Abstract
We recently described a strategy for intracellular delivery of macromolecules, utilizing pH-responsive ‘core-shell’ structured gel particles. These crosslinked hydrogel particles disrupt endosomes with low toxicity by virtue of physical sequestration of an endosome-disrupting ‘proton sponge’ core inside a nontoxic hydrophilic shell. Here we tested the efficacy of this system for cytosolic delivery of a broad range of macromolecular cargos, and demonstrate the delivery of proteins, whole viral particles, or siRNA oligonucleotides into the cytosol of dendritic cells and epithelial cells via core-shell particles. We assessed the functional impact of particle delivery for vaccine applications, and found that cytosolic delivery of protein antigens in dendritic cells via the core-shell particles promotes priming of CD8+ T-cells at 100-fold lower doses than soluble protein. Functional gene knockdown following delivery of siRNA using the particles was demonstrated in epithelial cells. Based on these findings, these materials may be of interest for a broad range of biomedical applications.
Introduction
Synthetic vectors for intracellular delivery of DNA, RNA, or protein drugs are of interest for their low cost, ease of large-scale production, and potential for improved safety relative to approaches such as viral vectors.1–4 To this end, a variety of polymeric systems for cytosolic drug delivery have been developed; prominent among these are pH-sensitive polycations, which disrupt endosomes in response to acidification of these intracellular compartments.4–9 A limitation of many such endosome-escape materials is that there is typically a positive correlation between the efficiency of cytosolic delivery and the degree of toxicity of the delivery agent. In addition, ‘polyplex’ approaches where cationic polymers are complexed with anionic macromolecular cargos are inherently complicated by the fact that the choice of cationic residues simultaneously influences both drug binding and endosomal escape properties of these materials. To address these issues, we recently developed crosslinked core-shell gel particles designed to physically segregate the function of cell/drug binding (mediated by the particle shell) from the function of endolysosomal disruption (mediated by the core).10 Cationic particles with a shell containing primary amines and a core composed of crosslinked poly(diethylaminoethyl methacrylate) (PDEAEMA) were shown to elicit highly efficient endolysosomal disruption via the ‘proton sponge’ effect while exhibiting minimal cytotoxicity.10
We are particularly interested in the application of such cytosolic delivery particles for the delivery of vaccine antigens and therapeutics to dendritic cells, the key antigen presenting cells involved in initiating primary adaptive immune responses.11 Delivery of protein to the cytosol of DCs permits loading of peptide fragments of these antigens onto class I MHC molecules for presentation to CD8+ T-cells. Such ‘cross presentation’ of exogenous antigen may be critical for the development of vaccines against HIV and cancer.12–14 In our initial work, we demonstrated that proteins with net negative charge could be adsorbed to the cationic shell of pH-responsive core-shell particles, allowing cross-presentation to antigen-specific CD8+ T-cells.
Building on these initial studies, we explored here in detail the efficiency and limitations of this electrostatic cargo-loading strategy for loading of several different classes of drug cargos on core-shell particles (CSPs). For protein delivery, we quantified the dose response of antigen cross-presentation elicited by the model protein antigen ovalbumin (ova) taken up by DCs in soluble form vs. bound to core-shell particles, and found that ova was presented at least ∼100-fold more efficiently than soluble protein antigen. To test the delivery of more complex antigens, we demonstrated the cytosolic delivery of whole inactivated influenza virus particles adsorbed to CSPs. Finally, electrostatic association of double-stranded RNA oligonucleotides with core-shell particles was demonstrated to promote cytosolic delivery of siRNA and gene knockdown in an epithelial cell line in vitro. Altogether, the data show that cytosolic delivery of a variety of macromolecular cargos is readily achieved via the simple approach of cargo adsorption to the shell layer of CSPs, providing a simple and robust strategy for drug binding and subsequent intracellular delivery.
Materials and Methods
Materials
All reagents were used as received without further purification. 2-diethylamino ethyl methacrylate (DEAEMA, 99%), methyl methacrylate (MMA, 99%), 2-aminoethyl methacrylate hydrochloride (AEMA, 90%), calcein, and ammonium peroxodisulfate (APS) were purchased from Sigma-Aldrich Chemical Co. Poly(ethylene glycol) dimethacrylate (PEGDMA, MWPEO=200 g/mol, cat # 00096–100, CAS # 25852–47–5) was purchased from Polysciences Inc. Cy5 mono-NHS ester was purchased from GE Healthcare UK Limited.
RPMI 1640, DMEM (with 4.5 g/L glucose), and Trypsin/EDTA (0.25% trypsin/2.21 mM EDTA in HBSS without sodium bicarbonate, calcium and magnesium) were purchased from Mediatech Inc. Fetal bovine serum (FBS) was purchased from Hyclone. Alexa Fluor® 488-conjugated ovalbumin (ova) was purchased from Invitrogen. Ovalbumin (chromatographically purified) was from Worthington Biomedical Corporation. Recombinant murine granulocyte macrophage colony stimulating factor (GM-CSF) was from Peprotech, Inc. Influenza A Antigen Strain Texas from embryonated chicken eggs (strain: Texas 1/77 (H3N2)) was purchased from Meridian Life Science, Inc. Sodium bicarbonate (pH 9.0, 1.0 M), Alexa Fluor-488 succinimidyl ester, and dimethyl sulfoxide (DMSO) were purchased from Molecular Probes. Recombinant human Interleukin 4 (rhIL-4), and recombinant human granulocyte macrophage colony stimulating factor (rhGM-CSF) were purchased from R&D Systems, Inc. Cyclophilin B siRNA, fluorophore-conjugated anti-GFP siRNA, DNA primers, and DharmaFECT1® transfection reagent were purchased from Dharmacon. TaqMan® Universal PCR Master Mix, TaqMan® Transcription Reagents, TagMan® Gene Expression Assays, and TagMan® primer-probes were from Applied Biosystems.
Core-shell particle synthesis and characterization
Core-shell gel particles with PDEAEMA or PMMA cores and AEMA-rich shells were synthesized as previously described.10 Briefly, 5 mmol DEAEMA or MMA was dispersed with 0.03 mmol PEGDMA in deionized water (9 mL) and polymerized at 70°C by addition of 0.01 g APS. The emulsion polymerization was allowed to proceed at 70°C for 3 hrs to grow the particle core, followed by injection of 0.24 mmol AEMA to grow the particle shells for an additional 1.5 hrs. The particles were purified as described10 and stored in PBS at 4°C. Two different batches of particles were prepared (batch 1 and 2) which had slightly different mean sizes. Batch 1 was used in the swelling characterization measurements (Table 1) and siRNA delivery studies; batch 2 was used in all other studies. These particles differed in mean size but behaved identically in our intracellular delivery assays. Hydrodynamic sizes of particles and zeta potentials were measured for dilute particle suspensions in pH 7.4 100 mM PBS and 5 mM NaCl aq. solutions, respectively, using a Brookhaven 90Plus light scattering instrument. SEM images of particles were obtained by drying particles from PBS aq. suspensions onto a substrate, coating the dried particles with 15 nm of Au to using an ion beam sputter coater (Gatan, Pleasanton, CA), and imaging the samples using an FEI/Philips XL30 FEG ESEM with 5 kV accelerating voltage.
Table 1.
Core-shell particle swelling behavior and surface charge
| Particles | Hydrodynamic diameter* (pH 7.4, 37°C) | ζ-potential (pH 7.4, 20°C) | swelling ratio** (pH 7.4, 37°C) | swelling ratio** (pH 5.5, 37°C) |
|---|---|---|---|---|
|
PDEAEMA (PDEAEMA-core/AEMA-shell) batch 1 |
208±4 nm | 37±1.8 mV | 3.7±1.6 | 23±1.9 |
|
PMMA (PMMA-core/AEMA-shell) |
310±10 nm | n.d. | 4.6±0.2 | 4.6±0.6 |
determined by dynamic light scattering
swelling ratio = hydrated mass/dry mass
Intracellular ova protein delivery and OT-I T-cell priming assay
Murine dendritic cell culture and OT-I T-cell isolation
Animals were cared for following institute, state, and federal guidelines under an IUCAC-approved protocol. DCs and T-cells were cultured (1×106 cells/mL/well in 24-well plate) in RPMI 1640 complete medium (10% FBS, 50 µM 2-mercaptoethanol, 5 mM L-glutamine, 10 mM HEPES, and penicillin/streptomycin). Bone marrow-derived dendritic cells (BMDCs) were prepared from the femur and tibias of C57Bl/6 female mice (Jackson Laboratory) following a procedure modified from Inaba et al.15 as previously described.16 DC2.4 cells, a dendritic cell clone originally derived by Shen et al.,17 were a gift from Prof. Kenneth Rock. Naïve CD8+ ova-specific T-cells were harvested from the spleens of OT-I TCR transgenic mice18, 19 (Jackson Laboratory) using a magnetic bead isolation kit (CD8+ T-cell negative selection kit, Miltenyi Biotec, Inc.) according to the manufacturer’s instructions.
