Abstract
Macrophages are important targets for HIV-1 and R5X4 strains play a central role in pathogenesis, especially in late-stage patients who may receive the fusion inhibitor T20. Sensitivity to T20 varies markedly among HIV-1 strains and is influenced by both viral and cellular factors that affect Env/CD4/coreceptor interactions. We addressed the relationship between T20 inhibition and pathway by which R5X4 HIV-1 infect primary macrophages, which express both coreceptors. In U87/CD4/coreceptor cells T20 sensitivity for entry through CCR5 and CXCR4 were correlated. In macrophages, the proportion of total entry mediated by each coreceptor differed among isolates. However, neither pathway was uniformly more or less sensitive to T20, nor did the proportion of entry mediated by each coreceptor predict T20 sensitivity. T20 sensitivity for macrophage infection overall correlated modestly with that for entry through CCR5 but not CXCR4, but unlike U87 cells, sensitivity of entry through CCR5 and CXCR4 were not correlated. These results suggest that strain-specific factors influence R5X4 T20 sensitivity regardless of coreceptor used; an absence of systematic differences in efficiency by which R5X4 strains use the two coreceptors; and that efficiency/kinetics of interactions with CCR5 are a central determinant of macrophage entry even when both pathways are utilized.
Keywords: HIV-1, chemokine receptor, viral entry, enfuvirtide, R5X4, CCR5, CXCR4, gp120, macrophage
Introduction
HIV-1 entry is initiated by binding of the viral Env surface glycoprotein gp120 to CD4 followed by interactions with a chemokine receptor, which trigger structural changes in the gp41 transmembrane glycoprotein of Env that lead to fusion. All HIV-1 strains use the chemokine receptor CCR5 (R5 variants), CXCR4 (X4 variants) or both (R5X4 variants). In nearly all individuals R5 variants are responsible for transmission and establishment of infection. Although disease may progress in the presence of R5 strains only, in 40–50% of infected individuals R5X4 variants emerge in place of or more frequently along side R5 variants, and their appearance in vivo is associated with accelerated disease progression. In some cases, X4 variants ultimately supplant R5X4 strains as disease progresses while in others R5X4 strains appear to persist1. Thus, although not essential, HIV-1 evolution from R5 to R5X4 or X4 is an important factor in accelerated pathogenesis.
T20 (Enfuvirtide) is a peptide derived from the HR2 heptad repeat sequence of gp41 that interacts with the HR1 domain of gp41 to block the HR1/HR2 association involved in the six-helix bundle formation necessary for virus-cell membrane fusion2. T20 was the first antiretroviral agent targeting entry in clinical use. The ability of T20 to block entry by prototype and primary HIV-1 strains, which is most frequently assayed in indicator cell lines that express CD4 plus one or the other coreceptor, varies markedly among isolates. Importantly, the efficiency with which T20 blocks entry is affected by the affinity of CD4-triggered gp120 for the coreceptor, fusion kinetics and receptor/coreceptor density, as well as other strain and cell-dependent factors3–5. Early reports suggested that T20 sensitivity might be greater for strains that use CXCR4 than those that use CCR5, although later reports did not support a clear dichotomy3, 6–8. While R5 and X4 strains have been extensively analyzed with respect to T20 inhibition, much less is known about R5X4 variants.
Studies of entry and entry inhibitors are most often carried out using indicator cell lines expressing one or the other coreceptor, which provide an efficient and convenient system. However primary cells differ from indicator cell lines in many characteristics including coreceptor expression levels and additional interactions that can affect entry and T20 sensitivity4. Macrophages and lymphocytes are the principal targets of infection in vivo. However, PBL are generally resistant to CCR5-mediated entry by R5X4 strains even though they are permissive for entry by R5 variants 9. In contrast, primary macrophages, which express both CCR5 and CXCR4, support entry of R5X4 variants through both coreceptor pathways9, 10. However, little is known about T20 inhibition of macrophage infection by R5X4 isolates or on entry through the different pathways.
In this study we asked how the sensitivity of R5X4 strains to the fusion inhibitor T20 differs depending on the pathway of infection using primary macrophages as targets. We addressed T20 inhibition of entry through each coreceptor separately for two reasons. First, because it blocks a step that represents a major distinguishing feature among HIV-1 variants, differential inhibition of R5X4 isolates through each pathway by T20 treatment would have the potential to shift the proportion of entry occurring through each pathway by these strains. Indeed, T20 is often used as salvage therapy in individuals with advanced disease who are may harbor R5X4 variants, and in whom viral replication may be incompletely suppressed. Secondly, in studying entry of an R5X4 strain via the two pathways the T20 binding site is invariant and thus differences in T20 sensitivity, if any, could not be ascribed to differences in drug binding. Since sensitivity to T20 is also profoundly affected by factors such as gp120/coreceptor affinity and fusion kinetics/triggering, sensitivity to this agent through each pathway therefore offers an indirect window into how these steps may differ between the two pathways utilized by R5X4 viruses.
