Abstract
Intratypic diversity of human papillomavirus (HPV) genome is generally characterized by point mutation, insertion, and/or deletion. Using PCR-based cloning and sequencing, we detected concurrent infection with 8 HPV16 variants in a woman enrolled in the ASCUS-LSIL Triage Study. The European variant was the major variant; each of the seven minor variants had partial DNA sequences identical to the European variant and another part identical to the African-2 variant. At a follow-up visit, only an HPV16 African-2 variant was detected. Results from the present study suggest presence of intratypic recombination of HPV genome in natural infection.
Keywords: human papillomavirus, recombination, variant
Introduction
Papillomavirus is an extremely diversified group of viruses. To date, more than 100 human papillomavirus (HPV) types have been characterized (de Villiers et al., 2004). For any given type of HPV, viral isolates that differ by less than 2% DNA sequence of the L1 open reading frame are further classified as variants. The intratypic diversity of HPV genome is generally documented as point mutation, insertion, and/or deletion. Inter- or intra-type recombination of HPV genome, although presumed theoretically, has not yet been reported from natural infections. By using phylogeny-based recombination detection methods, Varsani et al. (Varsani et al., 2006) suggested a possibility of exchange of HPV genes in a recombination model and delineated potential recombinant descendants of the ancestral sequences. Recent phylogenetic analyses further predicted intratypic recombination of HPV16 genome with potential breakpoints in the E7 gene (Angulo and Carvajal-Rodriguez, 2007; Carvajal-Rodriguez, 2008).
HPV16 is the type that confers the highest risk of invasive cervical cancer (Munoz et al., 2003). Investigations of diversity of HPV16 genome have identified five major phylogenetic lineages: European (E), Asian, Asian-American, African-1, and African-2 (Af2) variants (Eriksson et al., 1999; Yamada et al., 1997). Reported in this study are recombinant sequences of HPV16 variants detected in a woman who participated in the ASCUS-LSIL Triage Study (Schiffman and Adrianza, 2000).
Results
The study subject had HPV6, HPV16, HPV45, and HPV56 detected in her enrollment sample; 20 months later she had a follow-up sample positive for HPV16 alone. A polymerase chain reaction (PCR)-based subcloning and sequencing of the 751-bp fragments of HPV16 genome (from nucleotide position 7723 to 569) was performed on both cervical swab samples. As shown in Table 1, the initial analysis of 10 clones from the enrollment sample revealed that 8 clones contained sequences identical to HPV16 prototype (i.e., E variant); one clone had alterations of C-to-T, C-to-T, G-to-A, G-to-T, A-to-C, A-to-G, and G-to-A at position 7764, 7786, 7826, 7834, 7837, 7839, and 7868, respectively; and another clone had alterations of C-to-T, A-to-T, T-to-C, G-to-T, C-to-G, and G-to-T at position 31, 90, 109, 132, 143, and 145, respectively. Although these alterations were the same as those of HPV16 Af2 variant at the corresponding positions, the fragments in the latter 2 clones were not derived from the Af2 variant because sequences in the remaining part of the long control region and/or the E6 region were identical to the E variant. Given that nucleotide alterations across HPV genomes are often linked, the sequences with apparent disconnection of the alterations suggest a recent break and recombination. We therefore designated these 2 clones as HPV16 recombinants rather than new variants. Recombinant 1 had a potential breakpoint between position 7868 and 31 whereas recombinant 2 had potential breakpoints between position 7868 and 31 and between position 145 and 286.
Table 1.
