Abstract
The DEAD-box proteins CYT-19 in Neurospora crassa and Mss116p in Saccharomyces cerevisiae are broadly acting RNA chaperones that function in mitochondria to stimulate group I and group II intron splicing and activate mRNA translation. Previous studies showed that the S. cerevisiae cytosolic/nuclear DEAD-box protein Ded1p could stimulate group II intron splicing in vitro. Here, we show that Ded1p complements the mitochondrial translation and group I and II intron splicing defects in mss116Δ strains, stimulates the in vitro splicing of group I as well as group II introns, and functions indistinguishably from CYT-19 to resolve different non-native secondary and/or tertiary structures in the Tetrahymena thermophila LSU-ΔP5abc group I intron. The Escherichia coli DEAD-box protein SrmB also stimulates group I and II intron splicing in vitro, while the E. coli DEAD-box protein DbpA and vaccinia virus DExH-box protein NPH-II gave little if any group I or II intron splicing stimulation in vitro or in vivo. The four DEAD-box proteins that stimulate group I and II intron splicing unwind RNA duplexes by local strand separation and have little or no specificity, as judged by RNA-binding assays and stimulation of their ATPase activity by diverse RNAs. By contrast, DbpA binds group I and II intron RNAs non-specifically, but its ATPase activity is activated specifically by a helical segment of E. coli 23S rRNA, and NPH-II unwinds RNAs by directional translocation. The ability of DEAD-box proteins to stimulate group I and II intron splicing correlates primarily with their RNA-unwinding activity, which for the protein preparations used here was greatest for Mss116p, followed by Ded1p, CYT-19, and SrmB. Further, this correlation holds for all group I and II intron RNAs tested, implying a fundamentally similar mechanism for both types of introns. Our results support the hypothesis that DEAD-box proteins have an inherent ability to function as RNA chaperones by virtue of their distinctive RNA-unwinding mechanism, which enables refolding of localized RNA regions or structures without globally disrupting RNA structure.
Keywords: catalytic RNA, ribozyme, RNA chaperone, RNA-protein interaction, RNA structure
Introduction
DExH/D-box proteins, the collective designation of the five protein families of helicase superfamily II that work on RNA, are the largest class of enzymes in eukaryotic RNA metabolism.1–4 Commonly referred to as RNA helicases, they mediate ATP-dependent RNA and RNP structural rearrangements in a variety of cellular processes, including translation, ribosome assembly, RNA degradation, and pre-mRNA splicing. All DExH/D-box proteins have a helicase core, which consists of two tandem RecA-like domains with a series of at least eight conserved sequence motifs involved in RNA binding, ATP binding and ATP hydrolysis, and communication between the ATP- and RNA-binding sites.4 This helicase core is typically flanked by N- and/or C-terminal extensions, which differ among proteins and in some cases target the proteins to specific sites of action via protein-protein or protein-RNA interactions.5–8
DEAD-box proteins, named for the amino acid sequence of the conserved motif II, are the largest family of DExH/D-box proteins with 25 members in S. cerevisiae and at least 37 members in humans.9 By contrast to canonical helicases, which unwind duplexes by ATP-driven translocation on one of the helical strands, DEAD-box proteins employ a fundamentally different unwinding mechanism that does not involve translocation. DEAD-box proteins load directly onto helical regions, aided by surrounding RNA, and promote local unwinding of duplex strands in an ATP-dependent manner.10–12 This helicase mode enables partial denaturation of RNA secondary structure without globally unfolding the RNA. Recently, it has been shown that DEAD-box proteins require ATP binding but not hydrolysis to promote unwinding, which presumably occurs by a mechanism in which the ATP-bound form of the enzyme binds tightly to and displaces an RNA strand away from its partner in a duplex.13,14 ATP-hydrolysis promotes release of the bound strand and is thus necessary for enzyme recycling. In addition to RNA unwinding, DEAD-box proteins have other activities including ATP-independent RNA binding and strand annealing, and at present, it is not clear to what extent RNA unwinding as opposed to these other activities is critical for their physiological functions.4