Materials and Methods
Cells and viruses
Monocytes were isolated from heparinized blood of normal donors by selective adherence as previously described11, maintained in 10 cm dishes in RPMI with 10% fetal bovine serum and M-CSF (100 U/ml) for 6–7 days to allow differentiation into monocyte-derived macrophages (MDM), then re-plated in 48 well plates at 1.5×105 cells/well in DMEM with 10% FBS one day prior to infection.
The U87/CD4, U87/CD4/CCR5 and U87/CD4/CXCR4 cell lines were obtained through the NIH AIDS Research and Reference Reagent program12. Viruses used were the R5X4 prototypes 89.6 and DH12; R5X4 primary isolates 93BR020, 92HT594, 96USHIP9S obtained from the NIH AIDS Research and Reference Reagent program13; R5 prototype Bal; and macrophage-tropic X4 strain Tybe14. Virus stocks were grown in PHA/IL2-stimulated lymphocytes, titered by p24 antigen content, and treated with DNAse (50 units/ml) for 30 min. prior to infection.
Infections
Primary MDM and U87 indicator cells were seeded into 48- or 24-well plates, respectively, one day prior to infection. To analyze each pathway independently, MDM were incubated for 1 hour prior to infection with or without 2 ug/ml of the CXCR4 inhibitor AMD3100 (Sigma, St. Louis, MO) and/or 2 uM of the CCR5 inhibitor M65715 (a gift of M. Miller, Merck & Co., West Point, Pa), which are concentrations that fully block entry of these viruses through each pathway in macrophages. Different concentrations of T20 (Enfuvirtide; Fuzeon; Roche Pharmaceuticals, Nutley, NJ) were then added to wells prior to addition of virus. Cells were infected using 20 ng of p24 antigen of DNAse-treated virus, incubated for 72 hours, then harvested and analyzed for infection by quantitative PCR detection of viral reverse transcription products. To limit infection to only a single cycle, the protease inhibitors indinavir or nelfinavir (each at 10 ug/ml) were added at the time of infection and maintained thereafter.
Quantitative real-time PCR analysis
Entry and infection was quantified by assay of newly formed reverse transcription products. Three days after infection the cells were washed and then lysed in 50 ul of DNA lysis buffer. For each PCR reaction, 1.5 ul of lysate was added per 25 ul of reaction mixture and amplified using HIV-1 LTR and gag as well as cellular GAPDH primer/probe sets. Details of lysis conditions, primer/probe sets and amplification parameters have been described previously9. HIV-1 cDNA copy number was normalized to cellular GAPDH copy number.
Statistical Analysis
IC50 values were calculated from standard inhibition curves using GraphPad Prism software. IC50 values were found to be normally distributed following log transformation, which were therefore used for model based statistical analyses. Differences between pathways were analyzed using paired two-tailed T-tests. Both Pearson’s and partial correlation coefficients evaluated the relationship between EC50 values from entry mediated through different pathways, with partial correlations being used to adjust for the effects of different virus types. Additionally, interactions among virus, cell type and pathway were analyzed by ANOVA models.
Results
T20 blocking of R5X4 entry into U87/CD4/coreceptor indicator cells
To assess R5X4 HIV-1 sensitivity to the fusion inhibitor T20 we selected two R5X4 prototypes, HIV-1 strains 89.6 and DH12. Both of these strains are widely used in vitro, and are also used extensively in animal studies in the context of SHIV chimeras containing their respective env genes16, 17. We also analyzed three R5X4 primary isolates, 92HT594, 93BR020 and 96USHIP9S, to ensure that results were relevant to a range of R5X4 variants. These strains have been confirmed as R5X4 in various indicator cell lines, with similar entry efficiency through the two coreceptors in those assays9. In parallel we used the macrophage-tropic R5 prototype Bal, along with the X4 variant Tybe, which is an X4 primary isolate that infects macrophages through use of CXCR414. HIV/Env coreceptor interactions can be assessed using various approaches including cell-cell fusion assays, pseudotype reporter viruses, or infectious viral studies. Because we wanted to most closely reflect entry leading to productive infection, we utilized replication-competent infectious virus in an assay that detects initial steps of productive infection based on viral cDNA formation. A protease inhibitor was maintained in the cultures to ensure single-cycle infections and thus linear quantification of entry.
We first addressed coreceptor-specific entry using U87/CD4 indicator cells expressing CCR5 or CXCR4. Cells were infected with DNAse-treated virus in the presence or absence of T20 and lysed 3 days later for detection of viral reverse transcription products as an indication of infection. As shown in Table 1, the R5X4 strains exhibited a range of sensitivity to T20 in U87 indicator cells. EC50 values varied over an approximately 25-fold range for CCR5-dependent virus entry (0.006 ± 0.002 to 0.386 ± 0.171 ug/ml) and 65-fold range for CXCR4-dependent entry (0.028 ± 0.019 to 0.703 ± 0.388 ug/ml). This range is similar to variations in T20 sensitivity among other unrelated R5 and X4 viruses3, 6, 18. The R5 prototype Bal and X4 strain Tybe demonstrated EC50 values of 0.151 ± 0.054 and 0.033 ± 0.019 ug/ml on CCR5 and CXCR4-expressing cells, respectively, which were both within the range of sensitivity reflected by the R5X4 variants.