Analysis of endosome disruption by confocal microscopy
DC2.4 cells (1.2×105 cells/well) were plated in Lab-Tek™ chambered coverglasses (Nunc) for 18 hrs. Calcein (150µg/mL, 0.24mM) was added to the cells in complete medium (RPMI 1640 with 10% fetal bovine serum (FBS)) for 1 hr at 37°C. The cells were washed with warm medium and then incubated for a second hour with complete medium alone, PDEAEMA-core particles (25 µg/mL), or calcein and CSPs at 37°C. After incubation with dye/CSPs, cells were washed 3X with medium to remove extracellular calcein/particles and imaged live by confocal laser scanning microscopy (CLSM) at 37°C. CLSM was performed on a Zeiss LSM 510 using 40X, 63X, or 100X lenses; the microscope stage was enclosed by an environmental chamber maintaining 37°C during imaging.
Binding of ovalbumin to core-shell particles
Alexa Fluor® 488-conjugated ova (concentration as specified in the text) was mixed with PMMA or PDEAEMA core-shell particles for 5 min in 200 µL of serum-free RPMI medium to allow electrostatic binding of ova to the particles (CSPs). (Kinetics of binding were measured by incubating for times of 5–60 min). Unbound ova was removed by centrifugation of the particles at 15,000×g for 15 min, and the concentration of the ova remaining in the supernatant was determined from fluorescence measurements (exc. 488 nm/em. 530 nm) using a fluorescence microplate reader (SPECTRAmax™ GEMINI, Molecular Devices Corp.), calibrated to serial dilutions of a known ova standard. Ova bound to the particles was determined by subtracting the quantity of protein detected in the supernatant from the quantity measured in control vials containing protein solution but no particles. For cell delivery experiments, ova-loaded particles were resuspended in complete medium by bath sonication for 15 min. Sonication was employed to ensure complete dispersal of the pelleted protein-coated particles before addition to cells.
OT-I T-Cell Priming Assay
Freshly dissolved ova (0.1, 10, or 100 µg/mL) was mixed with 25 µg/mL PMMA or PDEAEMA core-shell particles for 5 min at 20°C to allow electrostatic binding of ova to particles as described above followed by washing of the particles to remove unbound protein. Soluble ova (0.1, 10 or 100 µg/mL) or ova-coated particles were then added to day 6 BMDCs (1×106 cells/well in 24-well plates) for 1 hour at 37 °C in complete medium. The ova- or CSP-pulsed BMDCs were washed 2X with complete medium, then replated in 96-well round-bottom plates in triplicate (1×105 cells/well/100 µL) for 3 hrs. OT-I CD8+ T cells were added to each well (5×105 cells/well/100 µL), and co-incubated with BMDCs for 3 days in complete medium. Interferon gamma (IFN-γ) in the supernatants of these cultures was then quantified using an ELISA kit (murine Interferon gamma (IFN-γ) Duoset ELISA Development Kit, R&D Systems) according to the manufacturer’s instructions.
Cytosolic delivery of Influenza A viral particles in human dendritic cells
Fluorescent labeling of Influenza A
To label Influenza A with fluorescent dye,20 50 µL of 1.0 M sodium bicarbonate (pH 9.0) was added to 500 µL (∼2 ×104 HA units) virus. Alexa Fluor® 488 succinimidyl ester (0.005 µg/HA unit) was dissolved in 2 µL DMSO and added to the virus solution. After stirring for 1 hr at 20°C in the dark, the labeled Influenza A was dialyzed (Slide-A-Lyzer® Mini Dialysis Units 3500 MWCO, Pierce) in PBS (1.0 M, pH7.4) at 4 °C overnight in the dark. The volume change was recorded to calculate the final concentration of the labeled virus.
Human monocyte-derived dendritic cell culture
Peripheral blood mononuclear cells (PBMCs) were isolated from fresh buffy coats of anonymous healthy volunteers (Research Blood Components, LLC) via centrifugation over a Ficoll-Paque® (GE Healthcare Ltd.) cushion. Monocytes were isolated from the PBMCs using MACS beads (CD14 monocytes-positive selection kit, Miltenyi Biotec, Inc.) according to the manufacturer’s instructions. The monocytes were suspended in RPMI 1640 complete medium containing human IL-4 (1,000 U/ml, 2.9× 04 U/ug) and GM-CSF (1,000 U/ml, 1.5×104 U/ug) in 24-well plates at 1×106 cells/well/mL. On day 2 and day 4, 80% of the medium was replaced by fresh medium containing IL-4 and GM-CSF to derive monocyte-derived dendritic cells (MDDCs). MDDCs were used on day 6–7.
Cytosolic delivery of influenza A
On Day 6, MDDCs were plated in Lab-Tek™ chambers (Nunc 8-well chambered coverglasses, 1.2×105 cells/well) and cultured in complete RPMI 1640 medium for 18 hr. Alexa Fluor® 488-labeled Influenza A (1657 HA U/mL) was mixed with 25 µg/mL PDEAEMA core-shell particles for 5 min in serum-free medium to allow electrostatic binding of Influenza A to particles. Unbound Influenza A was removed by centrifugation at 15,000×g for 15 min. Influenza A-decorated particles were resuspended in complete medium by sonicating for 15 min and added to cells at 25 µg/mL CSPs (corresponding to 1000 U/mL HA, based on fluorescence measurements quantifying the fraction of bound virus) for 1 hr at 37 °C; control cells were incubated with 1000 U/mL free influenza. After 3 washes with complete medium to remove extracellular Influenza A and particles, the cells were imaged live by Confocal Laser Scanning Microscope (CLSM) at 37 °C.
Cytosolic Delivery of siRNA
Epithelial cell culture
The BSC-40 cell epithelial cell line was purchased from American Type Culture Collection (ATCC). The cells were maintained in complete DMEM, supplemented with 10% heat-inactivated FBS, 100 IU/mL of penicillin and 100 IU/mL of streptomycin, and 2 mM L-glutamine.
Cytosolic delivery of siRNA
Fluorophore-labeled anti-luciferase siRNA ((sense) 5’-GUGCGCUGCUGGUGCCAACUU/36-FAM/-3' and (antisense) 3’-UUCACGCGACCACGGUUG-5') was used as a model oligonucleotide to measure the kinetics and efficiency of siRNA binding to PDEAEMA-core particles, following the same procedure described above for measuring fluorescent ova binding to the core-shell particles. Fluorophore-tagged anti-GFP siRNA ((sense) 5’-GGCUACGUCCAGGAGCGCAdTdT-3’ and (antisense) 3’-dTdTCCGAUGCAGGUCCUCGCGU-5’), an siRNA sequence targeting green fluorescence protein (GFP) mRNA, was used as a model oligo for monitoring cytosolic delivery of siRNA. The oligos were labeled with Cy5 at the 5’ end of the sense strand and/or Cy3 conjugated to the 3’ of the antisense strand. DC2.4 or BSC-40 cells were plated in Lab-Tek™ (Nunc) chambers (1.2×105 cells/ well) and cultured in RPMI1640 or DMEM complete medium, respectively. Labeled siRNA (2.6 µg/mL, 200 nM) was premixed with 25 µg/mL PDEAEMA CSPs in serum-free medium for 5 min to allow electrostatic binding of siRNA to particles. siRNA-decorated particles were then centrifuged and resuspended in complete medium by sonicating for 15 min and added to DC2.4 or BSC-40 cells for 1 hr at 37 °C. After three washes with complete medium to remove extracellular siRNA and particles, the cells were imaged live by CLSM at 37 °C.
Transfection of Epithelial Cells by siRNA
Cyclophilin B siRNA (Human/Mouse/Rat) was used as a model target for siRNA-mediated gene silencing due to its ubiquitous expression and non-essential role in cells. The siRNA sequences were: (sense) 5’-GGAAAGACUGUUCCAAAAAdTdT-3’ and (antisense) 5’-dTdTCCUUUCUGACAAGGUUUUU-5’. BSC-40 cells were plated in 6-well plate (2×105 cells/well) and cultured in DMEM complete medium overnight (∼ 12 hrs to reach approximately 70% confluence). Cyclophilin B siRNA (2.6 µg/well, 2 mL/well at 100 nM) was premixed with 25 µg/mL PDEAEMA core-shell particles for 5 min to allow electrostatic binding of siRNA to particles. The siRNA-decorated particles were centrifuged to remove unbound siRNA and resuspended in complete medium (2 mL/sample) by sonicating for 15 min. As a comparative positive control, DharmaFECT1® , a commercial cationic lipid-based transfection reagent was used to deliver the same amount of siRNA following the manufacturer’s suggested protocol: siRNA (2.6 µg/well, 100 nM) and DharmaFECT1® transfection reagent (4 µL/well) were separately diluted in 200 µL/well Opti-MEM® I (Invitrogen) medium, and incubated for 5 min at 20°C. The two solutions were mixed gently by pipetting, and incubated for 20 min at 20°C. 1600 µL/well of DMEM complete medium was added the to solution for the final desired volume of transfection medium (2 mL/sample). siRNA-coated particles or siRNA lipoplexes were then added to the cells at 37°C for 1 hr or 4 hrs, respectively. The cells were washed with PBS 3X, and then cultured in DMEM complete medium. Twenty-four hrs after the initial addition of transfection agents, the cells were collected by trypsinization and total cellular RNA was extracted using a RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The recovered RNA concentration was determined by UV absorbance at 260 nm and all samples had an A260/A280 ratio greater than 1.95. Functional siRNA delivery was measured by quantifying target mRNA levels using a two-step Real-Time Polymerase Chain Reaction (RT-PCR). Extracted total cellular RNA was converted into cDNA using random hexamers and TaqMan® reverse transcription reagents. The levels of cyclophilin B and actin mRNA (as a non-target control gene) were measured using a TaqMan® gene expression assay. Real-time PCR was carried out using 10 ng cDNA per sample on a PRISM 7700 sequence detection system (Perkin-Elmer Applied Biosystems). The target mRNA concentration was evaluated by the comparative Ct method.
