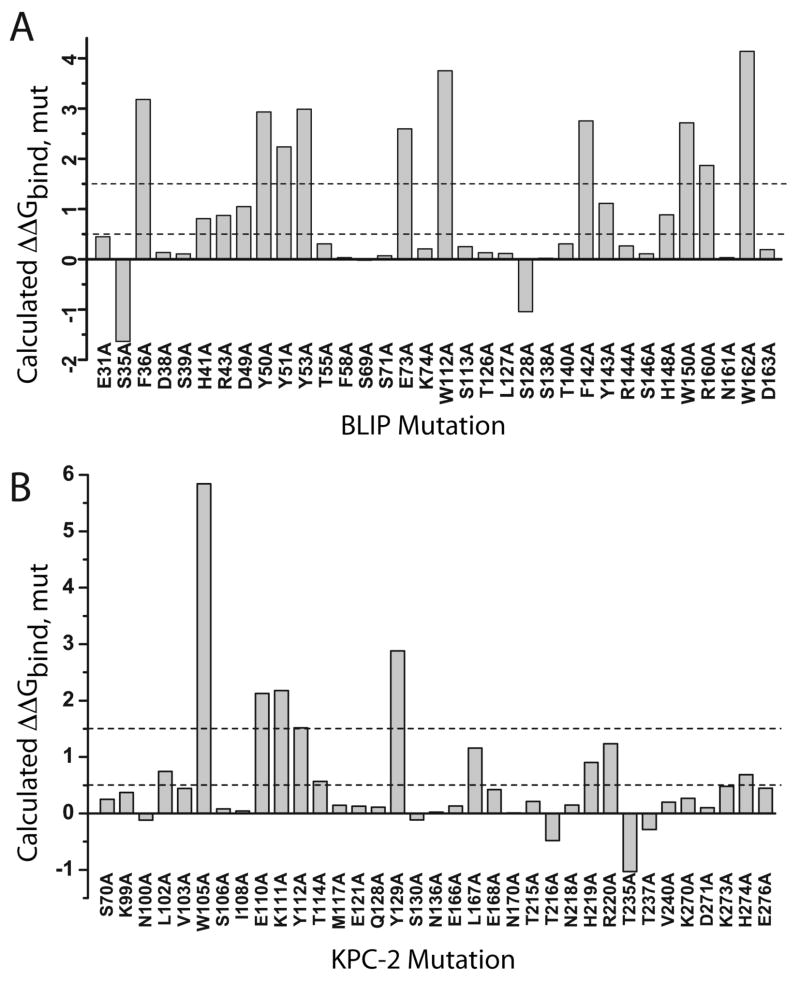
Results from mutagenesis studies have identified several BLIP positions as consensus hot spots for binding TEM-1, SHV-1, SME-1, and Bla1: F36, H41, D49, Y53, and W150 (5, 6). Additional BLIP residues (H148, R160, and W162) are hot spots in binding all but SHV-1. β-Lactamase position 104, and BLIP positions E73, K74, and Y50 have been described as specificity determinants, for which energetic contributions vary tremendously between binding partners (8). A computational alanine scan identified several BLIP positions as hot spots in the KPC-2/BLIP interface: F36, Y50, Y51, Y53, E73, W112, F142, W150, R160, and W162 (Figure 3A). Importantly, the computational results recapitulate much of the previously described experimental data; six of the ten positions identified by EGAD are conserved hot spots: F36, Y53, W112, W150, R160, and W162. The remaining predicted hot spots include two specificity determinants (Y50 and E73) and a TEM-1 hot spot/SME-1 warm spot (F142). It is likely, given the importance of these residues in other BLIP interactions, and our structural information, that they also have an energetic contribution for binding KPC-2. Little mutagenesis data has been reported for the β-lactamase side of BLIP interactions. However, applying computational alanine scanning to KPC-2 in the BLIP interface identified five KPC-2 hot spots (Trp105, Glu110, Lys111, Tyr112, and Tyr129) and six KPC-2 warm spots (L102, T114, L167, H219, R220, and H274) (Figure 3B). While future mutagenesis studies will be required to validate these results, these computational studies will serve as a useful guide
Figure 3.