Table 1.
Inhibition of R5X4 strains by T20 blocking in U87/CD4/coreceptor cells
| U87/CD4 | U87/CD4 | |
|---|---|---|
| CCR5 | CXCR4 | |
| 89.6 | 0.386 ± 0.171 | 0.703 ± 0.388 |
| DH12 | 0.159 ± 0.053 | 0.381 ± 0.191 |
| 93BR020 | 0.025 ± 0.010 | 0.109 ± 0.035 |
| 92HT594 | 0.006 ± 0.002 | 0.028 ± 0.019 |
| 96USHIP9S | 0.255 ± 0.103 | 0.598 ± 0.249 |
| Bal | 0.151 ± 0.054 | |
| Tybe | 0.033 ± 0.019 |
Values represent EC50 values (mean ± SEM; ug/ml) calculated from a minimum of three independent experiments.
P=0.035 for the R5X4 isolates in CCR5 versus CXCR4 indicator cells.
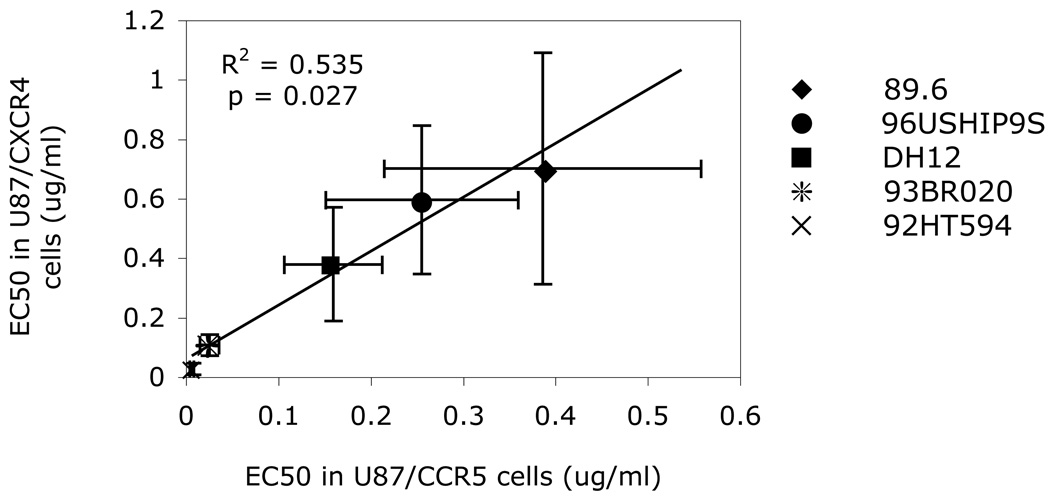
We next determined if T20 sensitivity for entry through CCR5 and through CXCR4 correlated with each other. As shown in Fig. 1, for the R5X4 strains, T20 EC50 values for entry through CCR5 was significantly correlated with those for entry through CXCR4. The fact that T20 sensitivity of each virus for entry through CCR5 and CXCR4 are linked indicates that strain-specific features determined sensitivity to the fusion inhibitor regardless of the coreceptor involved. EC50 values for entry into U87/CD4/CXCR4 cells were approximately two-fold higher than for CCR5-expressing cells (p=0.035; Table 1). Thus, the strong correlation between CCR5 and CXCR4 EC50 values indicates common mechanisms shared by the pathways for R5X4 strains. On the other hand, the higher EC50 values for entry through CXCR4 compared with CCR5 may well simply reflect the higher levels of CXCR4 than CCR5 on these indicator cells by FACS staining (data not shown).
FIG. 1. T20 blocking of R5X4 isolates in U87 indicator cell lines.
U87/CD4/CCR5 and U87/CD4/CXCR4 cells were treated with different concentration of T20 and infected with 20 ng of p24 of each virus. Entry and infection was assayed 72 hours later in cell lysates by qPCR using HIV-1 LTR & cellular GAPDH primer/probe sets, with HIV-1 copy number normalized to GAPDH. Data points represent EC50 values (means ± SEM of 5 replicate experiments) for each of the five R5X4 strains.
Contribution of each coreceptor to total macrophage entry by R5X4 strains
We previously showed that even though most R5X4 strains use only CXCR4 to enter primary human PBL, they use both coreceptors for entry into primary macrophages9. Therefore, as a prelude to analyzing inhibition of pathway-specific infection, we determined the proportional contribution of each coreceptor to total macrophage infection for each of the isolates. MDM were incubated with or without the CCR5 blocker M657 or CXCR4 blocker AMD3100 at concentrations that fully block entry through the respective pathway, infected with the various isolates in the presence of a protease inhibitor to prevent subsequent rounds of reinfection, and lysed three days later for qPCR quantification of reverse transcription products and entry.
