Abstract
Objective:
This study examined the genetic association between variation in the cannabinoid receptor 1 (CNR1) gene and cannabis dependence symptoms.
Method:
Adolescent and young adult subjects were recruited from three settings: a treatment program for youth with substance use disorders, the criminal justice system, and the community. A case-control sample consisted of 224 cases who endorsed at least one dependence symptom and 108 controls who tried cannabis but endorsed no symptoms. A family-based sample of 219 families was also analyzed.
Results:
Case-control analysis identified a nominal association between SNP rs1049353 and having one or more cannabis dependence symptoms (p = .029), but the association did not hold up in a combined sample. Family-based analysis found a trend for the same SNP (p = .07). We did not replicate a previous report that SNP rs806380 was associated with the development of cannabis dependence.
Conclusion:
These results provide inconclusive evidence of association between rs1049353/rs806380 and the development of cannabis dependence, and underscore the importance of replicating results of genetic association studies. Additional family-based studies are needed to clarify the role of the CNR1 gene, and its various SNPs, in the development of cannabis use disorders.
Keywords: cannabis dependence, CNR1, genetic association, cannabinoids
1.0 INTRODUCTION
Cannabis is the most commonly used illicit substance among adolescents and young adults. According to results from Monitoring the Future, in 2006 42% of 12th graders reported having tried cannabis at some point in their lifetime, 18% used within the past month, and 5% smoked cannabis daily (Johnston et al., 2007). In addition, cannabis dependence develops in approximately 10-14% of adolescent users (Chen & Anthony, 2003; K. Chen et al., 1997; Rey et al., 2004). Cannabis use disorders are increasing among young people (Compton et al., 2004), and according to the Substance Abuse and Mental Health Services Administration (SAMHSA), adolescents and young adults represent 66% of patients admitted to publicly funded treatment facilities for cannabis use problems (SAMHSA, 2006). Cannabis use is also associated with other serious consequences, including lower educational attainment (Lynskey & Hall, 2000), the development of other illicit drug use (Lynskey et al., 2006; Young et al., 2002), and development of serious psychiatric conditions (Lynskey et al., 2004). Thus, it is important to examine factors that put these populations at risk for cannabis dependence.
Twin studies examining the etiology of substance use disorders have found that genetic factors play a role in conferring risk for cannabis dependence (Agrawal & Lynskey, 2006; Kendler & Prescott, 1998; Lynskey et al., 2002; Tsuang et al., 1998). However, only two studies have examined candidate genes for their association with the risk for developing cannabis dependence (Hopfer et al., 2006; Tyndale et al., 2007). Both studies examined genes involved in the endocannabinoid system, and both found positive associations. Overall, more studies examining genes that influence cannabis dependence are needed.
There are two known receptors of the endocannabinoid system, cannabinoid receptor 1 (CNR1) and cannabinoid receptor 2 (CNR2). The CNR1 receptor is found throughout the brain and is expressed at high levels (Childers & Breivogel, 1998); the CNR2 receptor is found primarily in the periphery and appears to have an immune function. The cannabinoid receptor is the primary site of action of delta-9-tetrahydrocannabinol (THC), the principal psychoactive ingredient in cannabis (Childers & Breivogel, 1998; Onaivi et al., 2002). THC mimics the actions of endogenous cannabinoids and influences the action of major neurotransmitters such as dopamine and serotonin.
Genetic studies are beginning to examine the relation between genes involved in the endocannabinoid system and substance use disorders. Most of these studies have focused on the CNR1 gene. Several found a significant association between variation in CNR1 and a substance abuse phenotype, including polysubstance abuse or dependence (Comings et al., 1997; Zhang et al., 2004; Zuo et al., 2007), severe alcohol dependence (Schmidt et al., 2002), and cocaine addiction (Ballon et al., 2006). Although some studies have found no association between CNR1 and heroin dependence (Li et al., 2000) or alcohol/drug dependence (Comings et al., 1997; Covault et al., 2001), there is suggestive evidence that genetic variation in the endocannabinoid system is associated with substance use disorders. However, the above studies focused primarily on adult case-control samples, often examining phenotypes that combine various substances into a polysubstance category. Also, because the potential for spurious association is an important factor in genetic association studies, replication of any significant results is necessary.