Computational alanine scanning results for the KPC-2/BLIP interface using EGAD. Calculated ΔΔGbind, mut for each mutation in the interface are shown; positions with ΔΔGbind, mut > 1.5 kcal/mol (dashed line) are considered hot spots, and positions with ΔΔGbind, mut > 0.5 kcal/mol (dashed line) are considered “warm spots”. A) BLIP alanine mutants in the KPC-2/BLIP interface and B) KPC-2 alanine mutants in the KPC-2/BLIP interface.
Discussion
Comparisons with TEM-1 and other model systems
The close identity between TEM-1 and SHV-1 facilitated a direct comparison of interfacial contacts, which implicated β-lactamase position 104 in BLIP’S 1000-fold preference for TEM-1. BLIP inhibits TEM-1 (Ki value of 1 nM) approximately 13-fold more weakly than KPC-2 (Ki value of 0.08 nM). However, KPC-2 and TEM-1 are sufficiently different, that a determination of the exact interactions responsible for the subtly enhanced affinity is unclear (see Supporting Information for list of hydrogen bonds and vdW contacts). Furthermore, although changes in binding energy are usually attributed to interactions in the complex, destabilizaton or stabilization of either partner will also affect binding energy. Although future mutagenesis work will be critical towards identifying the interactions responsible for KPC-2/BLIP affinity, a few structural differences between the TEM-1/BLIP and KPC-2/BLIP interfaces are highlighted below.
In some cases, the amino acid substitutions from TEM-1 and KPC-2 result in different interfacial interactions, and appear to alter the interface networks as described by Reichmann et al. (22, 26). For example, the native TEM-1 residue Glu104 forms a favorable salt bridge with BLIP Lys74, in addition to contacts with the BLIP binding loop F142 in the center of cluster 2. The corresponding KPC-2 residue Pro104, instead, forms vdW interactions with BLIP Glu73 and Trp162, previously in cluster 4. Other residue substitutions result in little change of the interface interaction networks: for example, TEM-1 Tyr105 and KPC-2 Trp105 have similar roles. Both residues form vdW contacts with Ala47, Gly48, Glu73, Gly141, although KPC-2 Trp105 also contacts BLIP Tyr51, and TEM-1 Tyr105 contacts BLIP Lys74 and Phe142. Alternatively, the native KPC residue may contribute further interactions, for example, the substitution Asn99 to Lys99. In addition to making vdW contacts with BLIP His148 and Tyr150 similar to TEM-1 Asn99, KPC-2 Lys99 also contacts BLIP L127 and forms a hydrogen bond with BLIP Ser128. KPC-2 Ile108 contacts BLIP Ser35 and Phe36, whereas the native TEM-1 Val108 does not. KPC-2 Tyr129 makes vdW contact with BLIP Phe36 and Tyr50 similar to its counterpart, TEM-1 Met129, and furthermore makes a hydrogen bond with BLIP Glu31. Lastly, the TEM-1 residue may make interactions not present in KPC-2/BLIP. While both TEM-1 and KPC-2 contain Asn170, in TEM-1, this residue makes contact with BLIP Phe142, while in the KPC-2/BLIP interface, the residues are subtly adjusted and do not make vdW contacts.
Recently, BLIP-I, a BLIP homolog sharing 37% identity with BLIP, was structurally characterized in complex with TEM-1 (27). Though TEM-1/BLIP and TEM-1/BLIP-I complexes form with similar affinities, it was noted that although the specific residue-residue pairing interactions at the interface were variable, the number and types of interactions was comparable. Thus, it seems that β-lactamase/BLIP scaffolds show multiple examples of high affinity enzyme-inhibitor complexes.