Results and Discussion
Protein cargo loading on endosome-escaping core-shell particles via electrostatic adsorption on the shell
We recently reported the synthesis of core-shell particles (CSPs) via emulsion polymerization, comprised of a crosslinked poly(diethylaminoethyl methacrylate) (PDEAEMA) pH-responsive core and an aminoethyl methacrylate (AEMA)-rich shell.10 We demonstrated that upon internalization by cells, these particles rupture their confining endolysosomes in response to acidification of these compartments. In addition, we showed that adsorption of the negatively charged model protein antigen ovalbumin (ova) to the shell of these particles (Figure 1) followed by incubation of the particles with dendritic cells (DCs) allowed cytosolic delivery of the protein. Cytosolic delivery facilitated cross-presentation of peptides derived from ova and enhanced ova-specific CD8+ T-cell activation by these particle-loaded DCs (which we demonstrated for a single test dose of antigen).10
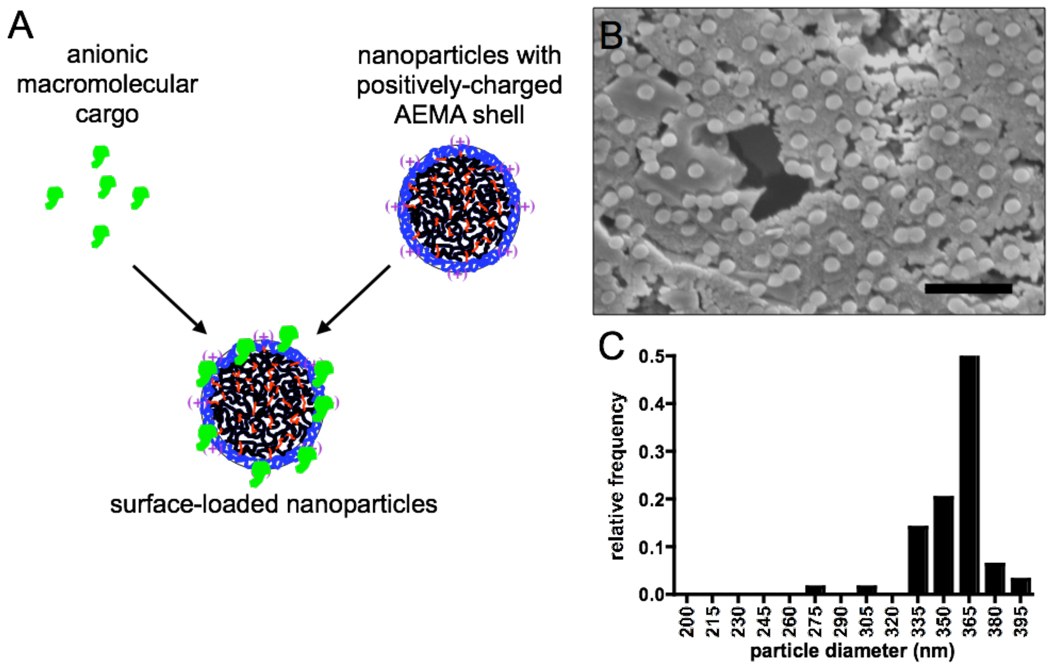
Figure 1. Core-shell particle surface loading and particle characterization.
(A) Schematic of drug cargo association with cationic core-shell PDEAEMA particles. (B, C) SEM image (B) and size histogram (C) of PDEAEMA-core particles (batch 2). Scale bar in B is 2 µm.
Building on these promising initial results, we sought to determine the efficiency of protein binding to core-shell particles via electrostatic association and explore in detail the mechanisms underlying enhanced T-cell priming elicited by CSPs, again using ova as a model protein antigen (46 KDa globular protein, pI 4.621). Monodisperse core-shell particles were synthesized by surfactant-free emulsion polymerization as previously described.10 Two particle types were prepared (Table 1): pH-responsive CSPs with a crosslinked PDEAEMA core/AEMA-rich shell, and control particles with a poly(methyl methacrylate) (PMMA) core/AEMA-rich shell. Both particle types have a zwitterionic surface due to the presence of both AEMA groups and sulfate groups from the APS free radical initiator in the shell layer.10, 22 Two batches of the pH-sensitive PDEAEMA-core particles prepared in the same manner were used in these studies: batch 1 was used in pH-response/swelling measurements (Table 1) and the siRNA delivery studies described below; batch 2 was used in all other experiments reported here. As shown in the DLS data of Table 1 and the SEM images and sizing data of Figure 1B and C, the particle batches differed in mean diameter (batch 2 particles were larger than batch 1) but both were relatively uniform in size, and we saw no differences in their endosome escape properties in our studies. Note that the diameters determined from SEM images of batch 2 particles (Figure 1C) are overestimates as the SEM samples were coated with gold for imaging, adding ∼30 nm to the diameter observed. As we previously reported, these particles exhibit a large change in swelling ratio as the pH of the medium drops from extracellular pH to endolysosomal pH (Table 1); the PMMA-core control particles exhibited no pH sensitivity.
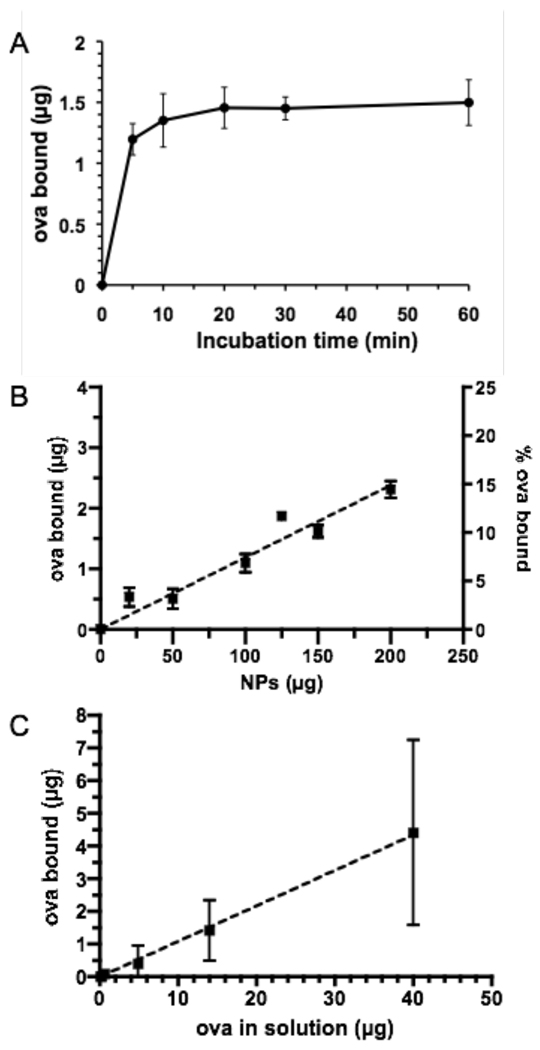
To assess the efficiency of electrostatic adsorption for loading of ova on the particle surfaces, we first quantified protein binding to the cationic particles as a function of time, protein concentration, and particle concentration. Soluble fluorophore-labeled ova was mixed with PDEAEMA core-shell particles in serum-free medium to allow protein binding, followed by removal of unbound ova by centrifugation, and the quantity of protein bound was assessed by fluorescence measurements on the particle supernatant. We found that in serum-free medium, ova binding equilibrated rapidly (binding already beginning to plateau by ∼5 min), as illustrated in Figure 2A for the case of 150 µg particles incubated with 80 µg/mL ova in 200 µL serum-free medium. We thus fixed the incubation time for protein binding at 5 min for subsequent experiments. We next measured binding of fixed concentrations of ova (80 µg/mL) to varying concentrations of particles (up to 1 mg/mL) following 5 min incubations. As shown in Figure 2B, the total amount of ova bound was approximately linear with particle concentration with ∼15% of the total ova bound to particles when 1 mg/mL CSPs were added. A linear regression of the data (dashed line) showed 12 µg ova bound per mg CSPs for this fixed concentration of soluble ova. When we instead fixed the concentration of particles at 625 µg/mL (125 µg in 200 µL medium) and varied the amount of ova added (Figure 2C), binding was again linear with ∼11% of ova added binding to the particles across a broad range of ova concentrations. The linear binding relationships suggest that ova loading is not saturated under these conditions, but maximum protein binding here (∼12 µg protein/mg CSPs) was comparable to ova loading typically achieved by protein encapsulation in biodegradable micro/particles23–26 and also compares well to other studies utilizing surface loading onto charged drug delivery particles.27 Notably, colloidal stability of the particles did not appear to be affected over this range of protein loading (no gross particle aggregation or sedimentation was observed). Thus, simple electrostatic adsorption allowed for rapid loading of the particles with substantial quantities of protein cargo.