Hopfer et al. (2006) conducted the first study to examine the association between CNR1 and a cannabis phenotype, and was the first to examine CNR1 in an adolescent sample. They examined four single nucleotide polymorphisms (SNPs) in the CNR1 gene for association with having developed one or more symptoms of cannabis dependence. Using a case-control design, the study included adolescent and young adult subjects who had used cannabis six or more times, where cases had developed cannabis dependence symptoms and controls had not. They found that SNP rs806380, located in intron 2 of the CNR1 gene, was significantly associated with developing dependence symptoms, with the G allele having a protective effect.
The current study sought to replicate the Hopfer et al. (2006) findings in a separate sample, while utilizing identical ascertainment, measures, SNPs, and phenotype. In addition, since population stratification is a concern in genetic association studies, we conducted a family-based association analysis to mitigate this confound. To our knowledge, no other genetic association studies examining the CNR1 gene have utilized a family-based design.
2.0 METHOD
2.1 Sample
Subjects included participants in ongoing studies of the genetics of adolescent and young adult substance abuse in Colorado, funded by the National Institute on Drug Abuse (NIDA; DA015522, DA011015). Youth who participated in these studies were recruited from treatment settings for youth with substance use disorders, criminal justice settings, and community-based twin, adoption, and family studies of adolescent substance use disorders. The consent forms for the original studies asked whether the information gathered could be used for analyses of “genetic tests related to substance use disorders.” Only those subjects whose parents consented and themselves assented to this statement were included in these analyses. The entire pool of potential subjects encompassed over 5,000 youth, and we selected those subjects who met the following criteria: 1) endorsed using cannabis at least six times, and 2) had at least one year between age of first cannabis use and age at testing.
This study examines subjects that were selected from the same pool as those subjects examined in the Hopfer et al. (2006) CNR1 study. However, none of the subjects overlap with those in the previous study, and we include independent cases as well as siblings of cases from the Hopfer study.
2.2 Measures
All youth were assessed with the Composite International Diagnostic Interview - Substance Abuse Module (CIDI-SAM). This instrument diagnoses abuse and dependence for ten drug classes. It primarily consists of a 30-to-60-minute interview that is designed for trained lay interviewers. The CIDI's reliability and validity (Cottler et al., 1989) made a version of it the main assessment for DSM-IV Substance Field Trials and for the National Comorbidity Study (Kessler et al., 1994). Its validity for use with substance dependent adolescents has been demonstrated (Crowley et al., 2001; Mikulich et al., 2001).
2.3 SNP Selection and Genotyping
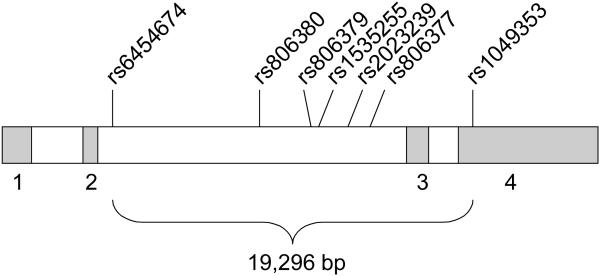
Using publicly available human genomics resources dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and the UCSC Genome Browser (http://genome.ucsc.edu/), we completed preliminary bioinformatics work examining the human CNR1 gene. This gene is located on chromosome 6q14 and spans 26,085 nucleotides. Seven SNPs were selected for genotyping: the four originally examined by Hopfer et al. (2006), plus three more. Table 1 lists information for each SNP, and Figure 1 displays the CNR1 gene structure and SNP locations. SNPs were selected based on validation status, a minor allele frequency (MAF) greater than 0.10, and location of the SNPs (to ensure some distribution across the length of the gene). None of these SNPs lead to amino acid changes.
Table 1.
SNP information
| SNP | alleles | Het. | MAF | MA | position |
|---|---|---|---|---|---|
| rs6454674* | T/G | .42 | .32 | G | 1654 |
| rs806380* | G/A | .44 | .33 | G | 9931 |
| rs806379 | A/T | .32 | .38 | A | 13317 |
| rs1535255 | C/A | .27 | .17 | C | 13376 |
| rs2023239 | G/A | .34 | .22 | G | 14102 |
| rs806377* | T/C | .51 | .48 | T | 15861 |
| rs1049353* | T/C | .37 | .24 | T | 20950 |
SNP examined in Hopfer et al. (2006) study
Het = heterozygosity, MAF = minor allele frequency, MA = minor allele
Figure 1.
CNR1 gene with genotyped SNP locations Note.