Structural Plasticity in KPC-2/BLIP
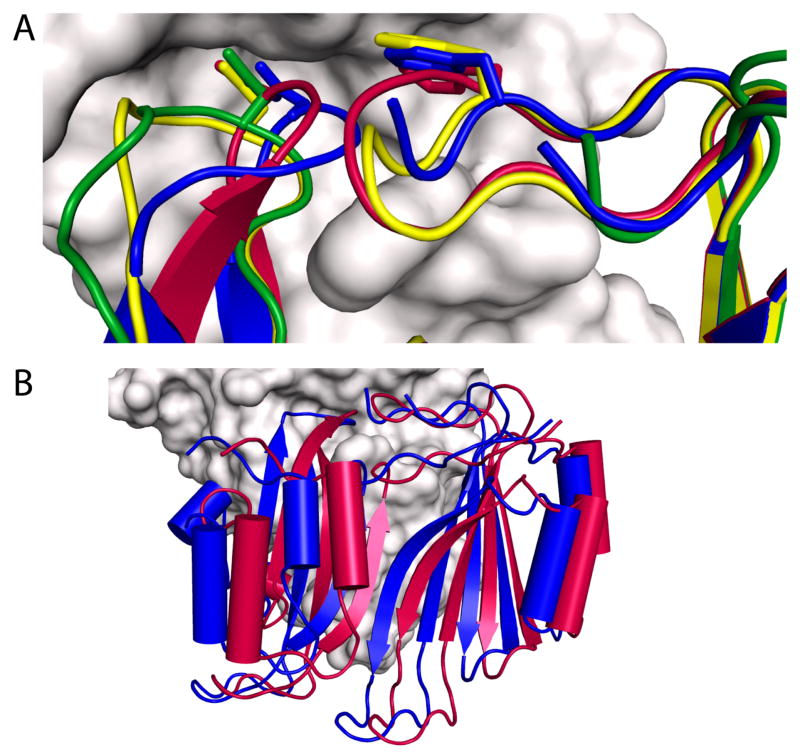
BLIP shows a small displacement from its KPC-2 bound position relative to its TEM-1 bound position (a rotation of 9° and translation of 2.5 Å between TEM-1 and KPC-2) (Figure 4B). Despite the overall offset of BLIP, the geometry of the interaction with BLIP D49 binding loop is closely conserved, which is consistent with the geometrical constraints of BLIP D49’s involvement in four intermolecular hydrogen bonds. Similarly, the homologous TEM-1/BLIP and TEM-1/BLIP-I structures differ by a 4.5 Å translation of BLIP-I with respect to BLIP (27). Another well-characterized enzyme/inhibitor system, the endonuclease colicins and cognate immunity proteins (Im9/E9 and Im7/E7), has been shown to differ by a rigid body rotation of 19° of Im7 compared to Im9 in binding their cognate enzymes (28). Similar to β-lactamases, the E colicin endonucleases (DNases) are bacterial enzymes involved in host cell survival. However, unlike β-lactamase enzymes, DNase toxins E7 and E9 are produced for the purpose of destroying competing cells. Host cell death is prevented by production of an immunity protein (Im7 or Im9) that potently inhibits the cognate enzyme (29). The rotation has the reported effect of presenting a slightly different face of the immunity protein’s specificity helix to the DNase. In the β-lactamase system, the rotation mode of BLIP appears to facilitate alternate hydrogen bonding networks and to accommodate the different surfaces of homologous partners.
Figure 4.
BLIP loop conformations in β-lactamase/BLIP complexes. A) The conformation of BLIP F142 and D49 binding loops are shown interacting with TEM-1(E104Y/Y105N) (yellow - PDB ID 2B5R), SHV-1(D104K) (green - PDB ID 2G2W), as well as wild type TEM-1 (red - PDB ID 1JTG), are aligned with BLIP (blue) from the KPC-2 (gray) complex. B) Structural alignment between the enzymes in the KPC-2/BLIP (PDB ID 3E2L) and TEM-1/BLIP (PDB ID 1JTG) complexes. BLIP in the complex with KPC-2 (blue) is displaced from its orientation in binding TEM-1 (red). This alignment differs from that shown in Figure 2A, in which the inhibitor proteins are aligned.