As shown in Fig. 2A, M657 completely blocked the R5 strain Bal but had no effect on X4 strain Tybe, while AMD3100 completely blocked Tybe but not Bal. In contrast, each single coreceptor inhibitor reduced entry by the R5X4 strains but only the combination of both blockers completely abrogated infection. For each strain the amount of newly formed viral cDNA when only CCR5 was available, relative to that when both coreceptors were available, varied from 53% to 94%, while the amount of entry that occurred when CXCR4 only was available ranged from 28% to 55% of total (Table 2). This result indicates that these R5X4 isolates use both CCR5 and CXCR4 on macrophages, and that the proportion of total macrophage entry mediated by each pathway differs for each isolate.
FIG. 2. Coreceptor-specific entry and T20 sensitivity of R5X4 strains in macrophages.
(A) MDM were treated with or without saturating concentrations of the CCR5 antagonist M657 (2 uM), the CXCR4 antagonist AMD3100 (2 ug/ml), or both, and infected using 20 ng of p24 of virus. Entry and infection was assayed 72 hours later in cell lysates by qPCR. Viral cDNA copy number was normalized to cellular GADPH, and percent entry through each pathway was determined on the basis of unblocked control macrophages. Data represent means ± SEM of replicate experiments using cells from 3 (primary isolates) or 4 (prototype strains) different donors. (B–F) MDM were treated with or without fully blocking concentrations of M657 or AMD3100 to isolate the CXCR4 and CCR5 pathways, respectively, exposed to T20, then infected using 20 ng of prototype and primary R5X4 strains. Cells were harvested 72 hour later and viral cDNA copy number assayed by qPCR, normalized to GAPDH. Percent entry through each pathway in the presence of different T20 concentrations was calculated relative to entry through that pathway (CCR5 only, CXCR4 only, or both pathway available) in the absence of T20. Data represent means ± SEM for 3–4 replicate experiments using cells from different donors.
Table 2.
Proportion of entry mediated by each coreceptor for R5X4 HIV-1 infection of primary macrophages
| 89.6 | DH12 | 93BR020 | 92HT594 | 96USHIP9S | |
|---|---|---|---|---|---|
| CCR5 | 77% | 65% | 53% | 78% | 94% |
| (60–95%) | (55–75%) | (50–60%) | (60–100%) | (80–120%) | |
| CXCR4 | 39% | 49% | 55% | 52% | 28% |
| (20–60%) | (45–55%) | (50–60%) | (35–60%) | (20–35%) |
Percent entry determined as the level of entry by quantitative PCR in the presence of single coreceptor blockers compared with that in the absence of blocker. Data represent means and range of 4–5 experiments using cells from different donors.
Because blocking each coreceptor alone reduced entry, we estimated the multiplicity of infection (MOI) for these experiments. Based on HIV to GAPDH quantification as well as direct TCID50 determination (9 and data not shown), MOIs were calculated to be less than 0.1, similar to relatively low MOIs used in most in vitro HIV-1 infection studies reported. Thus, at this MOI coreceptor availability should not be limiting for infection, and that if one pathway was blocked each infectious particle should have enough of the alternative coreceptor available for entry. The fact that single coreceptor antagonists reduced entry suggests that virus/receptor interactions are not highly plastic and when one pathway is blocked the CD4-bound virus is not completely free to engage the alternate coreceptor. On the other hand, the amount of entry that occurred when each pathway was assessed independently, when combined, was typically slightly greater than that occurring when both pathways were present together (108%–130%; Table 2), which suggests that some degree of compensatory increase in entry through the alternative coreceptor might take place.
Effect of coreceptor pathway on T20 inhibition of R5X4 entry into macrophages
We then used monocyte-derived macrophages as targets to assess coreceptor-dependent T20 inhibition of entry through each pathway independently. To assess the efficiency of T20 inhibition when macrophage infection was mediated through CCR5 alone compared with CXCR4 alone and when both pathways were present, MDM were incubated with or without fully blocking concentrations of the chemokine receptor antagonists (plus the protease inhibitor). Cells were then exposed to virus in the presence of varying concentrations of T20, and infection assayed 3 days later by quantitative PCR. For each coreceptor condition, entry in the presence of T20 treatment was determined relative to that in the absence of T20, under the same coreceptor blocking/availability conditions.