Numbered boxes represent exons; bp = base pairs
Genotyping was accomplished using Applied Biosystems TaqMan™ Assays-on-Demand. Genomic DNA was isolated from buccal cell swabs and preamplified using the method of Zheng et al. (2001). These methods yield high quality DNA and have been shown to be reliable for genotyping (Anchordoquy et al., 2003). TaqMan assays were performed under standard conditions using the ABI PRISM® 7900 instrument. All SNPs were tested on approximately 200 samples, which include some monozygotic twins to conduct accuracy checks. Also, a subset of the DNA samples was genotyped twice. Discordant rates were less than 0.5% for both of these quality control checks.
2.4 Analysis
For simplicity, the sample analyzed in the Hopfer et al. (2006) study will be referred to as the original sample, and the sample analyzed in this study will be referred to as the replication sample.
2.4.1 Case-control analysis
In order to replicate the original Hopfer et al. findings, we conducted case-control analyses on the entire replication sample, and then separately for our two largest ethnic groups, Caucasians and Hispanics. Analyses were conducted using Haploview (Barrett et al., 2005). As with the original sample, cases were defined by having endorsed one or more DSM-IV cannabis dependence symptoms, and controls were defined as those who experimented with cannabis at least six times but endorsed no cannabis dependence symptoms. Table 2 describes the ethnic, age, and gender distribution of the cases and controls, as well as the mean number of cannabis dependence symptoms. The total sample size was 332. Power analyses, based on a MAF of .29, an additive model, and an odds ratio of 1.7, indicated 80% power to detect an association.
Table 2.
Demographic characteristics for case-control sample
| N | Mean Age (SD) |
Sex | Ethnicity | Cannabis Symptoms (SD) |
|
|---|---|---|---|---|---|
| Cases | 224 | 18.4 (2.5) | M = 58% F = 42% |
Caucasian: 75% Hispanic: 17% Other/Biracial: 8% |
2.6 (1.7) |
| Controls | 108 | 18.8 (2.7) | M = 50% F = 50% |
Caucasian: 76% Hispanic: 21% Other/Biracial: 3% |
0.0 (0.0) |
2.4.2 Family-based analysis
To address concerns about ethnic stratification, we conducted a family-based association analysis with the combined original and replication samples. The sample for these analyses was selected from a pool that included those subjects analyzed in the case-control analyses (541 from the original sample, 332 from the replication sample), plus any full biological siblings and/or biological parents for whom we had phenotypic data. The dataset was trimmed down to include only a proband and his or her full biological siblings and/or parents. The final sample was comprised of 550 individuals (301 from the original sample and 249 from the replication sample) from 219 families. The families were 75% Caucasian, 15% Hispanic, and 10% Other/Biracial ethnicity. Excluding parents, the subjects were 64% male with a mean age of 18. For details about the original sample, see Hopfer et al. (2006).
Family data were analyzed using the Family Based Association Test (FBAT) as implemented in PBAT (Laird et al., 2000; Lange et al., 2002). The FBAT test is a score-based test that extends the variance components approach proposed by Fulker et al. to include complex within-family patterns of variation (Fulker et al., 1999). The score statistic represents the covariation between genotype and residual-trait, or within-family, deviation (Laird et al., 2000). FBAT analyses examined the same phenotype as the case-control analyses, and were conducted using an offset value of 12.9, which is the estimated population prevalence for the cannabis phenotype examined in this study. No Mendelian errors were found in the family data.
2.4.3 Haplotype analysis
Also, to replicate the original Hopfer et al. (2006) findings, we conducted a haplotype analysis using PLINK (Purcell et al., 2007), http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml. These analyses were conducted on the case-control replication dataset (n = 332), where all seven SNPs were included in analysis and PLINK was utilized to impute the haplotypes.
3.0 RESULTS
3.1 Hardy-Weinberg Equilibrium (HWE)
Six of the seven SNPs were in HWE, SNP rs806379 being the exception. Further examination showed that rs806379 was in HWE in Hispanics but not Caucasians. This SNP was not examined in the original Hopfer et al. (2006) sample. Overall, results for this SNP should be interpreted with caution.