Alternate loop conformations for β-lactamase/BLIP complexes have been reported for some mutations, illustrating BLIP’s plasticity in accommodating amino acid substitutions. For example, the BLIP F142 loop is displaced from the SHV D104K/BLIP interface (Figure 4A, PDB ID 2G2W). Briefly, because the SHV-1 D104K mutation is across the interface from BLIP K74, the new Lys104 conformer interferes with the canonical BLIP F142 loop placement (8). In addition, the BLIP D49 hairpin loop occupies alternate orientations in the presence of the SHV-1 D104K mutation, in the TEM-1 E104Y/Y105N mutation (PDB ID 2B5R), and in the TEM-1/BLIP W150A (PDB ID 3C7U) (22, 30). It has been suggested that significant backbone rearrangements are more likely in non-alanine mutations, as they introduce the need to avoid unfavorable interactions, as well as the potential for alternate favorable interaction modes (22). This is exemplified by structural analysis of the β-lactamase/BLIP system.
Consequences of the structural plasticity displayed by BLIP
The structural plasticity described above has biophysical implications for the mechanism of association and sources of affinity between the two binding partners. The first step in protein-protein association is the formation of the encounter complex. As partners dock together they retain their respective solvation shells and form few of the short-range interactions that are characteristic of the native complex (31). The encounter complex allows multiple conformations to be sampled before the annealing process, which is necessary to form the short-range hydrophobic, hydrogen bond, and electrostatic interactions, and solvent structure in the native complex (31). The dynamic properties of the intermediate appear to facilitate finding the most favorable conformation, especially important when multiple conformations may exist, for example, as in the highly similar modes of binding of immunity proteins to cognate and non-cognate colicins. Kinetic data suggests that colicin/immunity protein encounter complexes undergo rigid body rotation events to form the bound states (32). A similar mechanism is also possible for KPC-2/BLIP, although detailed association and dissociation kinetics have not yet been investigated to support this hypothesis.
Thermodynamically, the addition of a second accessible conformation for the complex increases the entropic energy component of the bound form(s). Additionally, assuming that the BLIP F142 loop is disordered upon binding KPC-2 in the second interface, the mobility should provide an entropic benefit to the complex, perhaps partially, or even completely, offsetting the interactions absent in the interface relative to TEM-1. In the case of bovine carbonic anhydrase II binding to a series of small molecule ligands, the enthalpic penalty due to decreased interactions was completely compensated for by the entropic benefit of increased flexibility, resulting in little change in overall binding energy (33). The flexibility of the BLIP F142 loop also has implications for the mechanisms of association with its β-lactamase binding partners. The possibility exists that the rate of exchange among F142 loop conformations is rate-limiting for protein association; or, alternatively, an “induced-fit” process occurs after formation of the encounter complex. Reported examples exist for both mechanisms of protein interaction (34, 35). Detailed analysis of the kinetics of association could provide key information into the events occurring upon binding in KPC-2/BLIP complexes.
BLIP D49 as an anchor residue
Rajamani et al. have extrapolated the known means of peptide binding to the major histocompatibility complex, where anchor residues dock into a binding pocket on the receptor protein, to a general mechanism for protein-protein association (36). Anchor residues in protein-protein interactions are generally on the smaller protein partner involved, have the highest change in solvent accessible surface area upon complex formation (ΔSASA), are experimentally observed hot spots of binding affinity, and often have decreased flexibility compared to other surface residues. They are proposed to facilitate a “lock-and-key” mechanism of binding by docking into recognition pockets to form the encounter complex. In β-lactamase/BLIP interactions, BLIP residue D49, and secondarily F142, appear to be anchored in the enzyme active site, positioning BLIP to form the complex. Rajamani et al. observe that a single or multiple anchors may be present, however, in cases where a single anchor is present, the residue’s ΔSASA exceeds 100 Å2. Accordingly, in the second KPC-2/BLIP interface lacking the BLIP F142 loop, D49 is sufficient as sole anchor (ΔSASA = 142 Å2).
Computational Prediction of BLIP Binding Hotspots
Computational techniques have become useful in guiding experimental efforts towards understanding protein binding determinants. Like other computational techniques, EGAD assumes a fixed backbone; therefore considering the documented flexibility in the BLIP backbone, specifically BLIP binding loops, it is acknowledged that many mutations may violate the fixed backbone assumption. However, it has been reported that even when computational techniques are not accurate in predicting precise local structure, they are useful in capturing global energetics (7).