The DEAD-box proteins CYT-19 in Neurospora crassa and Mss116p in Saccharomyces cerevisiae are broadly acting RNA chaperones, which function in mitochondria to stimulate the splicing of mitochondrial (mt) group I and group II introns, RNA-end processing, and mRNA translation.15,16 These proteins bind to diverse RNA substrates non-specifically and use ATP to resolve stable intermediate or non-native structures that slow RNA folding. In N. crassa, a mutation in the cyt-19 gene (cyt-19-1) inhibits the splicing of mt group I introns that require the CYT-18 protein, the mt tyrosyl-tRNA synthetase, for structural stabilization,15 while in S. cerevisiae, deletion of the MSS116 gene inhibits the splicing of all mt group I and group II introns.16,17 The latter depend upon a variety of different protein partners for structural stabilization, including group I and group II intron-encoded maturases, and the mt LeuRS, Cbp2p, and Mrs1p proteins, which bind specifically to different group I introns.18 Mss116p also functions in other RNA-processing reactions and activates translation of a subset of mt mRNAs, likely by disrupting RNA structures that occlude ribosome access.16 CYT-19 and Mss116p appear largely interchangeable, with all of the defects in mss116Δ strains being suppressed by the expression of CYT-1916 and the purified recombinant proteins promoting the splicing of many of the same group I and group II introns in vitro.19–21 For both proteins, maximal stimulation of splicing requires ATP, but in some cases, ATP-independent processes also contribute.21,22
The mechanism by which CYT-19 stimulates group I intron splicing has been studied by using derivatives of the Tetrahymena thermophila LSU group I intron, whose RNA-folding pathway has been well characterized. These studies showed that CYT-19 plus ATP can resolve a long-lived misfolded intermediate whose formation is favored by a non-native base-pairing interaction denoted Alt-P3.15 Detailed analysis showed that CYT-19 preferentially unwinds loosely associated, unstable duplexes and unfolds both the native and misfolded RNAs with subsequent refolding favoring the native RNA structure, which has higher stability.10,23 Thus, CYT-19 acts on these group I intron RNAs in a manner analogous to a classical protein chaperone in protein folding.
Mss116p and CYT-19 are thought to function similarly in group II intron splicing, but the nature of the RNA structural transitions are not well characterized. Biochemical studies have focused on reactions in which CYT-19 or Mss116p + ATP promotes the splicing of the yeast group II introns aI5γ and bI1 under near physiological conditions.19–22 For bI1, protease-digestion experiments showed that CYT-19 + ATP promotes the reverse (and presumably forward) splicing of bI1 by acting as an archetypical RNA chaperone, whose continued presence is not required after RNA folding has occurred.19 For aI5γ, however, analogous tests of protease sensitivity in an RNA cleavage reaction with a ribozyme derivative known as D135 RNA indicated that the continued presence of the DEAD-box protein is required, possibly reflecting that the DEAD-box protein accelerates a folding transition that occurs after substrate binding.21 This ATP-dependent folding transition may involve disruption of stable non-native structures and/or accelerated rearrangement of a rate-limiting on-pathway intermediate.21,22 In addition to this primary function, the DEAD-box protein could also stabilize aI5γ folding intermediates or promote strand annealing, which does not require ATP, before dissociating so that final folding can occur.19,21 One report suggested that Mss116p could promote the splicing of the aI5γ group II intron without unwinding RNA,20 but this does not appear to be the case.22
The broad range of action and interchangeability of CYT-19 and Mss116p raised the questions of whether other DEAD-box proteins might have similar non-specific RNA chaperone activity and whether this activity might be assayed by their effects on group I and II intron splicing reactions. We and others reported that Ded1p, a yeast DEAD-box protein that ordinarily acts in the nucleus and cytosol, functions similarly to CYT-19 and Mss116p in promoting the splicing of the aI5γ and bI1 group II introns in an ATP-dependent manner in vitro.20,21 Both CYT-19 and Mss116p have a basic C-terminal tail that contributes to non-specific RNA binding is thought to help tether the protein to RNA substrates for repeated rounds of RNA unwinding, and Ded1p contains an analogous C-terminal tail, which may function similarly.24,25 At this point, key questions are to what extent the mechanistic characteristics of CYT-19, Ded1p, and Mss116p are shared by other DExH/D-box proteins and how they relate to RNA chaperone activity.