As shown in Fig. 2B–F, T20 effectively inhibited macrophage infection by all 5 isolates under all conditions of coreceptor availability (CCR5 only, CXCR4 only, both CCR5 and CXCR4), with striking overlap in T20 inhibition curves for each isolate under the three conditions. EC50 values varied over a range of approximately 20-fold (0.033 ± 0.012 to 0.635 ± 0.417 ug/ml), with no evidence that inhibition was more efficient when only one pathway was available compared with both pathways (Table 3; p=ns). There was also no evidence to suggest that T20 inhibition was generally more efficient for R5X4 isolates when infection occurred through either CCR5 versus CXCR4 as single pathways (p=ns). The proportion of total macrophage entry mediated by each coreceptor also did not predict the relative sensitivity to T20 through that pathway. For example, four of the isolates were more easily blocked by T20 when entry was mediated by CXCR4 than by CCR5, whereas only one (92HT594) was more sensitive when entry was mediated by CCR5 (Table 3). However, 92HT594 was no more dependent on CXCR4 for macrophage entry than the other strains (Table 2), as might be expected if it differed in being more efficient at using macrophage CXCR4 than CCR5.
Table 3.
Coreceptor-specific blocking of R5X4 entry by T20 into primary macrophages
| MDM | MDM | MDM | |
|---|---|---|---|
| CCR5 & CXCR4 | CCR5 | CXCR4 | |
| 89.6 | 0.235 ± 0.189 | 0.147 ± 0.057 | 0.077 ± 0.052 |
| DH12 | 0.288 ± 0.194 | 0.635 ± 0.417 | 0.125 ± 0.051 |
| 93BR020 | 0.051 ± 0.015 | 0.074 ± 0.024 | 0.033 ± 0.012 |
| 92HT594 | 0.064 ± 0.046 | 0.048 ± 0.015 | 0.086 ± 0.016 |
| 96USHIP9S | 0.126 ± 0.022 | 0.243 ± 0.203 | 0.038 ± 0.015 |
| Bal | 0.086 ± 0.062 | ||
| Tybe | 0.057 ± 0.042 |
Values represent EC50 values (mean ± SEM; ug/ml) calculated from three or four independent experiments using cells from different donors. Dual-pathway entry was assessed in the absence of coreceptor inhibitor while CCR5-specific and CXCR4-specific was determined in the presence of fully blocking concentrations of CXCR4 or CCR5 antagonists AMD3100 or M657, respectively. P=ns for differences between any of the pathways for R5X4 isolates.
We then asked if there was a relationship between T20 potency during dual pathway-mediated and CCR5-mediated or CXCR4-mediated macrophage infection (Fig. 3). As shown in Fig. 3A, there was a modest but significant correlation between EC50 values for entry mediated through CCR5 alone and entry through both pathways (p=0.014). In contrast, T20 inhibition of entry through CXCR4 was not correlated with either dual-pathway entry or CCR5-mediated entry (Fig. 3B and 4C; p=ns). Thus, the overall ability of T20 to block macrophage infection appears to be linked most closely to inhibition of entry through CCR5, even when both CCR5 and CXCR4 are used by these isolates for entry.
FIG. 3. Relationship between T20 sensitivity in primary macrophages for different pathways of entry.
(A) EC50 values in the absence of coreceptor blocking (“both pathways”) compared with values in the presence of saturating concentrations of the CXCR4 inhibitor AMD3100 (“CCR5 pathway”). (B) EC50 values in the absence of coreceptor blocking (“both pathways”) compared with values in the presence of saturating concentrations of the CCR5 inhibitor M657 (“CXCR4 pathway”). (C) EC50 values measured in the presence of M657 (“CXCR4 pathway)” and in the presence of AMD3100 (“CCR5 pathway”). Data points reflect the EC50 values for each virus in individual experiments using cells from different macrophage donors. Partial correlations (R) between the EC50 values, adjusting for the virus effects, are shown with associated p-values
Discussion
This study compared CCR5 versus CXCR4-mediated infection of primary macrophages by R5X4 HIV-1 strains, and the sensitivity to T20 of entry through the two distinct pathways. We found that the proportion of total macrophage entry mediated by each coreceptor varied among the isolates, that T20 sensitivity in macrophage infection varied among strains but was largely independent of the coreceptor pathway used, and that overall T20 sensitivity for macrophage infection was more closely correlated with sensitivity of entry through CCR5 and not with entry through CXCR4. This result contrasted with U87 indicator cells, in which sensitivity to T20 for entry through the two pathways were closely correlated.
Many studies have evaluated the kinetics and efficiency of entry, as well as T20 sensitivity, for R5 versus X4 HIV-1 strains, but how CCR5 versus CXCR4-mediated entry differs for R5X4 strains has received little attention. This question is relevant for two reasons. Although strain-specific differences in coreceptor interactions means that R5X4 use of CCR5 and CXCR4 cannot be extrapolated to R5 and X4 single coreceptor variants, examining viruses that utilize either pathway, particularly in combination with cells that express both pathways, can minimize strain- and cell-specific differences and allow closer comparison of factors that are intrinsic to the specific pathway. Secondly, R5X4 strains typically emerge as disease progresses, and later stage patients are often those who have been heavily treated and thus may receive T20 as “salvage therapy”. In U87 indicator cells these R5X4 viruses were slightly more easily inhibited by T20 when entry was mediated by CCR5 than CXCR4. This result differs from the one previous report comparing CCR5 versus CXCR4-mediated R5X4 entry, which found the EC50 for strain 89.6 was higher on CCR5- than CXCR4-expressing U87 cells7. The relevance of cell lines to infection in vivo is uncertain, however, as the levels of coreceptor on engineered cells typically exceeds those on primary cells, and may even vary among different passages of indicator cells (data not shown). In contrast, monocyte-derived macrophages reflect a target cell important for infection in vivo and express similar levels of CCR5 and CXCR4 (19 and data not shown).