3.2 Case-Control Analysis
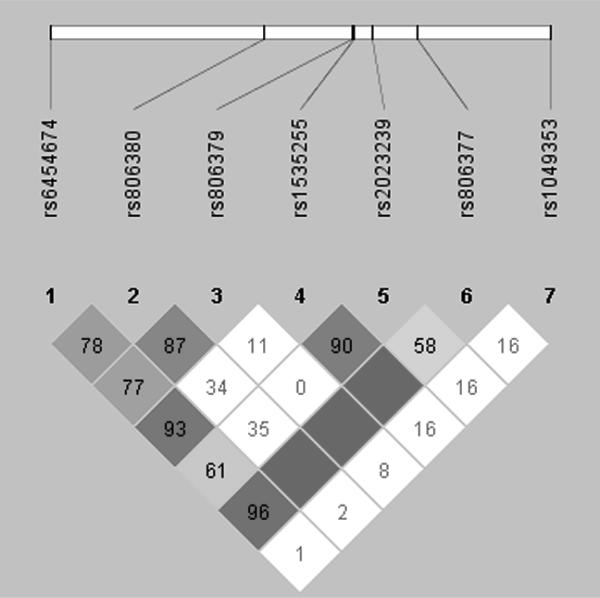
We found a nominal association between SNP rs1049353 in exon 4 and having one or more cannabis dependence symptoms (χ2 = 4.76, p = .029), where the C allele was significantly more frequent in cases. In addition, we did not find a significant association with SNP rs806380, the significant SNP from the original Hopfer et al. (2006) study. SNPs rs1049353 and rs806380 were not in linkage disequilibrium (LD; Figure 2).
Figure 2.
Pairwise linkage disequilibrium (D′) for the CNR1 SNPs
Note. Blank squares indicate D′ = 100%
Because of the ethnic diversity in our sample and the possibility of bias due to population stratification, we conducted separate case-control analyses for Caucasians (n = 249) and Hispanics (n = 61), our two largest ethnic groups. For the larger Caucasian sample, results were the similar to those for the entire sample, with a nominal association with the C allele of SNP rs1049353 (χ2 = 4.28, p = .039). We found no significant associations for the Hispanic sample.
In addition, due to differing results between the original and replication samples, and to increase power, we conducted case-control analyses after combining the original and replication samples. Since the original study only examined four SNPs, only those SNPs were examined in the combined analysis (see Table 3). Once related family members were removed from the combined file, the sample size was 730. Of the four SNPs examined, none were significant for the combined group or for Caucasian or Hispanic groups.
Table 3.
Case-control analysis results
| Replication sample | Combined sample | ||||||
|---|---|---|---|---|---|---|---|
| SNP | Assoc. allele |
Case/Control allele freq. |
χ2 | P | Case/Control allele freq. |
χ2 | P |
| rs6454674 | G | .335/.292 | 1.173 | 0.279 | .316/.318 | 0.010 | 0.920 |
| rs806380 | G | .327/.324 | 0.008 | 0.930 | .308/.334 | 1.071 | 0.301 |
| rs806379 | A | .384/.369 | 0.129 | 0.719 | NG | NG | NG |
| rs1535255 | C | .165/.165 | 0.000 | 0.999 | NG | NG | NG |
| rs2023239 | G | .217/.212 | 0.028 | 0.867 | NG | NG | NG |
| rs806377 | C | .530/.500 | 0.498 | 0.480 | .529/.505 | 0.759 | 0.384 |
| rs1049353 | C | .791/.705 | 4.762 | 0.029 | .762/.737 | 1.150 | 0.284 |
NG = SNP not genotyped in original sample (Hopfer et al., 2006)
3.3 Family-Based Analysis
FBAT results for combined original and replication samples found no significant associations (Table 4); however, we did find a trend for allele C of SNP rs1049353 (p = .07), the nominal SNP from the case-control analyses. Power analyses, based on a MAF of .294, an additive model, and an odds ratio of 1.7, indicated 42% power to detect an association.
Table 4.
Family-based analysis results
| SNP | Assoc. allele | Z | P |
|---|---|---|---|
| rs6454674 | G | 0.239 | 0.811 |
| rs806380 | G | 0.640 | 0.522 |
| rs806377 | C | 0.128 | 0.898 |
| rs1049353 | C | 1.786 | 0.074 |
A summary of results for all analyses is presented in Table 5. Analyses were not adjusted for multiple testing.
Table 5.