Here, we show that Ded1p can activate mt translation and stimulate the splicing of both group I and II introns in vivo as well as in vitro, and that CYT-19 and Ded1p can function indistinguishably as RNA chaperones to resolve diverse non-native secondary and tertiary structures in the T. thermophila LSU-ΔP5abc group I intron. We also show that the E. coli DEAD-box protein SrmB stimulates the splicing of group I and II introns in vitro, whereas the E. coli DEAD-box protein DbpA, whose unwinding activity is activated by binding a specific RNA, or the vaccinia virus DExH-box protein NPH-II, a processive RNA helicase, gave little if any stimulation of group I or II intron splicing in vivo or in vitro. Biochemical analysis indicates that unwinding by local strand separation and non-specific RNA binding, which together enable the remodeling of diverse RNA and RNP substrates, are critical for the ability of DEAD-box proteins to stimulate group I or group II intron splicing. Importantly, the ability of different DEAD-box proteins to stimulate group I and group II intron splicing in all cases examined correlates with their RNA-unwinding activity, indicating a pivotal role for this activity in RNA chaperone function.
Results
Ability of DExH/D-box proteins to complement the mt translation and RNA splicing defects in mss116Δ strains
We found previously that the defects in mt RNA splicing and translational activation in mss116Δ strains could be suppressed by the expression of CYT-19,16 and we wished to test whether Ded1p could similarly replace Mss116p in vivo. We also tested three other proteins: the E. coli DEAD-box proteins SrmB and DbpA, and the vaccinia virus DExH-box protein NPH-II. Each of these proteins differs from Ded1p in some respect: SrmB has relatively low ATPase and unwinding activity;26 DbpA’s ATPase and unwinding activity is stimulated by specific binding of a segment of E. coli 23S RNA;27,28 and NPH-II unwinds RNA by directional translocation along one of the duplex strands.29
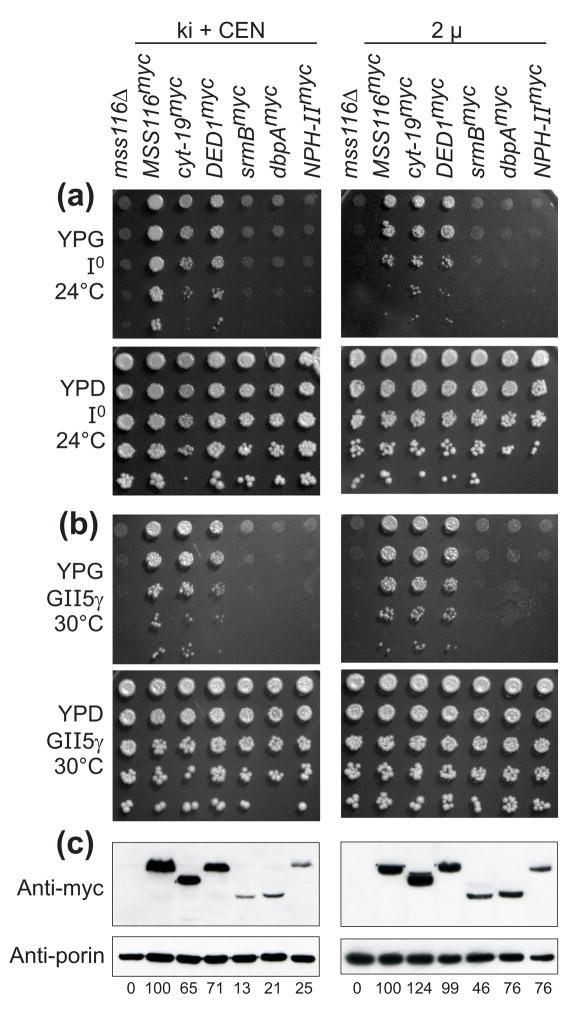
For each protein, we constructed a chimeric gene consisting of the promoter, 5′-untranslated region (UTR) and mt import sequence of MSS116 fused in-frame to the DExH/D-box protein coding sequence with a C-terminal myc tag followed by the MSS116 3′ UTR. These chimeric genes were tested for their ability to complement mss116Δ strains containing mtDNAs with different combinations of group I and II introns. In one set of strains (denoted knock-in + CEN strains), the chimeric genes were knocked in at the MSS116 locus and also expressed from the low copy centromere-based plasmid pRS416 (Figure 1, left). Immunoblots probed with an anti-myc antibody and normalized by the level of the outer membrane protein porin showed that CYT-19myc and Ded1pmyc were expressed in these strains at 65–71% the level of Mss116pmyc, while SrmBmyc, DbpAmyc, and NPH-IImyc were expressed at substantially lower levels (13–25% that of Mss116pmyc; Figure 