Figure 2. Ovalbumin adsorption to core-shell particles at pH 7.4.
(A) Kinetics of ova adsorption to PDEAEMA core-shell particles for 150 µg particles incubated with 80 µg/mL ova in 200 µL serum-free RPMI medium. (B) Particles at varying concentration were added to ova at a fixed concentration of 80 µg/mL for binding to particles for 5 min in 200 µL serum-free RPMI medium, followed by separation of particles by centrifugation. (C) Soluble ova at varying concentrations was mixed with 125 µg PDEAEMA core-shell particles in serum-free medium as in (B). Dashed lines in (B) and (C) are linear regressions to the data. Shown are the mean ± S.D. from triplicate samples.
Efficiency of cross-priming elicited by particle surface-bound antigen
Protein antigens in the cytosol of dendritic cells (DCs) are processed by the ‘classical’ MHC class I presentation pathway, and peptides derived from these antigens are presented by class I MHC at the cell surface to CD8+ T-cells.28 In contrast, exogenous soluble antigens trapped in endolysosomes following internalization are only very inefficiently presented on class I MHC (a process termed cross-presentation).13, 29–31 Thus, a promising application of these core-shell endosome-escaping particles is delivery of protein antigens into the cytosol of dendritic cells, amplifying presentation of antigenic peptides to CD8+ T-cells for in vitro screening of patient T-cell specificities and/or as vaccine delivery agents. Having established a dose curve delineating the amount of ova bound to particles as a function of protein concentration during particle loading, we next assessed the dose response for cross-presentation of ova-derived peptides to antigen-specific CD8+ T-cells, comparing DCs loaded with soluble ova, ova bound to PDEAEMA-core CSPs, or ova bound to PMMA-core CSPs. For soluble antigen pulsing, murine bone marrow-derived dendritic cells were incubated with different concentrations of ova in solution. For particle-mediated ova delivery, the same concentrations of ova were incubated for 5 min with 25 µg/mL PDEAEMA-core or PMMA-core particles for surface binding, followed by pelleting of the particles, resuspension in fresh medium, and addition of the ova-loaded particles to DCs. For all of the cell experiments described here, we fixed the final concentration of particles added to cells at 25 µg/mL, a dose determined in our prior studies to provide maximal endosome disruption in cells with minimal cytotoxicity following a 1 hr incubation of particles with cells.10 Soluble antigen- or ova/CSP-loaded DCs were washed into fresh medium after 1 hr, allowed to process internalized antigen for 3 hrs, and then co-cultured with naïve OT-1 CD8+ T cells (TCR-transgenic T-cells that recognize a peptide derived from ova presented in the context of H-2Kb class I MHC molecules).18 T-cell activation by ova-loaded DCs was detected on day 3 of the co-culture by measuring interferon-γ (IFN-γ), a cytokine secreted by activated T-cells.
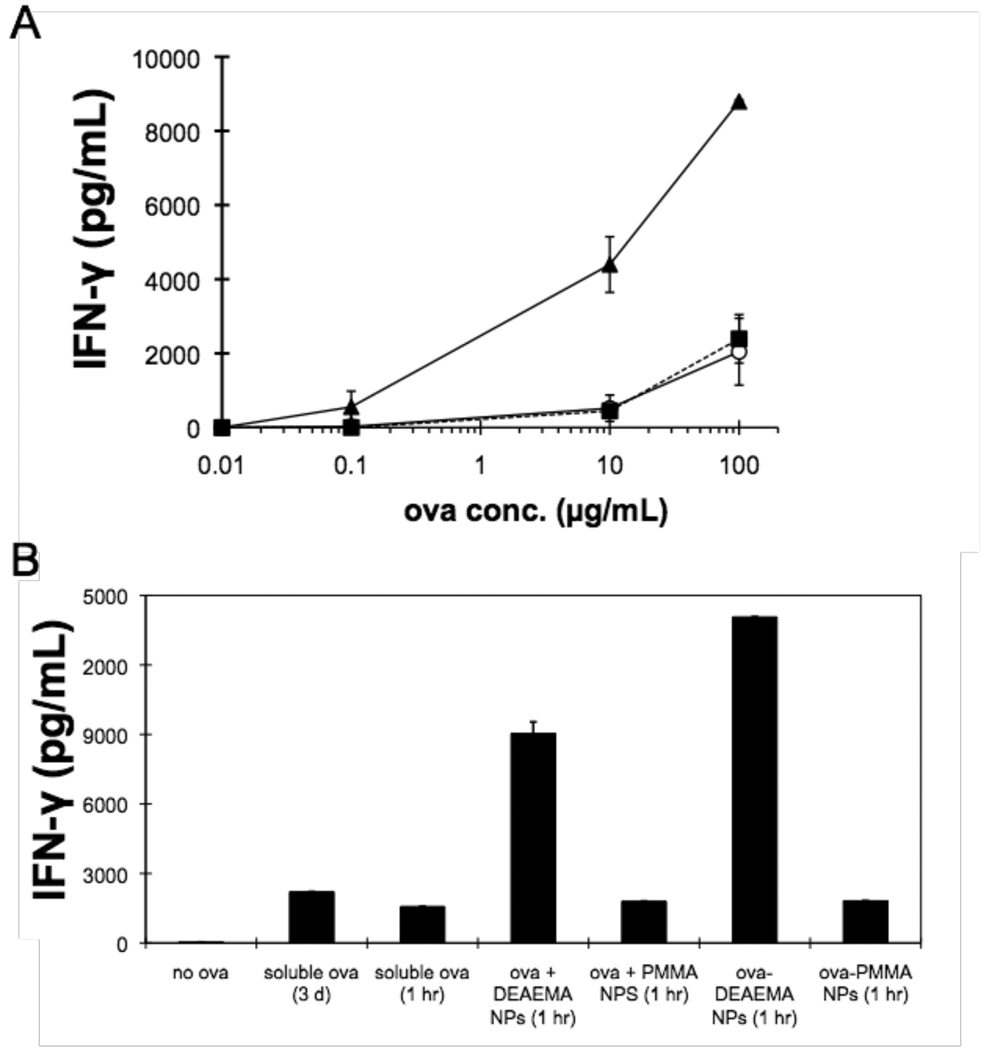
As shown in Figure 3A, DCs incubated with soluble ova protein only triggered IFN-γ secretion from T-cells above background levels when the ova concentration reached 100 µg/mL, consistent with prior data from our laboratory and others.10, 19, 32, 33 Particle-mediated delivery of ova reduced the dose of antigen required to elicit T-cell activation by at least ∼100-fold compared to soluble protein uptake by DCs, and DCs loaded with ova bound to pH-responsive CSPs elicited much higher IFN-γ levels (∼9–10 fold) from T-cells at any given ova dose. Particles alone at 25 µg/mL elicited no background T-cell activation (as illustrated by lack of response for 0.01 µg/mL ova dose in Figure 3A). Note that we have chosen to conservatively plot the ova dose for antigen bound to particles in terms of the concentration of ova mixed with particles during binding, because the amount of protein bound to the particles was too small to directly measure accurately. As shown in Figure 2, the amount of protein that actually binds to the particles during the particle loading step is likely ∼10% of the added protein or less, and thus the potency of PDEAEMA-core CSP delivery may be substantially greater than 100-fold increased relative to soluble ova. In contrast, DCs pulsed with ova bound to pH-insensitive PMMA-core CSPs elicited low levels of IFN-γ similar to the soluble ova control, confirming that endosomal escape triggered by the pH-responsive DEAEMA core was required for enhanced antigen presentation.
Figure 3. Cytosolic delivery of ova for OT-I T cell priming.
(A) BMDCs were incubated with different concentrations of soluble ova (○), ova-coated PDEAEMA core-shell particles (▲), or ova-coated PMMA core-shell particles (◾) in complete medium containing 10% FBS, before being washed and mixed with naïve OT-1 ova-specific CD8+ T cells. The concentration of particles was fixed at 25 µg/mL for all ova doses. IFN-γ secreted by the T-cells in response to antigen presentation by the DCs was measured by ELISA after 3 days. Shown are mean ± S.D. of triplicate samples. (B) BMDCs were pulsed with ova in different forms: soluble ova (10 µg/mL) for 1 hr or during an entire 3 day co-culture, ova mixed with PDEAEMA or PMMA core-shell particles (without pre-binding to particles, ova + CSPs), or ova adsorbed to PDEAEMA or PMMA core-shell particles prior to addition to DCs (ova-CSPs) in complete medium containing 10% FBS. DCs were washed following antigen pulsing and then co-cultured with OT-I T-cells. IFN-γ secreted by T-cells in response to antigen presentation by the DCs was measured by ELISA after 3 days. Shown are mean ± S.D. of triplicate samples.