EGAD’s ability to calculate changes in free energies of dissociation upon mutation in the β-lactamase/BLIP system has been evaluated using the TEM-1/BLIP interface (8). In a separate study, EGAD was employed to redesign BLIP to display higher affinity for one of its weakest targets, SHV-1 β-lactamase (7). However, the KPC-2/BLIP interaction is extremely tight, and may be close to optimal. Instead of utilizing protein design efforts, we use computation to dissect the contribution of individual residues in the interface. The success with recapitulating known BLIP hot spots provides evidence that EGAD captures the dominant energetic features of the interface. Furthermore, it reinforces that BLIP interacts with its targets using a conserved set of interactions, while additional residues show specificity for different targets.
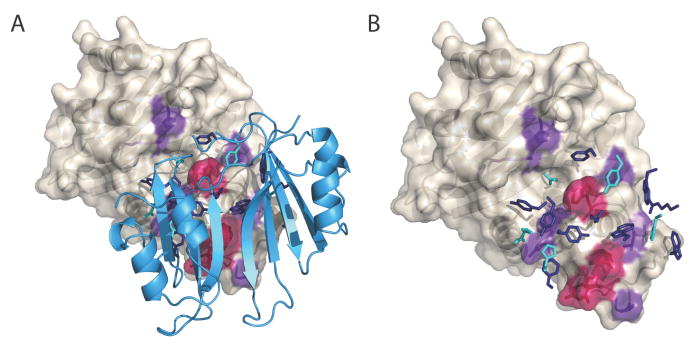
Although the surface area buried in the KPC-2/BLIP interface is large (>2500 Å2) involving ~50 residues, only a subset of residues make energetically important intermolecular interactions. BLIP hot spots are localized towards the top and middle of the protein, proximal to the enzyme’s active site (Figure 5). KPC hot/warm spots flank the active site, with additional hot/warm regions on the KPC-2 loop-helix region at the center of the interface. It has been shown that peptides or small molecule often interact through similar functional regions and hot spots, mimicking the native protein interaction (37, 38). The small protein inhibitor of α-thrombin, hirudine, has had success as a clinical anticoagulant (39, 40). Additionally, peptide mimics of protein-protein interactions show promise for designing therapeutics; for example, the N-terminus of the HIV-1 co-receptor, CCR5, binds to the gp120 envelope glycoprotein (41). Therefore, identification of KPC-2 hot spots for BLIP interaction has relevance towards the design of novel inhibitors.
Figure 5.
Hot residues (ΔΔGbind, Aia mut > 1.5 kcal/mol, red) and warm residues (1.5 < ΔΔGbind, Aia mut > 0.5 kcal/mol, purple) in the KPC-2/BLIP interface. A) The KPC-2 surface with BLIP cartoons. B) The BLIP backbone is removed for clarity.
Lessons from a promiscuous inhibitor
The TEM-1/BLIP interface has been shown to be composed of multiple clusters of interacting residues; Schreiber and co-workers have used site-directed mutagenesis to demonstrate cooperativity within, but additivity among different clusters (26). Recently, however, structural analysis of TEM-1/BLIP W150A (PDB ID 3C7U) showed a conformational rearrangement more than 25 Å from the mutation, demonstrating at least one instance of long distance cooperativity in BLIP interactions (30). Nevertheless, a general decomposability of interactions would be an obvious advantage that would allow a promiscuous protein to maintain affinity while accommodating amino acid substitutions between multiple partners. Backbone flexibility may be a feature at least partially enabling the cluster behavior of the interface.
