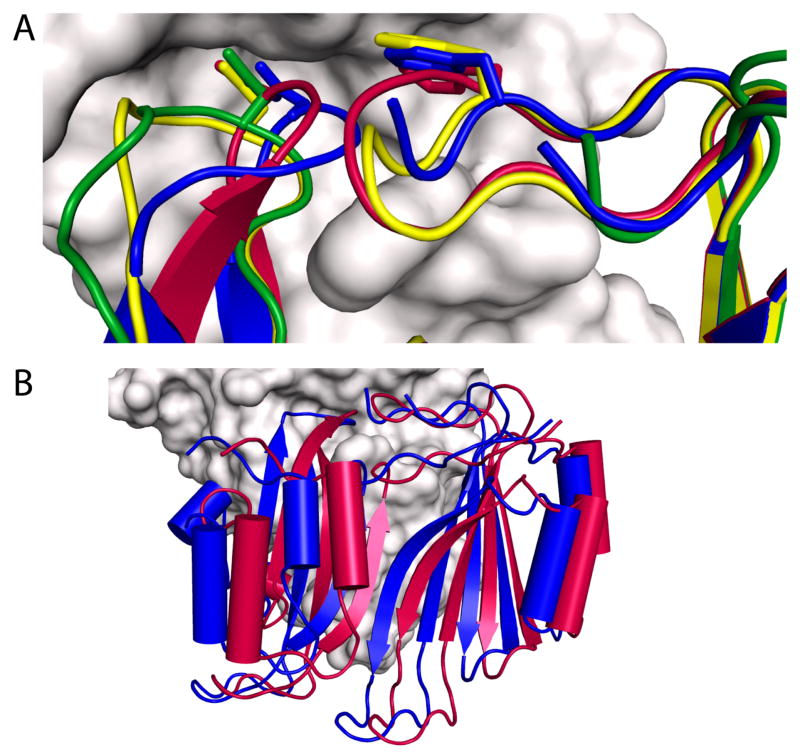
Figure 4.
BLIP loop conformations in β-lactamase/BLIP complexes. A) The conformation of BLIP F142 and D49 binding loops are shown interacting with TEM-1(E104Y/Y105N) (yellow - PDB ID 2B5R), SHV-1(D104K) (green - PDB ID 2G2W), as well as wild type TEM-1 (red - PDB ID 1JTG), are aligned with BLIP (blue) from the KPC-2 (gray) complex. B) Structural alignment between the enzymes in the KPC-2/BLIP (PDB ID 3E2L) and TEM-1/BLIP (PDB ID 1JTG) complexes. BLIP in the complex with KPC-2 (blue) is displaced from its orientation in binding TEM-1 (red). This alignment differs from that shown in Figure 2A, in which the inhibitor proteins are aligned.