In these dose response experiments, ova was incubated with DCs for only 1 hr in soluble or particle-bound form. Because ova bound to particles was internalized by DCs much faster than soluble ova (DJI and YH, unpublished observations), we next tested whether the enhanced antigen presentation achieved by PDEAEMA particle delivery of ova was simply a kinetic effect of more rapid ova uptake. One set of DCs was pulsed with ova for only 1 hr (10 µg/mL “soluble ova (1 hr)”), washed, and then co-cultured with T-cells as before. A second group of DCs was incubated with soluble ova (10 µg/mL) without washing during the entire 3-day T-cell stimulation period, to allow continuous antigen uptake. These two conditions were compared to DCs loaded with ova bound to PDEAEMA-core CSPs (25 µg/mL particles loaded for 5 min with 10 µg/mL ova, then washed and applied to DCs “ova-DEAEMA CSPs (1 hr)”). As shown in Figure 3B, we observed slightly higher secretion of IFN-γ from co-cultures where soluble ova was present in the medium for 3 days when compared to cultures where DCs were pulsed with soluble antigen for only 1 hr. However, the degree of cross-presentation/T cell stimulation measured was still much lower than that of detected for particle-delivered ova, demonstrating that the enhanced response to particle-bound ova was not simply due to a kinetic difference in antigen uptake/processing.
In addition, we compared the effectiveness of pulsing DCs for 1 hr with ova pre-adsorbed on CSPs (ova-CSPs) vs. simultaneous addition of soluble ova protein and CSPs to DCs without an explicit adsorption step to bind ova to the particles prior to addition to DCs (ova + CSPs). Soluble ova mixed with pH-sensitive particles was presented to T-cells less effectively than ova pre-adsorbed to the core-shell particles, and again, T-cell priming was much greater with the pH-sensitive particles than control PMMA particles lacking the pH-responsive DEAEMA core.
Pulse-chase analysis of endosome disruption
The cross-priming studies with protein antigen delivery above suggested that optimal cytosolic delivery of protein was achieved when the cargo was bound directly to the particles rather than simply being present in the medium with particles during cellular uptake. This result prompted us to examine whether core-shell particles are capable of lysing multiple endosomes in the cell, or whether the particles lyse only the endosome where they are initially confined. Our prior studies led us to the following model of core-shell particle endosome escape: (1) Protons and counterions such as chloride are pumped into the endosomal vesicle to acidify the compartment,34 (2) the DEAEMA groups in the particles absorb protons, becoming ionized, (3) association of counterions with these charged DEAEMA groups in the particle core drive osmotic swelling of the particle, (4) the endosome is disrupted either due to the osmotically-driven uptake of water into the endosome caused by counter ion buildup in the gel particle and/or the concomitant mechanical pressure of particle swelling. (These two responses are inherently linked in the swelling response of these polyelectrolyte gel particles). This mechanism is a classic proton sponge response and distinct from osmotic/membrane-fusion-based lysis of endosomes triggered in elegant work using polymersomes that solubilize or disassemble in response to endosomal pH/reductive capacity,35–37 or degradable polymersomes that undergo a vesicle-to-micelle transition in endosomes.38
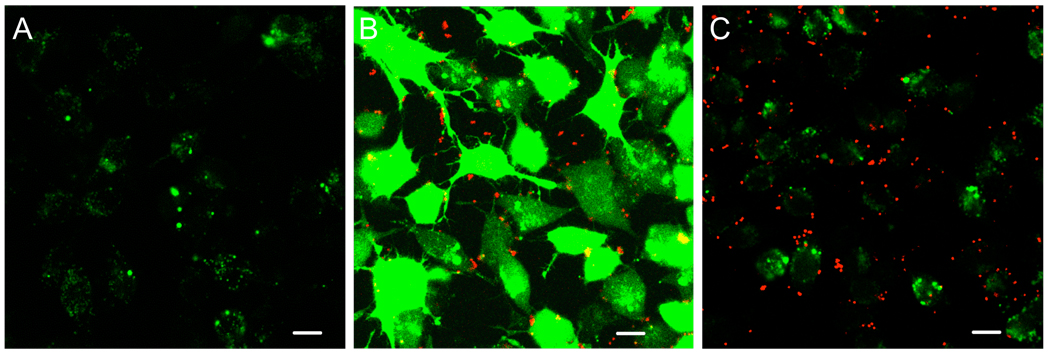
Further evidence for the importance of associating protein cargo with the particles to achieve efficient cytosolic delivery came from pulse-chase experiments with core-shell particles, using the membrane-impermeable fluorescent dye calcein as a tracer of endosome disruption. The DC cell line DC2.4 was incubated for 1 hr with calcein, washed, and then incubated for a second hour with medium alone, calcein mixed with PDEAEMA-core CSPs, or PDEAEMA-core CSPs alone. The cells were then washed and imaged live at 37°C by confocal microscopy. As shown in Figure 4A, when cells were incubated with calcein only for 1 hr, punctate green fluorescence was observed in the cells, corresponding to endosomes containing calcein internalized by the cells. When the cells were co-incubated with calcein and CSPs in the second hour, calcein was observed distributed throughout the cytosol, due to co-internalization of calcein molecules and CSPs into common endosomes, which were then ruptured by the pH-sensitive particles (Figure 4B).10 However, if DCs were serially incubated first with calcein, washed, and then incubated in a second step with PDEAEMA-core CSPs for 1 hr, both calcein and CSPs were observed in the DCs, but calcein remained punctate in endosomes distinct from the particles (Figure 4C). When particles were given to cells first followed by calcein, the results were qualitatively identical to Figure 4C (data not shown). This experiment demonstrated that CSPs are only able to trigger the cytosolic delivery of molecules initially co-localized in the same endosomes as the particles; the CSPs do not disrupt other vesicles in cells following escape from their initial endolysosomal compartment. Thus, maximal cargo delivery to the cytosol would only be expected if the CSPs and cargo are physically associated during uptake, to ensure that all cargo-containing endosomes are disrupted. Note that single confocal optical slices are shown in Figure 4, and these single-plane images visualize only those core-shell particles in the focal plane; generally, we find particles distributed at varying depths through the cells in z-sections. Thus, only a fraction of the total particles in any given cell will be captured in a single focal plane and these images underestimate the total number of particles per cell.
Figure 4. Cargo molecules must be localized in the same endolysosome as core-shell particles for cytosolic delivery to occur.
DC2.4 cells were incubated with calcein alone for 1 hr at 37 °C in medium containing 10% FBS, washed, and then incubated a second hour at 37°C with serum-containing medium alone (A), with calcein and 25 µg/mL PDEAEMA CSPs simultaneously in serum-containing medium (B), or with CSPs alone in serum-containing medium (C). Cells were washed after the second hr and imaged live at 37°C by CLSM. Images show fluorescence overlays of calcein (green) and Cy5-labeled particles (red). Scale bars 20 µm.