The extreme hydration of BLIP interfaces has also been implicated in its ability to recognize a range of enzymes with high affinity (4). Reichmann et al. have investigated the function of water in protein interfaces, and have concluded that it does not contribute to complex stability, but rather solvent molecules fill vacancies not occupied by amino acids (42). BLIP complexes are prime examples where this is the case: BLIP has global shape complimentarily to the β-lactamase fold, and the ability to accommodate variable sequences upon the scaffold is facilitated by the presence, or absence, of interfacial water molecules. Additional adaptability results from the sidechain atoms making up the majority of BLIP’s binding surface: sidechain atoms generally have more conformational flexibility than backbone atoms. The anchor functions of BLIP D49 and F142 may allow BLIP to sample different orientations, thus finding orientations with the best atomic shape complementarity and optimal hydrogen bonding network; even a shift of 1 Å may accommodate a different hydrogen bonding network. BLIP’s structural flexibility may be most central to BLIP’s promiscuity, as presented here in the KPC-2/BLIP co-structure, and described elsewhere. The plasticity of BLIP’s binding loops and orientation seems crucial to BLIP’s function as a potent inhibitor of a wide range of β-lactamases.
Supplementary Material
Acknowledgments
M.S.H would like to thank Jack F. Kirsch, Kimberly A. Reynolds, and members of the Handel laboratory for helpful discussions regarding the manuscript.
The abbreviations used are
- BLIP
β-lactamase inhibitor protein
- AU
asymmetric unit
- WT
wild type
- vdW
van der Waals
- RMSD
root mean square deviation
- SASA
solvent accessible surface area
Footnotes
The refined coordinates for the KPC-2/BLIP structures are deposited in the Protein Data Bank (entries 3E2K and 3E2L).
The KPC-2(G175S) sequence was initially reported as KPC-1. The G175S point mutation is not in the BLIP interface, nor does it alter binding affinity; we will refer to the structure as KPC-2/BLIP.
This research was supported by NSF grant 0344749 (T.M.H.), the U.S. Department of Veterans Affairs Merit Review Program, VISN 10 GRECC (R.A.B.), NIH 1R01 grant A1063517-01 (R.A.B.), the NCI grant CA077373 (J.M.B), and NIGMS grant GM071747 (J.M.B.).
Supporting Information Available
A list of interfacial hydrogen bonds is given in supporting information (Supporting Table 1), along with a list of interfacial vdW contacts (Supporting Table 2). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Walther-Rasmussen J, Hoiby N. Class A carbapenemases. J Antimicrob Chemother. 2007;60:470–482. doi: 10.1093/jac/dkm226. [DOI] [PubMed] [Google Scholar]
- 2.Mushtaq S, Ge Y, Livermore DM. Comparative activities of doripenem versus isolates, mutants, and transconjugants of Enterobacteriaceae and Acinetobacter spp. with characterized β-lactamases. Antimicrob Agents Chemother. 2004;48:1313–1319. doi: 10.1128/AAC.48.4.1313-1319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strynadka NCJ, Jensen SE, Johns K, Blanchard H, Page M, Matagne A, Frere J-M, James MNG. Structural and kinetic characterization of a β-lactamase-inhibitor protein. Nature. 1994;368:657–660. doi: 10.1038/368657a0. [DOI] [PubMed] [Google Scholar]
- 4.Strynadka NCJ, Jensen SE, Alzari PM, James MNG. A potent new mode of β-lactamase inhibition revealed by the 1.7 A X-ray crystallographic structure of the TEM-1-BLIP complex. Nat Struct Mol Biol. 1996;3:290–297. doi: 10.1038/nsb0396-290. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z, Palzkill T. Determinants of binding affinity and specificity for the interaction of TEM-1 and SME-1 β-lactamase with β-lactamase inhibitory protein. J Biol Chem. 2003;278:45706–45712. doi: 10.1074/jbc.M308572200. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Palzkill T. Dissecting the protein-protein interface between β-lactamase inhibitory protein and class A β-lactamases. J Biol Chem. 2004;279:42860–42866. doi: 10.1074/jbc.M406157200. [DOI] [PubMed] [Google Scholar]