While most of the strains tested were more easily blocked in macrophages by T20 when entry proceeded through CXCR4 than through CCR5, not all exhibited this pattern and no consistent or significant difference was evident between the two pathways. Similarly, the relative proportion of entry mediated by each coreceptor was not linked to the relative sensitivity to T20 for entry through that coreceptor. Thus, the overall coreceptor-independent T20 sensitivity of R5X4 strains in macrophages suggests that, for R5X4 strains as a group, there are unlikely to be systematic differences in how they interact with CCR5 and CXCR4 in macrophages in terms of entry efficiency, fusion kinetics, or other parameters known to influence T20 sensitivity. The window of sensitivity to T20 is believed to open upon gp120 binding to CD4 and close upon engagement of the coreceptor, which triggers formation of the T20-insensitive six-helix bundle20. CD4-triggered R5 gp120 typically has a higher affinity for CCR5 than CD4-triggered X4 gp120 for its cognate coreceptor21, and it is believed that other differences between individual envelopes, such as triggering kinetics, combine with coreceptor affinity to determine T20 sensitivity3. In contrast, since CD4 binding and subsequent conformational changes involved in R5X4 envelope triggering would not be affected by the coreceptor subsequently engaged, our result suggests indirectly that the efficiency/affinity with which CCR5 versus CXCR4 are engaged by CD4-triggered R5X4 gp120 are not systematically different. Of note, CD4-triggered R5X4 gp120 appears to have a lower equilibrium binding affinity to CCR5 than does CD4-triggered R5 gp12022, and greater sensitivity to CCR5 antagonists9, 23. Thus, the absence of systematic difference in R5X4 interaction with CCR5 versus CXCR4 suggested by these results implies that R5X4 gp120 are most likely “equally poor” in engaging each of the chemokine receptors.
The sensitivity of R5X4 strains to T20 for macrophage infection proceeding through both pathways correlated with the sensitivity when entry was mediated by CCR5 alone, whereas sensitivity to T20 for CXCR4-mediated macrophage entry did not correlate with either CCR5 or dual-pathway entry. This was true regardless of what proportion of overall macrophage entry was contributed by each coreceptor. Thus, even though both pathways contribute to entry, interactions with CCR5 appear to “dominate” in determining aspects of the entry process even when both pathways are present. The reasons for this are unclear, although it is possible that coreceptor antagonists targeting the chemokine receptor extracellular loop regions might still allow interactions with other domains such as the N-terminus24 that could enable the blocked coreceptor to affect Env interactions in a way that affects T20 sensitivity, even if fusion cannot ultimately proceed via that coreceptor.
T20 is most often used as “salvage” therapy in late stage patients, which is also the population most likely to harbor CXCR4-using variants. Therefore, coreceptor-dependent differences in T20 inhibition could have the potential to alter the pathway used by R5X4 variants in vivo. However, although most R5X4 strains were more sensitive to T20 when macrophage entry was mediated by CXCR4 compared with CCR5, one showed the opposite pattern and this difference did not reach statistical significance. Thus, while any individual strain may be inhibited differently for entry by each pathway, our data do not support the notion that T20 would preferentially alter coreceptor utilization by R5X4 isolates in general.
These R5X4 isolates use both entry pathways to infect primary macrophages, which differs from exclusive or nearly exclusive use of CXCR4 by R5X4 strains for primary lymphocyte infection9, 25, 26. Similar dependence on CXCR4 has been shown for R5X4 infection of lymphoid tissue ex vivo27 and, furthermore, that the majority of actively replicating (and thus lymphocyte-derived) plasma viremia in monkeys infected with R5X4 SHIV strains is insensitive to CCR5 inhibitors and dependent on CXCR417. Taken together these results suggest two distinct aspects of R5X4 HIV-1 replication: a lymphocyte replication cycle that is mainly or exclusively dependent on CXCR4, and a macrophage reservoir that is supported by both CCR5 and CXCR4. On the other hand, it has been reported that in animals infected with the R5X4 DH12-derived SHIV, both macrophages and lymphocytes are infected mainly through CXCR428, which differs from our findings in primary human macrophages in vitro that HIV-1 DH12 uses both pathways.