1(c), left). For that reason, we constructed a second set of strains in which the chimeric proteins were expressed from a multicopy 2 μ plasmid (Figure 1, right). In these strains, CYT-19myc and Ded1pmyc were expressed at or above the level of Mss116pmyc, while SrmBmyc, DbpAmyc, and NPH-IImyc were expressed at 46–76% of that level (Figure 1(c), right). For each expressed DExH/D-box protein, the immunoblots showed a single predominant band of the expected size after processing of the mt import sequence, suggesting that mt import has occurred and that most or all of the protein is present in mitochondria.
Figure 1.
Ability of different DExH/D-box proteins to complement the mt translation and RNA splicing defects in S. cerevisiae mss116Δ strains. (a) and (b) Growth complementation assays using (a) mss116Δ-I0, whose mtDNA lacks introns, and (b) mss116Δ-GII-5γ, whose mtDNA contains four group I introns and the group II intron aI5γ (see Materials and Methods). Each strain expresses a different myc-tagged DExH/D-box protein from a knock-in at the MSS116 locus plus a CEN plasmid (ki + CEN; left panels) or from a multicopy 2 μplasmid (2μ; right panels). After growing cells in YNB medium at 30°C with raffinose, serial dilutions were stamped onto plates containing glycerol (YPG) or glucose (YPD) and incubated at 24°C (a) or 30°C (b). (c) Immunoblots comparing levels of myc-tagged DExH/D-box proteins expressed in mss116Δ-GII-5γ ki + CEN (left) or 2 μ (right) strains at 30°C. After blotting, the membrane was divided at the position of the 40-kDa marker, and the upper and lower halves were probed with anti-myc and anti-porin monoclonal antibodies, respectively. Porin is a mt outer membrane protein used as a loading control. The numbers beneath the lanes indicate the levels of myc-tagged DExH/D-box proteins normalized for the level of porin and expressed as a percentage of the level of Mss116pmyc. The expected molecular weight of each protein after cleavage of the 4-kDa mt import sequence is: Mss116pmyc, 73 kDa; CYT-19myc, 65 kDa; Ded1pmyc, 67 kDa; SrmBmyc, 51 kDa; DbpAmyc, 49 kDa; NPH-IImyc, 79 kDa.
Because Mss116p is required for the synthesis of mt respiratory components, mss116Δ strains are unable to grow on media containing the non-fermentable carbon source glycerol and complementation can be scored readily by restoring growth on glycerol. To test the effect of the DExH/D-box proteins on mt translation, we used an mss116Δ-I0 strain, which lacks mtDNA introns. This strain grows at wild-type rates at 30°C, but is cold-sensitive at 24°C due to impaired translation of some mt mRNAs, particularly cytochrome oxidase subunit I (COX1) and subunit III (COX3) mRNAs.16 Plating assays showed that the translation defect in the mss116Δ I0 strain at 24°C is complemented by Mss116pmyc, CYT-19myc and Ded1pmyc in both the knock-in + CEN and 2 μ strains, while SrmBmyc, DbpAmyc or NPH-IImyc have little or no effect in either set of strains (Figure 1(a)).