Summary of results for all analyses
| Case-control | Case Control Replication |
Combined Case-control |
Family- based |
||||
|---|---|---|---|---|---|---|---|
| SNP | Original sample |
Replication sample |
Cauc. | Hisp. | Cauc. | Hisp. | |
| rs6454674 | -- | -- | -- | -- | -- | -- | -- |
| rs806380 | √ | -- | -- | -- | -- | -- | -- |
| rs806379 | NG | -- | -- | -- | -- | -- | -- |
| rs1535255 | NG | -- | -- | -- | -- | -- | -- |
| rs2023239 | NG | -- | -- | -- | -- | -- | -- |
| rs806377 | -- | -- | -- | -- | -- | -- | -- |
| rs1049353 | -- | √ | √ | -- | -- | -- | T |
Note. NG = not genotyped, Cauc = Caucasians, Hisp = Hispanics, √ = significant result, T = Trend
3.4 Haplotype analysis
The omnibus haplotype test, testing for association with the CNR1 locus, was not significant (χ2 (13) = 17.32, p = 0.185). Two haplotypes were significant: TATAATT (χ2 (1) = 4.96, p = 0.026) and TAACGCT (χ2 (1) = 5.03, p = 0.025). The frequencies of these haplotypes were 20% and 3%, respectively. The highlighted portions of the haplotypes represent the four SNPs that were tested in the original Hopfer et al. (2006) sample. When analyzing only these four SNPs, only the TATT haplotype was significant (χ2 (1) = 5.45, p = 0.020). Hopfer et al. found the GGCC, TACC, and GACC haplotypes significant, inconsistent with our results.
4.0 DISCUSSION
This study examined the association between variation in the CNR1 gene and cannabis dependence symptoms in adolescents and young adults. Using strict replication criteria, we attempted to replicate a previous result that SNP rs806380 was associated with developing one or more cannabis dependence symptoms (Hopfer et al., 2006). We did not replicate this previous finding. In addition, our haplotype analyses, while yielding two significant haplotypes, were not consistent with the results from the Hopfer et al. study. We did find a nominal association with the C allele of SNP rs1049353, and a trend for this SNP in a family-based analysis; however, a case-control analysis with combined original and replication samples did not achieve this result. Together, these studies represent the first to examine the association between variation in CNR1 and a cannabis phenotype.
Case-control analyses with our replication sample found a nominal association between SNP rs1049353 and cannabis dependence symptoms (p = 0.029, Table 3). When we conducted a case-control analysis with the combined original and replication samples, however, we found no significant associations (Table 3). A family-based analysis with the combined sample showed a trend for association with SNP rs1049353, but did not reach statistical significance (Table 4). Our power calculations showed that the family-based analysis was underpowered and would require a larger number of families (~ 291, rather than our 219) to achieve 80% power. However, our mixed results, combined with the fact that the analyses were not corrected for multiple tests, suggests that the associations we found with rs1049353 may be false positives.
To our knowledge, four studies have examined SNP rs1049353, also known as the silent mutation 1359G/A, for its association with a substance abuse phenotype. Hopfer et al. (2006), in the study preceding this one, as well as Zuo et al. (2007), found no significant result for rs1049353. Schmidt et al. (2002) compared cases displaying severe alcohol dependence symptoms to controls and found a significant association with rs1049353 in a Caucasian sample. However, their direction of effect was the opposite of that found in this study. Zhang et al. (2004) compared cases, identified as having abused multiple substances, to controls; they did not find a significant association with SNP rs1049353, but achieved significant results for three other SNPs examined in this study: rs806379, rs1535255, and rs2023239. SNP rs806379 is in high linkage disequilibrium (LD) with the significant SNP from the Hopfer et al. (2006) study (rs806380; r2 = .88). In our study, SNP rs1049353 shows low LD with all of these other SNPs. Overall, the pattern of results with specific CNR1 SNPs is unclear, but there is still some evidence that variation in CNR1 is associated with substance abuse.
This is the first study to utilize a family-based design to examine the association between variation in CNR1 and any phenotype. Unlike case-control studies, which are by far the most commonly seen in the field's literature, family-based studies are not susceptible to spurious results due to population admixture. Thus, despite marginal results for this SNP, it would be worthwhile to examine this SNP, and other CNR1 SNPs, in other family-based samples in order to clarify the role of any or all of these SNPs.
Finally, we did not find a significant association with SNP rs806380, and thus did not replicate the results from the Hopfer et al. study in any of our analyses, including the haplotype analyses. SNPs rs806380 and rs1049353 were not in linkage disequilibrium (Figure 2), and it is possible that either SNP is associated with cannabis dependence symptoms, or that multiple SNPs are associated. Future studies in independent samples will need to address this issue. Our replication attempt was notable, in that our methods included precisely the same subject ascertainment, measures, SNPs, and phenotype as the original study. Such strict replication criteria make it easier to come to conclusions about association study results, whether results are positive or negative. The results of this study, and their inconsistency with results of other studies, underscore the importance of replicating results found in candidate gene studies. Spurious associations are common in genetic association studies (Sullivan, 2007), and results that do not replicate a former finding but identify an association with a different SNP should be viewed with caution and followed up with other replication studies.