While macrophage infection by the R5X4 strains was completely blocked by the combination of CCR5 and CXCR4 antagonists, an unexpected finding was that even at quite low MOI infection mediated by a single pathway was reduced in comparison with entry mediated by both pathways. At such low MOI, virus would be expected to be the limiting factor and CD4 or coreceptor availability would not. This result, therefore, suggests that once bound to the cell each infectious virion is “committed” to that particular coreceptor and even if that pathway is blocked it is unable to enter through the other coreceptor instead, even though that pathway should be available. This restriction could result from fixed association between the coreceptors and CD4 or limited mobility within the membrane29, 30 that might restrict the ability of virus, once triggered by CD4, to freely choose whichever coreceptor is available and functional. Alternatively, it is possible that excess noninfectious virions might interact with and effectively inhibit utilization of the alternative pathway. Studies to clarify the basis for this coreceptor interference are currently under way.
In summary, this is the first study to address the T20 sensitivity and, indirectly, the efficiency of entry, by R5X4 strains through each coreceptor independently on a primary target cell where both pathways are present and support entry. We have shown that in monocyte-derived macrophages both CCR5 and CXCR4 support entry by R5X4 strains; that neither pathway is necessarily more or less sensitive to T20; that CCR5- and not CXCR4-dependent interactions determine T20 sensitivity overall; and that the close correlation between CCR5 and CXCR4 inhibition of entry in indicator cells is not evident in primary macrophages. These results provide insight into T20-mediated inhibition and how CCR5 and CXCR4 each function for entry a primary cell type important in vivo, and indicate that coreceptor-mediated entry in primary cells is more complex than that reflected by cell lines.
Acknowledgement
The authors thank M. Miller for M657; M. Kienzle for technical assistance; the Virus/Molecular, Immunology, and Biostatistics Cores of the Penn Center for AIDS Research; and R. Doms for critical reading of the manuscript. This work was supported by NIH grants to R.G.C. L.L. was supported by training grant T32 AI007632.
Source of support: PHS grants AI35502, MH61139, NS27405
References
- 1.Stalmeijer EH, Van Rij RP, Boeser-Nunnink B, et al. In vivo evolution of X4 human immunodeficiency virus type 1 variants in the natural course of infection coincides with decreasing sensitivity to CXCR4 antagonists. J Virol. 2004 Mar;78(6):2722–2728. doi: 10.1128/JVI.78.6.2722-2728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matthews T, Salgo M, Greenberg M, Chung J, DeMasi R, Bolognesi D. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat Rev Drug Discov. 2004 Mar;3(3):215–225. doi: 10.1038/nrd1331. [DOI] [PubMed] [Google Scholar]
- 3.Reeves JD, Gallo SA, Ahmad N, et al. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc Natl Acad Sci USA. 2002 Dec 10;99(25):16249–16254. doi: 10.1073/pnas.252469399. 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beausejour Y, Tremblay MJ. Susceptibility of HIV Type 1 to the Fusion Inhibitor T-20 Is Reduced on Insertion of Host Intercellular Adhesion Molecule 1 in the Virus Membrane. J Infect Dis. 2004 Sep 1;190(5):894–902. doi: 10.1086/422698. [DOI] [PubMed] [Google Scholar]
- 5.Heredia A, Gilliam B, DeVico A, et al. CCR5 density levels on primary CD4 T cells impact the replication and Enfuvirtide susceptibility of R5 HIV-1. Aids. 2007 Jun 19;21(10):1317–1322. doi: 10.1097/QAD.0b013e32815278ea. [DOI] [PubMed] [Google Scholar]
- 6.Derdeyn CA, Decker JM, Sfakianos JN, et al. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74(18):8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Zhang X, Matsuoka M, Hattori T. The possible involvement of CXCR4 in the inhibition of HIV-1 infection mediated by DP178/gp41. FEBS Lett. 2000 Dec 29;487(2):185–188. doi: 10.1016/s0014-5793(00)02336-x. [DOI] [PubMed] [Google Scholar]
- 8.Cilliers T, Patience T, Pillay C, Papathanasopoulos M, Morris L. Sensitivity of HIV type 1 subtype C isolates to the entry inhibitor T-20. AIDS Res Hum Retroviruses. 2004 May;20(5):477–482. doi: 10.1089/088922204323087714. [DOI] [PubMed] [Google Scholar]