Cytosolic delivery of particulate killed pathogen cargo
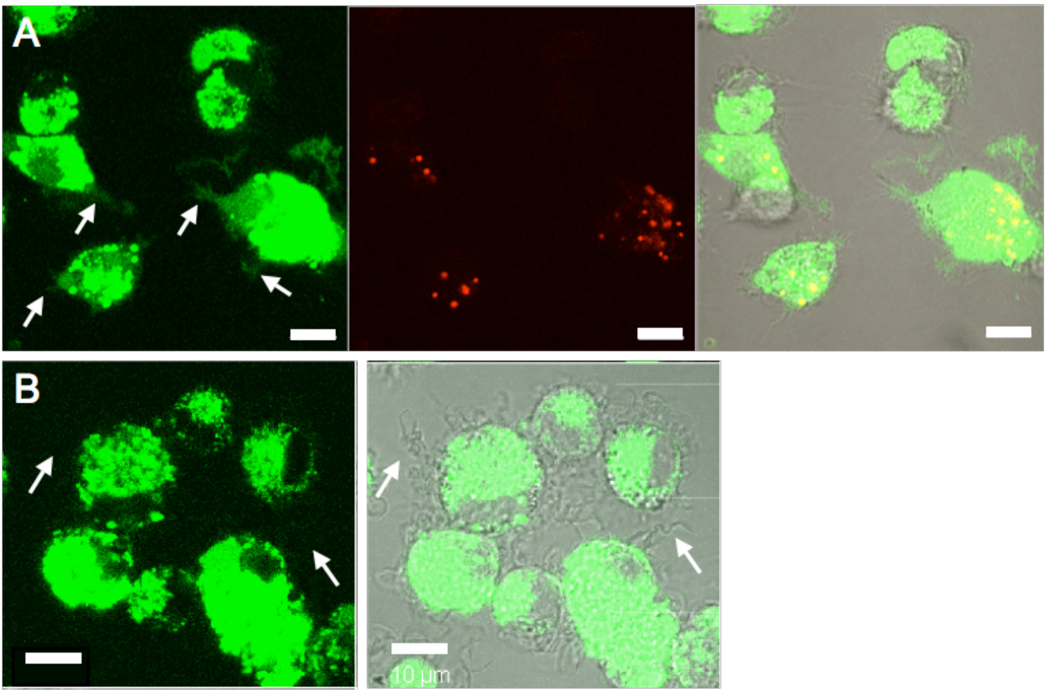
The enhanced cross presentation of protein antigens elicited by these core-shell particles could be exploited for in vitro screening of human immune responses to vaccination, or for direct stimulation of primary immune responses in vivo. However, many vaccines are based on particulate protein mixtures (such as inactivated viral particles39 or whole cell lysates40, 41) rather than single purified proteins. Antigen associated with large particles (500–1000 nm in diameter or greater) is known to be spontaneously cross-presented by dendritic cells following particle phagocytosis, via mechanisms that are still a matter of debate.32, 42–44 Thus, the endosome-disrupting core-shell particles studied here are likely irrelevant for promoting cross-presentation of very large particulate antigen cargos (e.g., whole yeast particle vaccines45, 46). However, antigen associated with particles < 500 nm in size (e.g., the size of typical viral particles) is not cross-presented efficiently (as demonstrated by the data in Figure 3 for PMMA particles that lack endosome escape capabilities).32 To test whether there is a fundamental limit to the size of cargos that can be delivered by the CSPs and to determine whether antigens which are themselves particles in the 100–300 nm size range can be delivered into the cytosol of DCs, we assessed the cytosolic delivery of whole inactivated influenza (H3N2) viral particles. Influenza virions are spherical or rod-like lipid-enveloped particles with mean diameters of ∼100–200 nm.47 For these experiments, we employed human monocyte-derived dendritic cells (MDDCs), to assess the efficacy of core-shell particle delivery in human DCs as opposed to the murine DCs tested previously. Fluorescently-labeled influenza virions were mixed with PDEAEMA particles in serum-free medium to permit adsorption to the CSPs, and the majority of free influenza A was then removed by centrifugation and aspiration of the supernatant. Virus-coated CSPs or virus alone were then resuspended in complete medium and added to MDDCs for 1 hr at 37 °C, followed by washing and confocal imaging of the live cells at 37°C (Figure 5). High levels of intracellular fluorescence were observed for DCs incubated with free influenza or virions bound to particles. High background internalization of flu particles alone was expected, due to binding of DC pattern recognition and scavenger receptors to the viral particles as part of the normal process of virus recognition.48, 49 Notably however, fluorescence of virions delivered by PDEAEMA-core CSPs was detected extending throughout the cytosol, including into the dendrites of DCs examined by 3D optical sectioning (arrows in Figure 5A), providing evidence for cytoplasmic delivery, while fluorescence was not detected in dendrites of DCs incubated with free virions (e.g., arrows in Figure 5B). Instead, virion fluorescence was confined to a dense punctate distribution in the cell body, suggestive of endolysosomal compartmentalization of the free viral particles. These results suggest that the pH-responsive core-shell particles are capable of promoting cytoplasmic delivery of complex biological cargos such as particulate pathogens, which are much larger than individual proteins. Whether the virions are intact following endosomal escape cannot be resolved by confocal microscopy, but for the purposes of antigen delivery, intact viral particles are not necessarily needed since the virions must be proteolyzed by the cytosolic antigen processing machinery for antigen presentation to occur.
Figure 5. Cytosolic delivery of influenza A virions in MDDCs.
Human monocyte-derived DCs were incubated for 1 hr in complete medium containing 10% FBS with inactivated whole influenza virions adsorbed to CSPs (A) or flu virions alone (B), washed, and imaged live at 37°C by CLSM. Shown are fluorescence images of labeled influenza A (green), particles (red), and brightfield/fluorescence overlays. White arrows in A highlight flu fluorescence accumulated in dendrites of DCs that have internalized particles, while arrows in B illustrate a corresponding lack of fluorescence signature in the dendrites of DCs internalizing flu in the absence of CSPs. Scale bars 10µm.
Electrostatic shell loading and intracellular delivery of siRNA
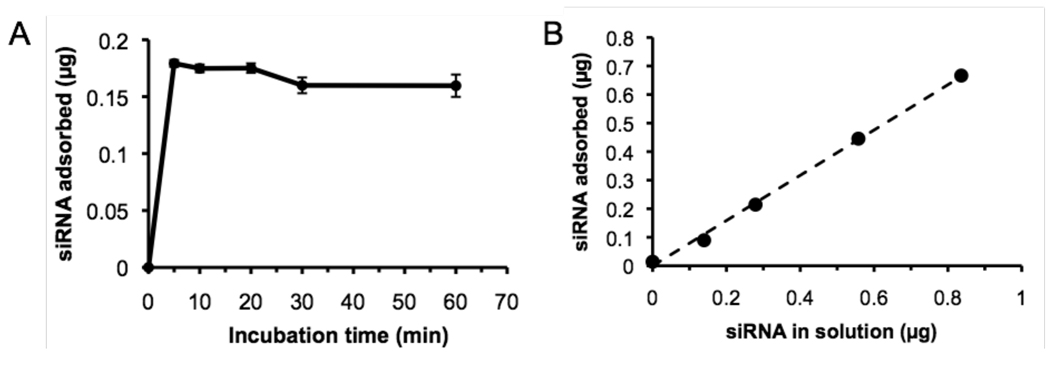
In addition to protein/antigen delivery for vaccine purposes, cytosolic delivery of RNA oligonucleotides is of significant interest, for gene knockdown by RNA interference or to stimulate pro-inflammatory interferon responses in immunotherapy treatments.50–53 Thus, in a final series of experiments we tested whether electrostatic binding of double-stranded RNA oligos could be used to promote cytosolic delivery of siRNA for gene silencing. The kinetics and efficiency of siRNA binding to particles were first tested with a model fluorescently-tagged 21-nucleotide dsRNA. Similar to the adsorption of ova to core-shell particles, siRNA binding to particles in serum-free medium was very rapid, plateauing within 5 min (Figure 6A). In a second set of experiments, a fixed quantity of particles (150 µg) was incubated with increasing concentrations of siRNA and the oligo binding was quantified. Similar to the ova protein binding data of Figure 2C, binding was approximately linear vs. siRNA dose and did not saturate over the concentration range tested (Figure 6B). At the highest siRNA concentration tested (300 nM), ∼80% of the added siRNA was adsorbed to the particles, giving ∼4.4 µg siRNA bound per mg CSPs.
Figure 6. Kinetics and efficiency of siRNA binding to pH-sensitive core-shell particles.
(A) PDEAEMA-core particles (150 µg) were incubated with 200 nM fluorescently-labeled siRNA in 200 µL serum-free RPMI medium, then separated by centrifugation and siRNA binding was quantified by measuring fluorescence remaining in the particle supernatants. (B) Soluble siRNA at varying concentrations was mixed with 150 µg PDEAEMA core-shell particles in 200 µL serum-free medium for 5 min followed by separation of particles from free siRNA by centrifugation. Dashed line is a linear regression to the data. Shown are the mean ± S.D. from triplicate samples; error bars in B are smaller than the point sizes.
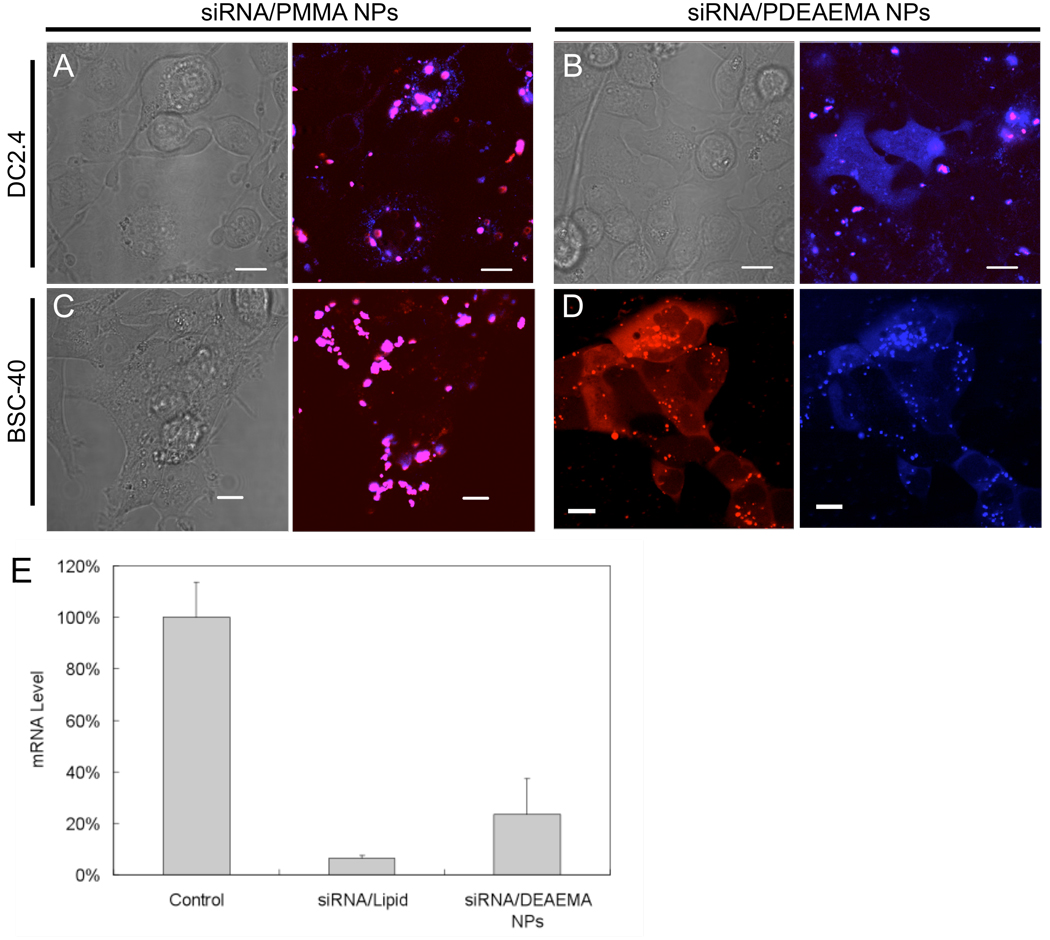
We employed DC2.4 cells and an epithelial cell BSC-40 (for comparison with dendritic cell behavior), to determine whether PDEAEMA particles could deliver siRNA to the cell cytosol efficiently. Fluorescently-labeled siRNA was adsorbed to labeled PDEAEMA or PMMA core-shell particles as before for protein loading, and then the siRNA-loaded particles were added to DC2.4 cells for 1 hour at 37 °C, followed by washing and confocal imaging. Labeled siRNA was colocalized in punctate spots with PMMA-core CSPs in DCs (Figure 7A), but observed throughout the cytosol and nucleus in DC2.4 cells treated with PDEAEMA-core CSPs (Figure 7B). The fraction of cells exhibiting cytosolic siRNA fluorescence when treated with PDEAEMA CSPs was 43.0 ± 2.3% (S.D. of three individual samples, n = 150 cells scored).