To test the effect of the DExH/D-box proteins on splicing of group I and II introns, we first used the strain mss116Δ-GII-5γ, which contains four group I introns (aI3 and aI4 in the COX1 gene and bI4 and bI5 in the cytochrome b (COB) gene) and one group II intron (aI5γ in COX1). This strain is unable to grow on glycerol due to impaired splicing of these introns at 30°C, a temperature at which mt translation is not dependent upon Mss116p function. Plating assays showed that the growth defect in mss116Δ GII-5γ at 30 °C is also complemented by Mss116pmyc, CYT-19myc, and Ded1pmyc, but not by SrmBmyc, DbpAmyc, or NPH-IImyc in either the ki + CEN or 2 μstrains (Figure 1(b)).
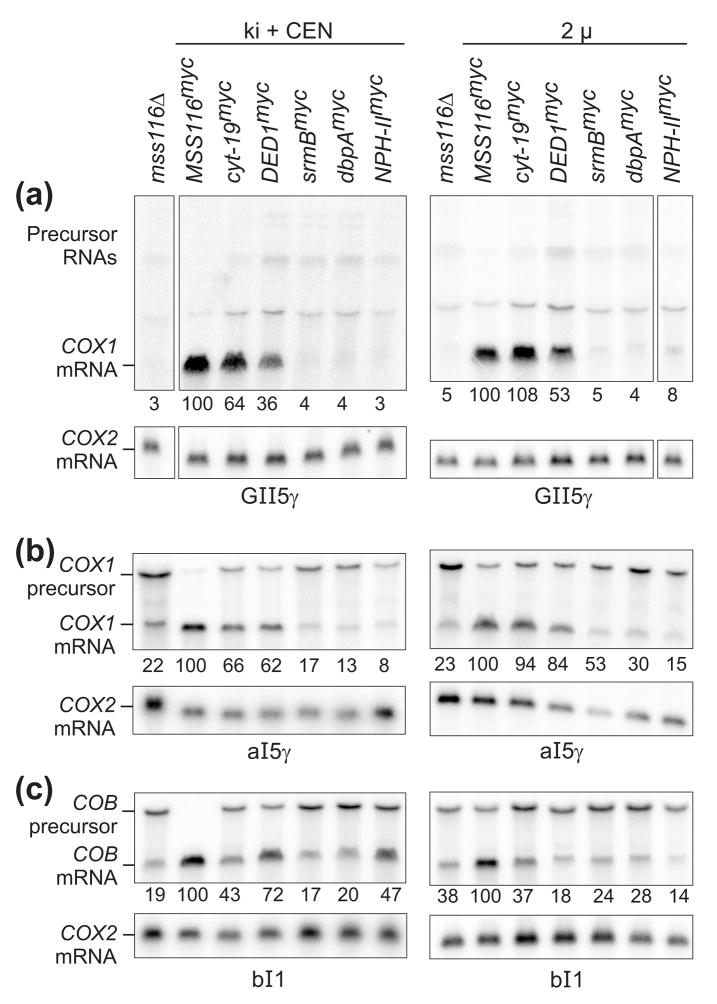
To confirm complementation of group I and II intron splicing defects in mss116Δ-GII-5γ strains, we carried out Northern hybridizations in which mt RNAs were hybridized with COX1 exon probes to detect COX1 mRNA and precursor RNAs (Figure 2(a)). The levels of COX1 mRNA (numbers beneath the blots) were normalized by the level of COX2 mRNA, whose synthesis does not require group I or II intron splicing, and expressed as a percent of that in the presence of Mss116p. The blots show that the mss116Δ-GII-5γ strain produced low levels of COX1 mRNA due to defective group I and II intron splicing, and these levels were increased by the expression of Mss116pmyc, CYT-19myc, and Ded1pmyc, but not by expression of SrmBmyc, DbpAmyc, and NPH-IImyc. The low and varying levels of COX1 precursor RNAs presumably reflect a combination of multiple precursor RNA species due to random order of intron removal and instability of large precursor RNAs containing multiple group I and II introns.