This study did have some limitations. First, there are numerous factors that can influence association study results, including sampling, linkage disequilibrium patterns, and effect size (Cardon & Bell, 2001). Studies that show different SNPs in the same gene to be associated with a particular phenotype need further examination before conclusions can be made about the role of those SNPs in the disease of interest. Second, a portion of our sample is comprised of youth with polysubstance dependence and conduct problems, and results from this study may differ from those of studies examining youth with cannabis problems without other comorbid conditions. However, studies examining population-based samples have shown that those who display cannabis problems often display high rates of conduct problems and alcohol dependence as well (Stinson et al., 2006). Third, our sample was selected for youth with substance and conduct problems, and thus is not representative of the general population. Examining the association between variation in CNR1 and cannabis problems in a large, population-based sample would be very informative.
In summary, we did not replicate an association previously found with SNP rs806380 and having one or more cannabis symptoms. We found inconclusive evidence for an association between SNP rs1049353, suggesting that the result may be a false positive. This result, coupled with the results for other studies, suggests that variation in the CNR1 gene may be associated with cannabis dependence symptoms, but that these and others' results are in need of replication, particularly in family-based samples. Overall, more studies examining the CNR1 gene are necessary to clarify the role of this gene in the development of substance use disorders.
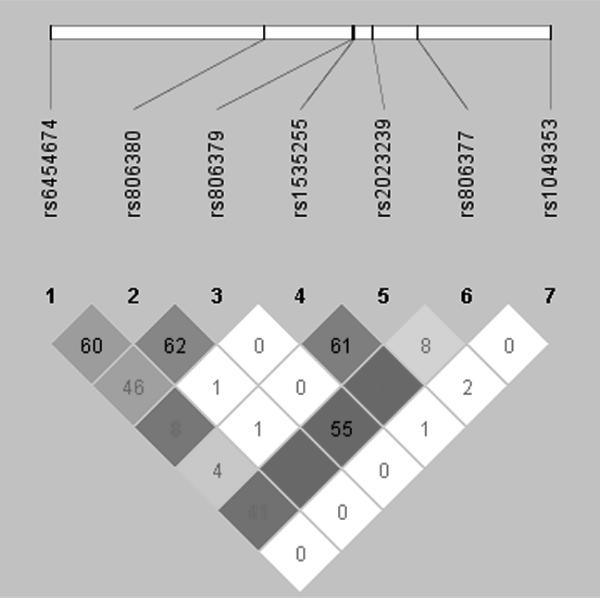
Figure 3.
Pairwise linkage disequilibrium (R2) for the CNR1 SNPs
Note. Blank squares indicate R2 = 100%
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agrawal A, Lynskey MT. The genetic epidemiology of cannabis use, abuse and dependence. Addiction. 2006;101(6):801–812. doi: 10.1111/j.1360-0443.2006.01399.x. [DOI] [PubMed] [Google Scholar]
- Anchordoquy H, McGeary C, Liu L, Krauter KS, Smolen A. Genotyping of three candidate genes after whole-genome preamplification of DNA collected from buccal cells. Behav Genet. 2003;33:73–78. doi: 10.1023/a:1021007701808. [DOI] [PubMed] [Google Scholar]
- Ballon N, Leroy S, Roy C, Bourdel MC, Charles-Nicolas A, Krebs M, Poirier MF. (AAT)n repeat in the cannabinoid receptor gene (CNR1): association with cocaine addiction in an African-Caribbean population. Pharmacogenomics. 2006;6(2):126–130. doi: 10.1038/sj.tpj.6500352. [DOI] [PubMed] [Google Scholar]
- Barrett J, Fry B, Maller J, Daly M. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2(2):91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- Chen CY, Anthony JC. Possible age-associated bias in reporting of clinical features of drug dependence: epidemiological evidence on adolescent-onset marijuana use. Addiction. 2003;98(1):71–82. doi: 10.1046/j.1360-0443.2003.00237.x. [DOI] [PubMed] [Google Scholar]
- Chen K, Kandel DB, Davies M. Relationships between frequency and quantity of marijuana use and last year proxy dependence among adolescents and adults in the United States. Drug Alcohol Depend. 1997;46(12):53–67. doi: 10.1016/s0376-8716(97)00047-1. [DOI] [PubMed] [Google Scholar]
- Childers SR, Breivogel CS. Cannabis and endogenous cannabinoid systems. Drug Alcohol Depend. 1998;51(12):173–187. doi: 10.1016/s0376-8716(98)00075-1. [DOI] [PubMed] [Google Scholar]
- Comings DE, Muhleman D, Gade R, Johnson P, Verde R, Saucier G, MacMurray J. Cannabinoid receptor gene (CNR1): association with I.V. drug use. Molec Psychiatry. 1997;2:161–168. doi: 10.1038/sj.mp.4000247. [DOI] [PubMed] [Google Scholar]