DNA sequence variations of HPV16 variants from nucleotide position 7722 to 567 detected by polymerase chain reaction-based cloning and sequencing
| Nucleotide alterations at position* |
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 7 | 7 | 7 | 7 | 7 | 7 | 7 | 3 | 9 | 1 | 1 | 1 | 1 | 2 | 2 | 3 | 4 | ||||
| 7 | 7 | 8 | 8 | 8 | 8 | 8 | 1 | 0 | 0 | 3 | 4 | 4 | 8 | 8 | 3 | 0 | ||||
| 6 | 8 | 2 | 3 | 3 | 3 | 6 | 9 | 2 | 3 | 5 | 6 | 9 | 5 | 3 | ||||||
| 4 | 6 | 6 | 4 | 7 | 9 | 8 | ||||||||||||||
| Sample | No. of clones | C | C | G | G | A | A | G | C | A | T | G | C | G | T | A | C | A | HPV16 Prototype |
|
| Initial test | Enrollment | 8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E variant |
| Enrollment | 1 | T | T | A | T | C | G | A | - | - | - | - | - | - | - | - | - | - | Recombinant 1 | |
| Enrollment | 1 | - | - | - | - | - | - | - | T | T | C | T | G | T | - | - | - | - | Recombinant 2 | |
| Follow-up | 10 | T | T | A | T | C | G | A | T | T | C | T | G | T | A | G | T | G | Af2 variant | |
| Retest | Enrollment | 8 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | E variant |
| Enrollment | 1 | T | T | A | T | C | G | A | T | T | C | T | G | T | - | - | - | - | Recombinant 3 | |
| Enrollment | 1 | - | - | - | - | - | - | - | - | T | C | T | G | T | A | G | T | G | Recombinant 4 | |
| Follow-up | 10 | T | T | A | T | C | G | A | T | T | C | T | G | T | A | G | T | G | Af2 variant | |
The nucleotide positions are numbered according to those documented in Human Papillomavirus 1997 Compendium (Meissner, 1997) and correspond to those of the original HPV16 sequence (Seedorf et al., 1985). For each variant sequence, positions that are identical to the prototype sequence are marked with a dash in the alignment. HPV = human papillomavirus; E = European; Af2 = African 2.
Considering that nucleotide alterations detected in a single clone might result from PCR errors, the cervical sample was reassayed by PCR-based cloning and sequencing. Again, the prototype sequences were detected in 8 clones of the enrollment sample and recombinant sequences were detected in 2 clones that were designated as HPV16 recombinant 3 and 4, respectively (Table 1). Although these recombinants differed in positions of breakpoints, 7 out of 13 nucleotide alternations between recombinant 1 and 3, and 5 out of 10 nucleotide alternations between recombinant 2 and 4 overlapped. An additional analysis of 85 clones from another PCR-based cloning of enrollment sample revealed that 82 clones contained sequences identical to the prototype; one had alterations of C-to-T at position 335 and A-to-G at position 403; one had alterations of T-to-A, A-to-G, C-to-T, and A-to-G at position 286, 289, 335 and 403, respectively; and the third displayed sequences the same as recombinant 4 except for an additional change of C-to-T at position 31. Sequences of 20 clones from the follow-up sample (10 from the initial test and 10 from the repeat) were identical to sequences of HPV16 Af2 variant.
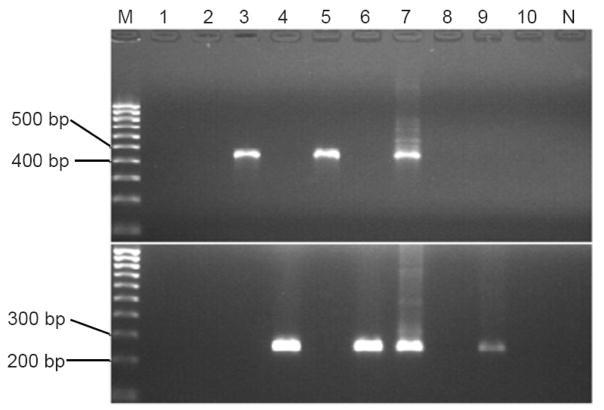
To examine whether these recombinant sequences resulted from artifacts of PCR- or cloning-mediated recombination, we designed a recombinant-specific (RS)-PCR according to the known sequences. The specificity of the assay was confirmed by testing a group of plasmids. As shown in Figure 1, plasmid samples with HPV16 recombinant 1 or 3 were positive by RS-PCR with primer set A; those with HPV16 recombinant 2 or 4 were positive by RS-PCR with primer set B; and those with HPV16 E or Af2 variant were negative by RS-PCR with either primer set A or primer set B. The PCR products of the enrollment sample were positive by RS-PCR with either set of primers (lane 7). This suggests that these recombinant sequences were present prior to the cloning procedure. When the RS-PCR was performed on the enrollment sample, only the one with primer set B displayed a visible band (lane 9), suggesting presence of the recombinant sequences in natural infection.
Figure 1.
Products of recombinant-specific polymerase chain reaction (RSP) with primer set A (upper part) or B (lower part) on 1.5% ethidium bromide-stained agarose gel.