Figure 7. pH-sensitive core-shell particles enable cytosolic siRNA delivery and gene knockdown.
(A, B) CLSM images of labeled siRNA (blue) and CSP (red) delivery in DC2.4 cells. DC2.4 cells were incubated for 1 hr in complete medium (10% FBS) with siRNA adsorbed to control PMMA CSPs (A) or pH-responsive DEAEMA CSPs (B), washed, and imaged live at 37°C. Scale bars in A and B 10µm. (C, D) CLSM images of labeled siRNA (blue, Cy5-labeled sense strand; red, Cy3-labled anti-sense strand) and CSP delivery in BSC-40 epithelial cells. Epithelial cells were incubated in serum-containing medium with siRNA adsorbed to PMMA CSPs (C) or DEAMA CSPs (D) for 1 hr, washed, and imaged live at 37°C. Scale bars in C and D 20 µm. (E) RT-PCR analysis of Cyclophilin B mRNA downregulation at 24 hrs following siRNA delivery into BSC-40 cells via DharmaFECT1® transfection reagent (siRNA/Lipid, 4 hrs) or siRNA adsorbed to CSPs (siRNA/DEAEMA CSPs, 1hr), relative to untreated control cells.
We next tested the intracellular delivery of fluorescent siRNA to BSC-40 epithelial cells in a similar protocol. In these experiments, each strand of the double-stranded siRNA was labeled with a different fluor (cy3 or cy5) while the CSPs were left unlabeled, and uptake of siRNA on PMMA or PDEAEMA CSP carriers was again assessed after 1 hr by confocal microscopy. As shown in Figure 7C, fluorescence from the two siRNA strands remained co-localized and displayed a punctuate distribution in cells receiving siRNA adsorbed to PMMA CSPs. However, when BSC-40 cells were incubated with siRNA-coated PDEAEMA core-shell particles (Figure 7D), Cy3 and Cy5 fluorescence was observed throughout the cytosol of ∼40% of cells, indicating intracellular delivery of siRNA. To evaluate whether the observed delivery was effective for gene silencing, real-time PCR (RT-PCR) was used to detect the mRNA level of siRNA-transfected cells. BSC-40 cells were transfected with Cyclophilin B-targeting siRNA electrostatically adsorbed to PDEAEMA particles or delivered using a commercial lipid-based transfection reagent (DharmaFECT1®). Lipoplexes were added to cells for 4 hrs per the manufacturer’s optimal conditions, while siRNA-coated particles were added for 1 hr to cells, then cells were exchanged into fresh medium to minimize toxicity from either transfection agent. Note that the 1 hr time for the particles was chosen based on our prior experience that substantial particle uptake (as illustrated in Figure 4, Figure 5, and Figure 7) occurs within this time while viability remains > 90% for the cells we have tested. Viability of the cells was comparably high in both conditions for this comparison. Twenty-four hrs after the initial addition of siRNA to cells, total cellular RNA was isolated and cyclophilin B mRNA levels were assessed. As shown in Figure 7E, siRNA delivered by the commercially available cationic lipid reduced mRNA levels to 6.3±1.2% of control cell levels, while mRNA expression in cells treated with PDEAEMA CSPs reached 23.3 ± 14.0 % of untreated cells. The slightly lower degree of gene knockdown observed for the CSPs compared to the lipid reagent in this assay may relate to the incomplete cytosolic distribution of siRNA observed by microscopy after 1 hr, though a higher fraction of cells appears to become transfected over time once the cells are washed at 1 hr. We currently believe that this incomplete transfection is due to relatively slow unbinding of siRNA from the CSPs, as the particles escape endosomes extremely efficiently (we previously showed endosome disruption occurs in > 95% cells at these particle doses10) but may bind the electrostatically-bound oligos with high avidity and release them very slowly in the cytosol. Modulating the siRNA release rate via altered shell chemistry is a subject of ongoing research. Nonetheless, the instability of lipoplexes make many lipid-based transfection reagents of limited value for in vivo siRNA delivery, and core-shell particles may provide an approach to overcome this limitation.
Conclusions
We applied pH-sensitive PDEAEMA-core/PAEMA-shell particles as an intracellular drug delivery system, and demonstrated that these particles facilitate cytosolic delivery of a broad range of membrane-impermeable macromolecules such as ova protein, influenza A, or siRNA mediated by simple electrostatic adsorption of these diverse cargos to the charged surfaces of the CSPs. Cytosolic delivery of protein antigens by these materials dramatically lowered the dose of antigen required to elicit naïve CD8+ T-cell priming by dendritic cells, by at least ∼100-fold compared to soluble antigen uptake by DCs. In addition, potent gene knockdown via siRNA delivery by these particles was observed. These particles offer the advantage of allowing cytosolic delivery in the presence of serum (unlike unstable lipoplexes). In addition, the uniform size and crosslinked particle structure allow the particle physical characteristics to be stable and better-defined under varying environmental conditions, unlike dynamic assemblies such as vesicles or physically-bonded polyplexes. However, rational design of cargo release from such core-shell particles once endosomal escape is achieved is an area for future optimization of these materials. The ability of these monodisperse particles to deliver a broad range of macromolecular cargos with low intracellular toxicity may be of interest for in vitro immune response screening and RNA transfection, as well as in vivo gene modulation and vaccine delivery.
Acknowledgments
This work was supported by the Institute of Soldier Nanotechnology, Team 2.2.2, Contract Number DAAD19-02-D-0002, the Human Frontier Science Program, and the National Science Foundation (award 0348259). J.J.L. and J.C. were supported in part by NIH grants CA112967 (to D. Lauffenburger), and CA119349 (to R.L./R. Weissleder). J.J.L. was partly supported by a postdoctoral fellowship from the National Science and Engineering Research Council of Canada. D.J. I. is an investigator of the Howard Hughes Medical Institute.