- Compton WM, Grant B, Colliver J, Glantz MD, Stinson FS. Prevalence of marijuana use disorders in the United States: 1991-1992 and 2001-2002. JAMA. 2004;291(17):2114–2121. doi: 10.1001/jama.291.17.2114. [DOI] [PubMed] [Google Scholar]
- Cottler LB, Robins LN, Helzer JE. The reliability of the CIDI-SAM: a comprehensive substance abuse interview. Br J Addiction. 1989;84(7):801–814. doi: 10.1111/j.1360-0443.1989.tb03060.x. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Kranzler H. Association study of cannabinoid receptor gene (CNR1) alleles and drug dependence. Molec Psychiatry. 2001;6:501–502. doi: 10.1038/sj.mp.4000925. [DOI] [PubMed] [Google Scholar]
- Crowley TJ, Mikulich SK, Ehlers KM, Whitmore EA, MacDonald MJ. Validity of structured clinical evaluations in adolescents with conduct and substance problems. J Am Acad Child Adolesc Psychiatry. 2001;40(3):265–273. doi: 10.1097/00004583-200103000-00005. [DOI] [PubMed] [Google Scholar]
- Fulker DF, Cherny SS, Sham P, Hewitt JK. Combined linkage and association sib-pair analysis for quantitative traits. Am J Hum Genet. 1999;64:259–67. doi: 10.1086/302193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer CJ, Young SE, Purcell S, Crowley TJ, Stallings MC, Corley R, Rhee SH, Smolen A, Krauter K, Hewitt JK, Ehringer MA. Cannabis receptor haplotype protective against developing cannabis dependence symptoms in adolescents. Am J Med Genet. 2006;141B:895–901. doi: 10.1002/ajmg.b.30378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LD, O'Malley PM, Bachman JG, Schulenberg JE. Monitoring the Future national results on adolescent drug use: Overview of key findings, 2006. National Institute on Drug Abuse; Bethesda, MD: 2007. NIH Publication No. 07-6202. [Google Scholar]
- Kendler KS, Prescott CA. Cannabis use, abuse, and dependence in a population-based sample of female twins. Am J Psychiatry. 1998;155(8):1016–1022. doi: 10.1176/ajp.155.8.1016. [DOI] [PubMed] [Google Scholar]
- Kessler RC, McGonagle KA, Zhao S, Nelson CB, Hughes M, Eshleman S, Wittchen HU, Kendler KS. Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States. Results from the National Comorbidity Survey. Arch Gen Psychiatry. 1994;51(1):8–19. doi: 10.1001/archpsyc.1994.03950010008002. [DOI] [PubMed] [Google Scholar]
- Laird N, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epi. 2000;19(Suppl):S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lange C, DeMeo D, Laird N. Power and design consideration for a general class of family-based association tests: Quantitative traits. Am J Hum Genet. 2002;71:1330–1341. doi: 10.1086/344696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu X, Zhu Z, Zhao J, Hu X, Ball DM, Sham PC, Collier DA. No association between (AAT)n repeats in the cannabinoid receptor gene (CNR1) and heroin abuse in a Chinese population. Molec Psychiatry. 2000;5:128–130. doi: 10.1038/sj.mp.4000670. [DOI] [PubMed] [Google Scholar]
- Lynskey MT, Glowinski AL, Todorov A, Bucholz KK, Madden PA, Nelson E, Statham DJ, Martin NG, Heath AC. Major depressive disorder, suicidal ideation, and suicide attempt in twins discordant for cannabis dependence and early-onset cannabis use. Arch Gen Psychiatry. 2004;61(10):1026–1032. doi: 10.1001/archpsyc.61.10.1026. [DOI] [PubMed] [Google Scholar]
- Lynskey MT, Hall W. The effects of adolescent cannabis use on educational attainment: a review. Addiction. 2000;95(11):1621–1630. doi: 10.1046/j.1360-0443.2000.951116213.x. [DOI] [PubMed] [Google Scholar]