Lane M, 100-bp DNA Marker; lanes 1–6, plasmids with human papillomavirus type 16 European variant, African 2 variant, recombinants 1, 2, 3, and 4, respectively; lane 7, PCR products of enrollment sample; lane 8, PCR products of follow-up sample; lane 9, enrollment sample; and lane 10, follow-up sample; lane N, no DNA template control.
One concern was whether the positive bands generated by RS-PCR resulted from potential plasmid contamination. To address this, the cervical sample and its PCR products were assayed by PCR with M13 forward and reverse primers. This set of primers is able to amplify sequences of the vector we used. No positive signal was seen on the agarose gel (data not shown). A direct DNA sequencing of the RS-PCR products revealed that sequences generated with primer set A were identical to HPV16 recombinant 1 and those with primer set B were identical to the recombinant 4. Neither the follow-up sample (lane 10) nor its PCR product (lane 8) was positive by RS-PCR. To determine whether we were able to detect the Af2 variant from the enrollment sample and the E variant from the follow-up sample, both samples and their 751-bp PCR products were PCR-assayed with a pair of primers for the E variant (forward primer B and reverse primer A) and a pair of primers for Af2 variant (forward primer A and reverse primer B). Consistent with the sequencing results, the enrollment sample and its PCR products were positive by PCR with the first but not the second set of primers, vice versa for the follow-up sample and its PCR products (data not shown).
In view of a change of the predominant variants between the enrollment and follow-up visits, another concern was whether there was a sample switch or sample-to-sample contamination. To address this, both samples were tested for polymorphisms of short tandem repeats (STRs) by PCR-based combined DNA index system. The genotypic profile is summarized in Table 2. We initially tested 2 diluted samples (0.1ng/ul) in triplicate. All 3 replicates of diluted follow-up sample were allele-matched exactly. All replicates of diluted enrollment sample were also consistent except for locus vWA of replicate 2. The enrollment sample was reassayed in undiluted form (1ng/ul). The patterns were exactly the same as those seen in the follow-up sample. Since the inconsistency was restricted to a single locus on one out of the 4 replicates (3 diluted and 1 undiluted), it is deemed insignificant. Because the probability of two different samples matching-by-chance for polymorphisms of all 9 STRs is only 1.36 × 10−12 (Sacchetti et al., 1999), both samples should be from the same person.
Table 2.
Profiling of short tandem repeats in 9 loci and Amelogenin
| D35S1358 |
vWA |
FGA |
Amelogenin |
D8S1179 |
D21S11 |
D18S51 |
D5S818 |
D13S317 |
D7S820 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Follow-up sample | replicate 1 | 15 | 16 | 15 | 18 | 26 | X | 14 | 15 | 28 | 32.2 | 14 | 16 | 11 | 14 | 12 | 13 | 10 | 11 | |
| replicate 2 | 15 | 16 | 15 | 18 | 26 | X | 14 | 15 | 28 | 32.2 | 14 | 16 | 11 | 14 | 12 | 13 | 10 | 11 | ||
| replicate 3 | 15 | 16 | 15 | 18 | 26 | X | 14 | 15 | 28 | 32.2 | 14 | 16 | 11 | 14 | 12 | 13 | 10 | 11 | ||
| Enrollment sample | replicate 1 | 15 | 16 | 15 | 18 | 26 | X | 14 | 15 | 28 | 32.2 | 14 | 16 | 11 | 14 | 12 | 13 | 10 | 11 | |
| replicate 2 | 15 | 16 | 12 | 15 | 18 | 26 | X | 14 | 15 | 28 | 32.2 | 14 | 16 | 11 | 14 | 12 | 13 | 10 | 11 | |
| replicate 3 | Insuf* | 15 | 15 | 18 | 26 | X | 14 | 15 | 28 | 32.2 | 14 | 16 | 11 | 14 | 12 | 13 | 10 | 11 | ||
| undiluted | 15 | 16 | 15 | 18 | 26 | X | 14 | 15 | 28 | 32.2 | 14 | 16 | 11 | 14 | 12 | 13 | 10 | 11 | ||
Insufficient for evaluation because it had a call that fell out of the bin.