- 7.Reynolds KA, Hanes MS, Thomson JM, Antczak AJ, Berger JM, Bonomo RA, Kirsch JF, Handel TM. Computational redesign of the SHV-1 β-lactamase/β-lactamase inhibitor protein interface. J Mol Biol. 2008;382:1265–1275. doi: 10.1016/j.jmb.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reynolds KA, Thomson JM, Corbett KD, Bethel CR, Berger JM, Kirsch JF, Bonomo RA, Handel TM. Structural and computational characterization of the SHV-1 β-lactamase/β-lactamase inhibitor protein interface. J Biol Chem. 2006;281:26745–26753. doi: 10.1074/jbc.M603878200. [DOI] [PubMed] [Google Scholar]
- 9.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CWJ, Sweet RM, editors. Methods in Enzymology: Macromolecular crystallogr A. 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 10.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 11.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 12.Adams PD, Grosse-Kunstleve RW, Hung LW, loerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 13.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 14.Kleywegt G. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr D. 1996;52:842–857. doi: 10.1107/S0907444995016477. [DOI] [PubMed] [Google Scholar]
- 15.Cheng YC, Prusoff WH. Relationship between the inhibition constant (KI) and the concentration of inhibitor which causes 50 per cent inhibition (150) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 16.Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swaren P, Maveyraud L, Raquet X, Cabantous S, Duez C, Pedelacq JD, Mariotte- Boyer S, Mourey L, Labia R, Nicolas-Chanoine MH, Nordmann P, Frere JM, Samama JP. X-ray analysis of the NMC-A β-lactamase at 1.64-A resolution, a class A carbapenemase with broad substrate specificity. J Biol Chem. 1998;273:26714–26721. doi: 10.1074/jbc.273.41.26714. [DOI] [PubMed] [Google Scholar]
- 18.Ke W, Bethel CR, Thomson JM, Bonomo RA, van den Akker F. Crystal structure of KPC-2; insights into carbapenemase activity in class A β-lactamases. Biochemistry. 2007;46:5732–5740. doi: 10.1021/bi700300u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majiduddin FK, Palzkill T. Amino Acid Sequence Requirements at Residues 69 and 238 for the SME-1 {beta}-Lactamase To Confer Resistance to {beta}-Lactam Antibiotics. Antimicrob Agents Chemother. 2003;47:1062–1067. doi: 10.1128/AAC.47.3.1062-1067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith CA, Caccamo M, Kantardjieff KA, Vakulenko S. Structure of GES-1 at atomic resolution: insights into the evolution of carbapenamase activity in the class A extended-spectrum β-lactamases. Acta Cryst D. 2007;63:982–992. doi: 10.1107/S0907444907036955. [DOI] [PubMed] [Google Scholar]
- 21.Raquet X, Lamotte-Brasseur J, Bouillenne F, FrËre J-M. A disulfide bridge near the active site of carbapenem-hydrolyzing class A β-lactamases might explain their unusual substrate profile. Proteins. 1997;27:47–58. doi: 10.1002/(sici)1097-0134(199701)27:1<47::aid-prot6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 22.Reichmann D, Cohen M, Abramovich R, Dym O, Lim D, Strynadka NCJ, Schreiber G. Binding hot spots in the TEM1-BLIP interface in light of its modular architecture. J Mol Biol. 2007;365:663–679. doi: 10.1016/j.jmb.2006.09.076. [DOI] [PubMed] [Google Scholar]
- 23.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 24.Pokala N, Handel TM. Energy functions for protein design: adjustment with protein-protein complex affinities, models for the unfolded state, and negative design of solubility and specificity. J Mol Biol. 2005;347:203–227. doi: 10.1016/j.jmb.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 25.Chowdry AB, Reynolds KA, Hanes MS, Voorhies M, Pokala N, Handel TM. An object-oriented library for computational protein design. J Comput Chem. 2007;28:2378–2388. doi: 10.1002/jcc.20727. [DOI] [PubMed] [Google Scholar]