References
- 1.Juliano R, Alam MR, Dixit V, Kang H. Nuc. Acids Res. 2008;36(12):4158–4171. doi: 10.1093/nar/gkn342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Remaut K, Sanders NN, De Geest BG, Braeckmans K, Demeester J, De Smedt SC. Mater. Sci. & Eng. R-Reports. 2007;58(3–5):117–161. [Google Scholar]
- 3.Ghosh P, Han G, De M, Kim CK, Rotello VM. Adv Drug Deliv Rev. 2008;60(11):1307–1315. doi: 10.1016/j.addr.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Pack DW, Hoffman AS, Pun S, Stayton PS. Nat Rev Drug Discov. 2005;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- 5.Murthy N, Campbell J, Fausto N, Hoffman AS, Stayton PS. Bioconjug Chem. 2003;14(2):412–419. doi: 10.1021/bc020056d. [DOI] [PubMed] [Google Scholar]
- 6.Thomas M, Klibanov AM. Proc Natl Acad Sci U S A. 2002;99(23):14640–14645. doi: 10.1073/pnas.192581499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Little SR, Lynn DM, Ge Q, Anderson DG, Puram SV, Chen J, Eisen HN, Langer R. Proc Natl Acad Sci U S A. 2004;101(26):9534–9539. doi: 10.1073/pnas.0403549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park TG, Jeong JH, Kim SW. Adv Drug Deliv Rev. 2006;58(4):467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Wightman L, Kircheis R, Rossler V, Carotta S, Ruzicka R, Kursa M, Wagner E. J Gene Med. 2001;3(4):362–372. doi: 10.1002/jgm.187. [DOI] [PubMed] [Google Scholar]
- 10.Hu Y, Litwin T, Nagaraja AR, Kwong B, Katz J, Watson N, Irvine DJ. Nano Lett. 2007;7(10):3056–3064. doi: 10.1021/nl071542i. [DOI] [PubMed] [Google Scholar]
- 11.Steinman RM, Banchereau J. Nature. 2007;449(7161):419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- 12.Pardoll DM. Nat Rev Immunol. 2002;2(4):227–238. doi: 10.1038/nri774. [DOI] [PubMed] [Google Scholar]
- 13.Melief CJ. Eur J Immunol. 2003;33(10):2645–2654. doi: 10.1002/eji.200324341. [DOI] [PubMed] [Google Scholar]
- 14.Letvin NL. Nat Rev Immunol. 2006;6(12):930–939. doi: 10.1038/nri1959. [DOI] [PubMed] [Google Scholar]
- 15.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. J Exp Med. 1992;176(6):1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stachowiak AN, Wang Y, Huang YC, Irvine DJ. J Immunol. 2006;177(4):2340–2348. doi: 10.4049/jimmunol.177.4.2340. [DOI] [PubMed] [Google Scholar]
- 17.Shen ZH, Reznikoff G, Dranoff G, Rock KL. J Immunol. 1997;158(6):2723–2730. [PubMed] [Google Scholar]
- 18.Clarke SRM, Barnden M, Kurts C, Carbone FR, Miller JF, Heath WR. Immunol Cell Biol. 2000;78(2):110–117. doi: 10.1046/j.1440-1711.2000.00889.x. [DOI] [PubMed] [Google Scholar]
- 19.Jain S, Yap WT, Irvine DJ. Biomacromolecules. 2005;6(5):2590–2600. doi: 10.1021/bm0503221. [DOI] [PubMed] [Google Scholar]
- 20.Kumari K, Gulati S, Smith DF, Gulati U, Cummings RD, Air GM. Virol J. 2007;4 doi: 10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiku H, Matsui M, Murakami S, Kiyozumi Y, Mizukami F, Sakaguchia K. Anal Biochem. 2003;318(1):80–85. doi: 10.1016/s0003-2697(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 22.Amalvy JI, Unali GF, Li Y, Granger-Bevan S, Armes SP, Binks BP, Rodrigues JA, Whitby CP. Langmuir. 2004;20(11):4345–4354. doi: 10.1021/la035921c. [DOI] [PubMed] [Google Scholar]
- 23.Zhang XQ, Dahle CE, Baman NK, Rich N, Weiner GJ, Salem AK. J Immunother. 2007;30(5):469–478. doi: 10.1097/CJI.0b013e31802fd8c6. [DOI] [PubMed] [Google Scholar]
- 24.Uchida T, Martin S, Foster TP, Wardley RC, Grimm S. Pharm Res. 1994;11(7):1009–1015. doi: 10.1023/a:1018987404751. [DOI] [PubMed] [Google Scholar]
- 25.Coombes AG, Yeh MK, Lavelle EC, Davis SS. J Contr Rel. 1998;52(3):311–320. doi: 10.1016/s0168-3659(98)00006-6. [DOI] [PubMed] [Google Scholar]
- 26.Yeh MK, Coombes AGA, Jenkins PG, Davis SS. Journal of Contr Rel. 1995;33(3):437–445. [Google Scholar]
- 27.Kazzaz J, Neidleman J, Singh M, Ott G, O'Hagan DT. Journal of Contr Rel. 2000;67(2–3):347–356. doi: 10.1016/s0168-3659(00)00226-1. [DOI] [PubMed] [Google Scholar]
- 28.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 29.Heath WR, Carbone FR. Nat Rev Immunol. 2001;1(2):126–134. doi: 10.1038/35100512. [DOI] [PubMed] [Google Scholar]
- 30.Gilboa E. J Clin Invest. 2007;117(5):1195–1203. doi: 10.1172/JCI31205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, Davey GM, Sutherland RM, Kurts C, Lew AM, Hirst C, Carbone FR, Heath WR. J Immunol. 2001;166(10):6099–6103. doi: 10.4049/jimmunol.166.10.6099. [DOI] [PubMed] [Google Scholar]
- 32.Kovacsovics-Bankowski M, Clark K, Benacerraf B, Rock KL. Proc Natl Acad Sci U S A. 1993;90(11):4942–4946. doi: 10.1073/pnas.90.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murthy N, Xu M, Schuck S, Kunisawa J, Shastri N, Frechet JM. Proc Natl Acad Sci U S A. 2003;100(9):4995–5000. doi: 10.1073/pnas.0930644100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonawane ND, Szoka FC, Jr, Verkman AS. J Biol Chem. 2003;278(45):44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 35.Lomas H, Massignani M, Abdullah KA, Canton I, Lo Presti C, MacNeil S, Du J, Blanazs A, Madsen J, Armes SP, Lewis AL, Battaglia G. Faraday Discuss. 2008;139:143–159. doi: 10.1039/b717431d. discussion 213–28, 419–20. [DOI] [PubMed] [Google Scholar]
- 36.Lomas H, Canton I, MacNeil S, Du J, Armes SP, Ryan AJ, Lewis AL, Battaglia G. Adv Mater. 2007;19(23) 4238-+ [Google Scholar]
- 37.Cerritelli S, Velluto D, Hubbell JA. Biomacromolecules. 2007;8(6):1966–1972. doi: 10.1021/bm070085x. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed F, Pakunlu RI, Srinivas G, Brannan A, Bates F, Klein ML, Minko T, Discher DE. Mol Pharm. 2006;3(3):340–350. doi: 10.1021/mp050103u. [DOI] [PubMed] [Google Scholar]
- 39.Stephenson I, Nicholson KG, Gluck R, Mischler R, Newman RW, Palache AM, Verlander NQ, Warburton F, Wood JM, Zambon MC. Lancet. 2003;362(9400):1959–1966. doi: 10.1016/S0140-6736(03)15014-3. [DOI] [PubMed] [Google Scholar]
- 40.Fields RC, Shimizu K, Mule JJ. Proc Natl Acad Sci U S A. 1998;95(16):9482–9487. doi: 10.1073/pnas.95.16.9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SY, Doh HJ, Ahn JS, Ha YJ, Jang MH, Chung SI, Park HJ. Vaccine. 1999;17(6):607–616. doi: 10.1016/s0264-410x(98)00241-2. [DOI] [PubMed] [Google Scholar]
- 42.GuermoCSPrez P, Saveanu L, Kleijmeer M, Davoust J, Van Endert P, Amigorena S. Nature. 2003;425(6956):397–402. doi: 10.1038/nature01911. [DOI] [PubMed] [Google Scholar]
- 43.Houde M, Bertholet S, Gagnon E, Brunet S, Goyette G, Laplante A, Princiotta MF, Thibault P, Sacks D, Desjardins M. Nature. 2003;425(6956):402–406. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- 44.Reis e Sousa C, Germain RN. J Exp Med. 1995;182(3):841–851. doi: 10.1084/jem.182.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stubbs AC, Martin KS, Coeshott C, Skaates SV, Kuritzkes DR, Bellgrau D, Franzusoff A, Duke RC, Wilson CC. Nat Med. 2001;7(5):625–629. doi: 10.1038/87974. [DOI] [PubMed] [Google Scholar]
- 46.Howland SW, Tsuji T, Gnjatic S, Ritter G, Old LJ, Wittrup KD. J Immunother. 2008;31(7):607–619. doi: 10.1097/CJI.0b013e318181c87f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. Proc Natl Acad Sci U S A. 2006;103(50):19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thitithanyanont A, Engering A, Ekchariyawat P, Wiboon-ut S, Limsalakpetch A, Yongvanitchit K, Kum-Arb U, Kanchongkittiphon W, Utaisincharoen P, Sirisinha S, Puthavathana P, Fukuda MM, Pichyangkul S. J Immunol. 2007;179(8):5220–5227. doi: 10.4049/jimmunol.179.8.5220. [DOI] [PubMed] [Google Scholar]
- 49.McGreal EP, Miller JL, Gordon S. Curr Opin Immunol. 2005;17(1):18–24. doi: 10.1016/j.coi.2004.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novina CD, Sharp PA. Nature. 2004;430(6996):161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- 51.Aigner A. J Biotechnol. 2006;124(1):12–25. doi: 10.1016/j.jbiotec.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 52.Hamm S, Heit A, Koffler M, Huster KM, Akira S, Busch DH, Wagner H, Bauer S. Int Immunol. 2007;19(3):297–304. doi: 10.1093/intimm/dxl146. [DOI] [PubMed] [Google Scholar]
- 53.Agrawal S, Kandimalla ER. Nat Biotechnol. 2004;22(12):1533–1537. doi: 10.1038/nbt1042. [DOI] [PubMed] [Google Scholar]