Discussion
In efforts to examine the genomic diversity of HPV16 variants, we found recombinant sequences of the E and Af2 variants in one specimen. These sequences, although detected only in a single clone, were unlikely due to PCR errors because all of the nucleotide alterations matched to the Af2 variant at the corresponding positions and the majority of them were overlapped between the clones. Also, it is unlikely that the result was an artifact of cloning-mediated recombination, because the recombinant sequences were detected from the PCR products prior to the cloning procedure. We are aware of a possibility of potential PCR-mediated recombination which can arise due to presence of multiple related sequence templates, incomplete primer extension, and crossover events in the PCR cycle (Judo, Wedel, and Wilson, 1998; Shafikhani, 2002; Wu et al., 2007). However, there was no detectable Af2 variant template in this specimen. More importantly, the recombinant sequences were detected directly from the cervical sample by RS-PCR with primer set B.
These recombinant sequences differed in number of copies. As suggested by direct DNA sequencing of the RS-PCR products, recombinant 1 and 4 were more abundant than recombinant 3 and 2, respectively. The copy number of recombinant 4 was larger than that of recombinant 1 because only the former was detected by RS-PCR of the enrollment sample. The presence of multiple recombinants in a single sample is not totally unexpected, given that parental sequences can be disrupted at different sites. It is possible that more recombinants might have been found, had a whole genome been examined.
It should be pointed out that recombination after diversification of HPV16 variants might be a rare event because it has not been reported before. In this study, it was detected only in 1 of 80 samples analyzed. We do not know how these recombinant sequences were formed. If the recombinants result from a recent homologous recombination, a concurrent presence of both E and Af2 variants is required. However, no Af2 variant was detected from this sample, even by PCR with a set of primers specific for Af2 variant. It is unclear whether the Af2 variant was latent at the time of sample collection. Alternatively, the study subject might have acquired different HPV16 recombinants concurrently or separately from the same or different sex partners.
Neither HPV16 prototype nor recombinant was detected, but Af2 variant was in the follow-up sample. It remains unanswered whether the disappearance of the prototype and recombinants at a follow-up visit was due to a clearance of the infection or suppression to a level below the detectable threshold. One may think that the recombinant sequences might be intermediates of the change from the prototype to Af2 variant. However, this does not seem biologically plausible because of a slow evolutionary process of HPV (Bernard, Chan, and Delius, 1994), More likely, the Af2 variant detected in the follow-up sample represented reactivation of a latent infection or a separate infection. Given that HPV16 variants that are abundant initially usually remain predominant at subsequent visits (van Belkum et al., 1995; Xi et al., 1995), a change of the predominant variants found in this study was a rare event. The reason for this is not clear.
To the best of our knowledge, this is the first report showing HPV16 recombinant sequences in natural infection. Our data suggest that similar to other viruses (Nimonkar and Boehmer, 2003; Thiry et al., 2005), DNA recombination might play a role in diversification of HPV genome. It may occur after diversification of HPV variants and continue to drive the evolution of current HPV types and variants. This may in part explain extraordinary diversity of this family of viruses.
In summary, the present study provided evidence of HPV16 recombinant sequences in natural infection but no evidence of sustained replication of these recombinants.
Materials and methods
Specimen
Cervical swab samples in Specimen Transport Medium (Digene Corporation, Silver Spring, MD) were from women enrolled in the ASCUS-LSIL Triage Study. Of 40 pairs of HPV16-positive enrollment and follow-up samples analyzed, only one displayed recombinant sequences of HPV16 variants. This pair of samples was collected froma 21-year-old woman on May 27, 1997 (enrollment) and January 28, 1999 (follow-up), respectively.
PCR-based cloning and sequencing
The 751-bp DNA fragments (corresponding to the 3′ part of HPV16 long control region and the entire E6 region) were generated by PCR with a pair of primers (forward, nucleotide position 7701–7722, 5′-AGGCACATATTTTTGGCTTGTT; reverse, nucleotide position 591-568, 5′-TTCATGCAATGTAGGTGTATCTCC). PCR products were cloned into plasmids using a TOPO TA Cloning kit according to the protocol recommended by the manufacturer (Invitrogen, Carlsbad, CA). Clones containing target inserts were identified by digestion with appropriate restriction enzymes followed by electrophoreses on 1.5% agarose gel. Plasmid DNAs were purified with a QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA). The purified DNA templates were sequenced by Northwoods (Northwoods DNA Inc., Solway, MN). To minimize PCR error-introduced misclassification, DNA fragments for cloning were generated by two independent PCR assays and 20 clones per sample were selected for sequencing. To verify the recombinant sequences, another PCR-based cloning was performed on the enrollment sample and additional 85 clones were sequenced. DNA sequences were analyzed using Sequencer™ software (Gene Codes Corp., Ann Arbor, MI).