- 26.Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G. The modular architecture of protein-protein binding interfaces. Proc Natl Acad Sci. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gretes M, Lim DC, de Castro L, Jensen SE, Kang SG, Lee KJ, Strynadka NCJ. Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the β-Lactamase Inhibitory Proteins BLIP, BLIP-I, and BLP. J Mol Biol. 2009;389:289–305. doi: 10.1016/j.jmb.2009.03.058. [DOI] [PubMed] [Google Scholar]
- 28.Kuhlmann UC, Pommer AJ, Moore GR, James R, Kleanthous C. Specificity in protein-protein interactions: the structural basis for dual recognition in endonuclease colicin-immunity protein complexes. J Mol Biol. 2000;301:1163–1178. doi: 10.1006/jmbi.2000.3945. [DOI] [PubMed] [Google Scholar]
- 29.Kleanthous C, Kuhlmann UC, Pommer AJ, Ferguson N, Radford SE, Moore GR, James R, Hemmings AM. Structural and mechanistic basis of immunity toward endonuclease colicins. Nat Struct Mol Biol. 1999;6:243–252. doi: 10.1038/6683. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Palzkill T, Chow DC. Structural insight into the kinetics and ΔCp of interactions between TEM-1 β-lactamase and β-lactamase inhibitory protein (BLIP) J Biol Chem. 2009;284:595–609. doi: 10.1074/jbc.M804089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor MG, Rajpal A, Kirsch JF. Kinetic epitope mapping of the chicken lysozyme. HyHEL-10 fab complex: Delineation of docking trajectories. Protein Science. 1998;7:1857–1867. doi: 10.1002/pro.5560070902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keeble AH, Kleanthous C. The kinetic basis for dual recognition in colicin endonuclease-immunity protein complexes. J Mol Biol. 2005;352:656–671. doi: 10.1016/j.jmb.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 33.Krishnamurthy VM, Bohall BR, Semetey V, Whitesides GM. The paradoxical thermodynamic basis for the interaction of ethylene glycol, glycine, and sarcosine chains with bovine carbonic anhydrase II; an unexpected manifestation of enthalpy/entropy compensation. J Am Chem Soc. 2006;128:5802–5812. doi: 10.1021/ja060070r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki N, Tsumoto K, Hajicek N, Daigo K, Tokita R, Minami S, Kodama T, Hamakubo T, Kozasa T. Activation of leukemia-associated RhoGEF by Gα13 with significant conformational rearrangements in the interface. J Biol Chem. 2008:M804073200. doi: 10.1074/jbc.M804073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 36.Rajamani D, Thiel S, Vajda S, Camacho CJ. Anchor residues in protein-protein interactions. Proc Natl Acad Sci. 2004;101:11287–11292. doi: 10.1073/pnas.0401942101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wrighton NC, Farrell FX, Chang R, Kashyap AK, Barbone FP, Mulcahy LS, Johnson DL, Barrett RW, Jolliffe LK, Dower WJ. Small Peptides as Potent Mimetics of the Protein Hormone Erythropoietin. Science. 1996;273:458–463. doi: 10.1126/science.273.5274.458. [DOI] [PubMed] [Google Scholar]
- 38.Livnah O, Stura EA, Johnson DL, Middleton SA, Mulcahy LS, Wrighton NC, Dower WJ, Jolliffe LK, Wilson IA. Functional Mimicry of a Protein Hormone by a Peptide Agonist: The EPO Receptor Complex at 2.8 Å. Science. 1996;273:464–471. doi: 10.1126/science.273.5274.464. [DOI] [PubMed] [Google Scholar]
- 39.Bates SM, Weitz JI. Emerging anticoagulant drugs, Arterioscler. Thromb Vasc Biol. 2003;23:1491–1500. doi: 10.1161/01.ATV.0000084827.77945.66. [DOI] [PubMed] [Google Scholar]
- 40.Rydel TJ, Ravichandran KG, Tulinsky A, Bode W, Huber R, Roitsch C, Fenton JW., 2nd The structure of a complex of recombinant hirudin and human α-thrombin. Science. 1990;249:277–280. doi: 10.1126/science.2374926. [DOI] [PubMed] [Google Scholar]
- 41.Huang C-c, Lam SN, Acharya P, Tang M, Xiang S-H, Hussan SS-u, Stanfield RL, Robinson J, Sodroski J, Wilson IA, Wyatt R, Bewley CA, Kwong PD. Structures of the CCR5 N Terminus and of a Tyrosine-Sulfated Antibody with HIV-1 gp120 and CD4. Science. 2007;317:1930–1934. doi: 10.1126/science.1145373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichmann D, Phillip Y, Carmi A, Schreiber G. On the contribution of water-mediated interactions to protein-complex stability. Biochemistry. 2008;47:1051–1060. doi: 10.1021/bi7019639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.