- 9.Yi Y, Shaheen F, Collman RG. Preferential use of CXCR4 by R5X4 human immunodeficiency virus type 1 isolates for infection of primary lymphocytes. Journal of Virology. 2005;79(3):1480–1486. doi: 10.1128/JVI.79.3.1480-1486.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi Y, Rana S, Turner JD, Gaddis N, Collman RG. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–779. doi: 10.1128/jvi.72.1.772-777.1998. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collman R, Hassan NF, Walker R, et al. Infection of monocyte-derived macrophages with human immunodeficiency virus type 1 (HIV-1). Monocyte-tropic and lymphocyte-tropic strains of HIV-1 show distinctive patterns of replication in a panel of cell types. J.Exp.Med. 1989;170:1149–1163. doi: 10.1084/jem.170.4.1149. 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjorndal A, Deng H, Jansson M, et al. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao F, Morrison SG, Robertson DL, et al. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. The WHO and NIAID Networks for HIV Isolation and Characterization. J Virol. 1996 Mar;70(3):1651–1667. doi: 10.1128/jvi.70.3.1651-1667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi Y, Chen W, Frank I, et al. An unusual syncytia-inducing human immunodeficiency virus type 1 primary isolate from the central nervous system that is restricted to CXCR4, replicates efficiently in macrophages, and induces neuronal apoptosis. J Neurovirol. 2003 Aug;9(4):432–441. doi: 10.1080/13550280390218706. 2003. [DOI] [PubMed] [Google Scholar]
- 15.Finke PE, Caldwell C, Dorn C, et al. The discovery of potent human CCR5 antagonists; Paper presented at: 10th National Conference of the Inflammation Research Association; Hot Springs, VA: 2000. Sep 24–28, [Google Scholar]
- 16.Shibata R, Igarashi T, Haigwood N, et al. Neutralizing antibody directed against the HIV-1 envelope glycoprotein can completely block HIV-1/SIV chimeric virus infections of macaque monkeys. Nature Med. 1999;5:204–210. doi: 10.1038/5568. 1999. [DOI] [PubMed] [Google Scholar]
- 17.Veazey RS, Klasse PJ, Ketas TJ, et al. Use of a small molecule CCR5 inhibitor in macaques to treat simian immunodeficiency virus infection or prevent simian-human immunodeficiency virus infection. J Exp Med. 2003 Nov 17;198(10):1551–1562. doi: 10.1084/jem.20031266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heil ML, Decker JM, Sfakianos JN, Shaw GM, Hunter E, Derdeyn CA. Determinants of human immunodeficiency virus type 1 baseline susceptibility to the fusion inhibitors enfuvirtide and T-649 reside outside the peptide interaction site. J Virol. 2004 Jul;78(14):7582–7589. doi: 10.1128/JVI.78.14.7582-7589.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallo SA, Puri A, Blumenthal R. HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry (Mosc) 2001 Oct 16;40(41):12231–12236. doi: 10.1021/bi0155596. [DOI] [PubMed] [Google Scholar]
- 21.Doranz BJ, Baik SSW, Doms RW. Use of a gp120 binding assay to dissect the requirements and kinetics of human immunodeficiency virus fusion events. J Virol. 1999;73:10346–10358. doi: 10.1128/jvi.73.12.10346-10358.1999. 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baik SSW, Doms RW, Doranz BJ. HIV and SIV gp120 binding does not predict coreceptor function. Virology. 1999;259:267–273. doi: 10.1006/viro.1999.9779. 1999. [DOI] [PubMed] [Google Scholar]
- 23.Simmons G, Reeves JD, Hibbitts S, et al. Co-receptor use by HIV and inhibition of HIV infection by chemokine receptor ligands. Immunol Rev. 2000;177:112–126. doi: 10.1034/j.1600-065x.2000.17719.x. [DOI] [PubMed] [Google Scholar]
- 24.Melikyan GB, Platt EJ, Kabat D. The role of the N-terminal segment of CCR5 in HIV-1 Env-mediated membrane fusion and the mechanism of virus adaptation to CCR5 lacking this segment. Retrovirology. 2007 Aug 8;4(1):55. doi: 10.1186/1742-4690-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghezzi S, Menzo S, Brambilla A, et al. Inhibition of R5X4 dualtropic HIV-1 primary isolates by single chemokine co-receptor ligands. Virology. 2001;280(2):253–261. doi: 10.1006/viro.2000.0753. 2001. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Lou B, Lal RB, Gettie A, Marx PA, Moore JP. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J Virol. 2000;74:6893–6910. doi: 10.1128/jvi.74.15.6893-6910.2000. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glushakova S, Yi Y, Grivel JC, et al. Preferential coreceptor utilization and cytopathicity by dual-tropic HIV-1 in human lymphoid tissue ex vivo. J Clin Invest. 1999 Sep;104(5):R7–R11. doi: 10.1172/JCI7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Igarashi T, Donau OK, Imamichi H, et al. Macrophage-tropic simian/human immunodeficiency virus chimeras use CXCR4, not CCR5, for infections of rhesus macaque peripheral blood mononuclear cells and alveolar macrophages. J Virol. 2003 Dec;77(24):13042–13052. doi: 10.1128/JVI.77.24.13042-13052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lapham CK, Zaitseva MB, Lee S, Romanstseva T, Golding H. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nature Med. 1999;5:303–308. doi: 10.1038/6523. 1999. [DOI] [PubMed] [Google Scholar]
- 30.Lee S, Lapham CK, Chen H, et al. Coreceptor competition for association with CD4 may change the susceptibility of human cells to infection with T-tropic and macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 2000 Jun;74(11):5016–5023. doi: 10.1128/jvi.74.11.5016-5023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]