Recombinant-specific PCR
The RS-PCR was used to examine whether the recombinant sequences were PCR- or cloning-related artifacts. The internal primers listed in Table 3 were designed to specifically bind to the known recombinant sequences. If the nucleotide at the 3′ end of the forward primer is identical to the E variant, the nucleotide at the 3′ end of the reverse primer is identical to the Af2 variant, and vice versa. The specificity of the RS-PCR was assessed by testing a set of plasmid samples with known inserted sequences. An aliquot of 2 μl PCR products or cervical sample DNA extracts was assayed by RS-PCRin a reaction volume of 25 μl containing 1 × PCR buffer, 3mM MgCl2, 200mM dNTP, 10mM primers, and 1.25 U AmpliTag Gold™ (Roche Molecular Systems, Branchburg, NJ). Amplification was carried out with a cycling program of 30 seconds at 95°C, 1 minute at 58°C, and 1 minute at 72°C for 35 cycles. The RS-PCR products were visualized on 1.5% ethidium bromide-stained agarose gel and verified by direct DNA sequencing. To determine whether there was an Af2 variant in the enrollment sample and an E variant in the follow-up sample, we further paired these internal primers differently to make them specifically target the Af2 or E variant.
Table 3.
Primers for recombinant-specific polymerase chain reaction
| Sequence* (5′-3′) | Nucleotide position | |
|---|---|---|
| Primer set A | ||
| Forward A | TAGTCATACATTaTTCATTTtTAcAg | 7814–7839 |
| Reverse A | TTCCATACAAACTATAACAATAATG | 335–359 |
| Primer set B | ||
| Forward B | TAGTCATACATTGTTCATTTGTAAAA | 7814–7839 |
| Reverse B | TTGTTTGCAGCTCTGTGCATAAa | 145–167 |
Letters highlighted in gray indicate positions where the nucleotides differ between the HPV16 prototype and Af2 variant: capital letters, identical to the prototype; small letters, identical to the Af2 variant.
Primer set A and B were designed to target recombinants 1 and 3 and recombinants 2 and 4, respectively.
Authentication of sample identified by PCR-based combined DNA index system
To determine whether the enrollment and follow-up samples were from a same person, DNA fragments of STRs necessary for combined DNA index system (CODIS) typing in 9 loci (Sacchetti et al., 1999) and Amelogenin on the sex chromosomes (Mannucci et al., 1994) were PCR-generated using the AmpFLSTR Profiler Plus kit (Applied Biosystems, Foster City, CA). Allele assignment was done by Genemapper software (Applied Biosystems) based on AmpFLSTR program parameters. The software also assigns the peak number labels in accordance with an AmpFLSTR provided ladder to indicate the number of STRs at the loci. Two samples are identical if all alleles match across loci.