- Lynskey MT, Heath AC, Nelson EC, Bucholz KK, Madden PA, Slutske WS, Statham DJ, Martin NG. Genetic and environmental contributions to cannabis dependence in a national young adult twin sample. Psychol Med. 2002;32(2):195–207. doi: 10.1017/s0033291701005062. [DOI] [PubMed] [Google Scholar]
- Lynskey MT, Vink JM, Boomsma DI. Early onset cannabis use and progression to other drug use in a sample of Dutch twins. Behav Genet. 2006;36(2):195–200. doi: 10.1007/s10519-005-9023-x. [DOI] [PubMed] [Google Scholar]
- Mikulich SK, Hall SK, Whitmore EA, Crowley TJ. Concordance between DSM-III-R and DSM-IV diagnoses of substance use disorders in adolescents. Drug Alcohol Depend. 2001;61(3):237–248. doi: 10.1016/s0376-8716(00)00143-5. [DOI] [PubMed] [Google Scholar]
- Onaivi E, Leonard C, Ishiguro H, Zhang L, Lin Z, Akinshola B, Uhl GR. Endocannabinoids and cannabinoid receptor genetics. Prog Neurobiol. 2002;66(5):307–344. doi: 10.1016/s0301-0082(02)00007-2. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly M, Sham P. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81 doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey J, Martin A, Krabman P. Is the party over? Cannabis and juvenile psychiatric disorder: the last ten years. J Am Acad Child Adolesc Psychiatry. 2004;43:1194–1205. doi: 10.1097/01.chi.0000135623.12843.60. [DOI] [PubMed] [Google Scholar]
- Substance Abuse and Mental Health Services Administration (SAMHSA) Treatment Episode Data Set (TEDS). Highlights - 2004. National Admissions to Substance Abuse Treatment Services. (DASIS Series: S-31, DHHS Publication # (SMA) 06-4140) Substance Abuse and Mental Health Services Administration; Rockville, MD: 2006. [Google Scholar]
- Schmidt LG, Samochowiec J, Finckh U, Fiszer-Piosik E, Horodnicki J, Wendel B, Rommelspacher H, Hoehe MR. Association of a CB1 cannabinoid receptor gene (CNR1) polymorphism with severe alcohol dependence. Drug Alcohol Depend. 2002;65:221–224. doi: 10.1016/s0376-8716(01)00164-8. [DOI] [PubMed] [Google Scholar]
- Stinson FS, Ruan W, Pickering R, Grant B. Cannabis use disorders in the USA: prevalence, correlates, and comorbidity. Psychol Med. 2006;36:1447–1460. doi: 10.1017/S0033291706008361. [DOI] [PubMed] [Google Scholar]
- Sullivan P. Spurious Genetic Associations. Biol Psychiatry. 2007;61:1121–1126. doi: 10.1016/j.biopsych.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, True W, Lin N, Toomey R, Eaves L. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55(11):967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- Tyndale R, Payne J, Gerber A, Sipe J. The fatty acid amide hydrolase C385A (P129T) missense variant in cannabis users: studies of drug use and dependence in Caucasians. Am J Med Genet. 2007;144B:660–666. doi: 10.1002/ajmg.b.30491. [DOI] [PubMed] [Google Scholar]
- Young SE, Corley RP, Stallings MC, Rhee SH, Crowley TJ, Hewitt JK. Substance use, abuse and dependence in adolescence: prevalence, symptom profiles and correlates. Drug Alcohol Depend. 2002;68(3):309–322. doi: 10.1016/s0376-8716(02)00225-9. [DOI] [PubMed] [Google Scholar]
- Zhang P, Ishiguro H, Ohtski R, Hess J, Carillo F, Walther D, Onaivi ES, Arinami T, Uhl GR. Human cannabinoid receptor 1: 5′ exons, candidate regulatory regions, polymorphisms, haplotypes, and assocation with polysubstance abuse. Molec Psychiatry, 2004;9(10):916–931. doi: 10.1038/sj.mp.4001560. [DOI] [PubMed] [Google Scholar]
- Zheng S, Ma X, Buffler P, Smith M, Wiencke J. Whole genome amplification increases the efficiency and validity of buccal cell genotyping in pediatric populations. Cancer Epidemiol Biomarkers Prev. 2001;10:697–700. [PubMed] [Google Scholar]
- Zuo L, Kranzler H, Luo X, Covault J, Gelernter J. CNR1 variation modulates risk for drug and alcohol dependence. Biol Psychiatry. 2007;62(6):616–626. doi: 10.1016/j.biopsych.2006.12.004. [DOI] [PubMed] [Google Scholar]