Acknowledgments
The authors would like to thank the ALTS Group for providing the biological specimens and HPV typing results, and the members of the Research Cell Biorepository at Fred Hutchinson Cancer Research Center for help with the combined DNA index system typing. This work was supported by Public Health Service grant CA84396 to L.F.X from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angulo M, Carvajal-Rodriguez A. Evidence of recombination within human alpha-papillomavirus. Virol J. 2007;4:33. doi: 10.1186/1743-422X-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard HU, Chan SY, Delius H. Evolution of papillomaviruses. Curr Top Microbiol Immunol. 1994;186:33–54. doi: 10.1007/978-3-642-78487-3_3. [DOI] [PubMed] [Google Scholar]
- Carvajal-Rodriguez A. Detecting recombination and diversifying selection in human alpha-papillomavirus. Infect Genet Evol. 2008;8 (5):689–92. doi: 10.1016/j.meegid.2008.07.002. [DOI] [PubMed] [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324 (1):17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Eriksson A, Herron JR, Yamada T, Wheeler CM. Human papillomavirus type 16 variant lineages characterized by nucleotide sequence analysis of the E5 coding segment and the E2 hinge region. J Gen Virol. 1999;80 (Pt 3):595–600. doi: 10.1099/0022-1317-80-3-595. [DOI] [PubMed] [Google Scholar]
- Judo MS, Wedel AB, Wilson C. Stimulation and suppression of PCR-mediated recombination. Nucleic Acids Res. 1998;26 (7):1819–25. doi: 10.1093/nar/26.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannucci A, Sullivan KM, Ivanov PL, Gill P. Forensic application of a rapid and quantitative DNA sex test by amplification of the X-Y homologous gene amelogenin. Int J Legal Med. 1994;106(4):190–3. doi: 10.1007/BF01371335. [DOI] [PubMed] [Google Scholar]
- Meissner J. Sequencing errors in reference HPV clones. In: Myers G Baker C, Munger K, et al., editors. Human Papillomaviruses 1997: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. 3. Los Alamos: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1997. pp. 110–23. [Google Scholar]
- Munoz N, Bosch FX, de Sanjose S, Herrero R, Castellsague X, Shah KV, Snijders PJ, Meijer CJ. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348(6):518–27. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Boehmer PE. Reconstitution of recombination-dependent DNA synthesis in herpes simplex virus 1. Proc Natl Acad Sci U S A. 2003;100(18):10201–6. doi: 10.1073/pnas.1534569100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti L, Calcagno G, Coto I, Tinto N, Vuttariello E, Salvatore F. Efficiency of two different nine-loci short tandem repeat systems for DNA typing purposes. Clin Chem. 1999;45 (2):178–83. [PubMed] [Google Scholar]
- Schiffman M, Adrianza ME. ASCUS-LSIL Triage Study. Design, methods and characteristics of trial participants. Acta Cytol. 2000;44 (5):726–42. doi: 10.1159/000328554. [DOI] [PubMed] [Google Scholar]
- Seedorf K, Krammer G, Durst M, Suhai S, Rowekamp WG. Human papillomavirus type 16 DNA sequence. Virology. 1985;145 (1):181–5. doi: 10.1016/0042-6822(85)90214-4. [DOI] [PubMed] [Google Scholar]
- Shafikhani S. Factors affecting PCR-mediated recombination. Environ Microbiol. 2002;4 (8):482–6. doi: 10.1046/j.1462-2920.2002.00326.x. [DOI] [PubMed] [Google Scholar]
- Thiry E, Meurens F, Muylkens B, McVoy M, Gogev S, Thiry J, Vanderplasschen A, Epstein A, Keil G, Schynts F. Recombination in alphaherpesviruses. Rev Med Virol. 2005;15 (2):89–103. doi: 10.1002/rmv.451. [DOI] [PubMed] [Google Scholar]
- van Belkum A, Juffermans L, Schrauwen L, van Doornum G, Burger M, Quint W. Genotyping human papillomavirus type 16 isolates from persistently infected promiscuous individuals and cervical neoplasia patients. J Clin Microbiol. 1995;33 (11):2957–62. doi: 10.1128/jcm.33.11.2957-2962.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varsani A, van der Walt E, Heath L, Rybicki EP, Williamson AL, Martin DP. Evidence of ancient papillomavirus recombination. J Gen Virol. 2006;87 (Pt 9):2527–31. doi: 10.1099/vir.0.81917-0. [DOI] [PubMed] [Google Scholar]
- Wu L, Tang T, Zhou R, Shi S. PCR-mediated recombination of the amplification products of the Hibiscus tiliaceus cytosolic glyceraldehyde-3-phosphate dehydrogenase gene. J Biochem Mol Biol. 2007;40 (2):172–9. doi: 10.5483/bmbrep.2007.40.2.172. [DOI] [PubMed] [Google Scholar]
- Xi LF, Demers GW, Koutsky LA, Kiviat NB, Kuypers J, Watts DH, Holmes KK, Galloway DA. Analysis of human papillomavirus type 16 variants indicates establishment of persistent infection. J Infect Dis. 1995;172 (3):747–55. doi: 10.1093/infdis/172.3.747. [DOI] [PubMed] [Google Scholar]
- Yamada T, Manos MM, Peto J, Greer CE, Munoz N, Bosch FX, Wheeler CM. Human papillomavirus type 16 sequence variation in cervical cancers: a worldwide perspective. J Virol. 1997;71 (3):2463–72. doi: 10.1128/jvi.71.3.2463-2472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]