Figure 2.
Northern hybridizations comparing the ability of different DExH/D-box proteins to complement group I and group II intron splicing defects in S. cerevisiae mss116Δ strains. The mss116Δ strains contain (a) GII-5γ mtDNA, (b) mtDNA containing only aI5γ, or (c) mtDNA containing only bI1, and they express the different DExH/D-box proteins from a knock-in at the MSS116 locus plus a CEN plasmid (ki + CEN; left panels) or from a multicopy 2 μ plasmid (right panels). Strains were grown at 30°C, and RNA was isolated, run in a 1% agarose gel containing 0.24% guanidinium thiocyanate, blotted to a nylon membrane, and hybridized with 32P-labeled oligonucleotides for COX1 exon 6 to monitor splicing of introns aI3α, aI4α, and aI5γ; COB exon 6 to monitor splicing of bI1; and COX2 mRNA used as a loading control. The same blots were stripped and reprobed sequentially. The numbers beneath the lanes indicate levels of COX1 or COB mRNA normalized for the amount of COX2 mRNA and expressed as a percentage of the level of COX1 or COB mRNA in the MSS116myc lane.
Because we were particularly interested in whether the DExH/D-box proteins could stimulate splicing of the group II introns aI5γ and bI1, we did additional Northern hybridizations with mss116Δ-aI5γ and mss116Δ-bI1 strains, whose mtDNAs contain only the group II intron aI5γ or bI1, respectively. As shown in Figure 2(b) and (c), Mss116pmyc, CYT-19myc, and Ded1pmyc stimulated the splicing of both group II introns, as judged both by the ratio of precursor to mRNA and by the levels of mRNA normalized to COX2 mRNA. Notably, CYT-19myc and Ded1pmyc appeared to stimulate the splicing of bI1 in the knock-in + CEN strain, but not in the 2 μ strain, suggesting that an optimal level of DEAD-box protein expression may be required for splicing stimulation. SrmBmyc and DbpAmyc had little or no effect on the level of COX1 or COB mRNA in either the knock-in + CEN or 2 μ strain, while NPH-IImyc had no effect on the level of COX1 mRNA in the 2 μ strains, but increased the level of COB mRNA in the knock-in + CEN strain. This increase in COB mRNA levels was observed in Northern blots with four independent RNA preparations from bI1 and GII-5γ knock-in + CEN or CEN strains expressing NPH-II (not shown). However, in vitro splicing assays below show that NPH-II tested over a range of protein concentrations does not promote the splicing of bI1. Thus, the increase in COB mRNA levels in Northern blots could be due to an effect on some other process, such as an increased rate of transcription or a decreased rate of mRNA turnover.
For all introns tested, the cognate DEAD-box protein Mss116pmyc complemented the RNA splicing defect to a greater degree than the non-cognate DEAD-box proteins CYT-19myc and Ded1pmyc, even when expressed at similar levels as in the 2 μ strains, in agreement with previous results for CYT-19myc.16 The ability of Ded1p to complement the mt translation and group I and II intron splicing defects in mss116Δ strains demonstrates that it can act on native RNP complexes in vivo. Because SrmBmyc, DbpAmyc, and NPH-IImyc are expressed at lower levels than Mss116pmyc, CYT-19myc, and Ded1pmyc, we cannot distinguish whether their smaller effects in vivo are due to inability to function efficiently in S. cerevisiae mt translation and RNA splicing or to their lower expression levels. Also, as alluded to above, although increased steady-state mRNA levels in vivo are consistent with complementation of an RNA splicing defect, without supporting biochemical data we cannot exclude that effects on other processes, such as transcription or mRNA turnover, contribute to the increase in mRNA levels.
Ability of different DExH/D-box proteins to stimulate group I intron splicing in vitro
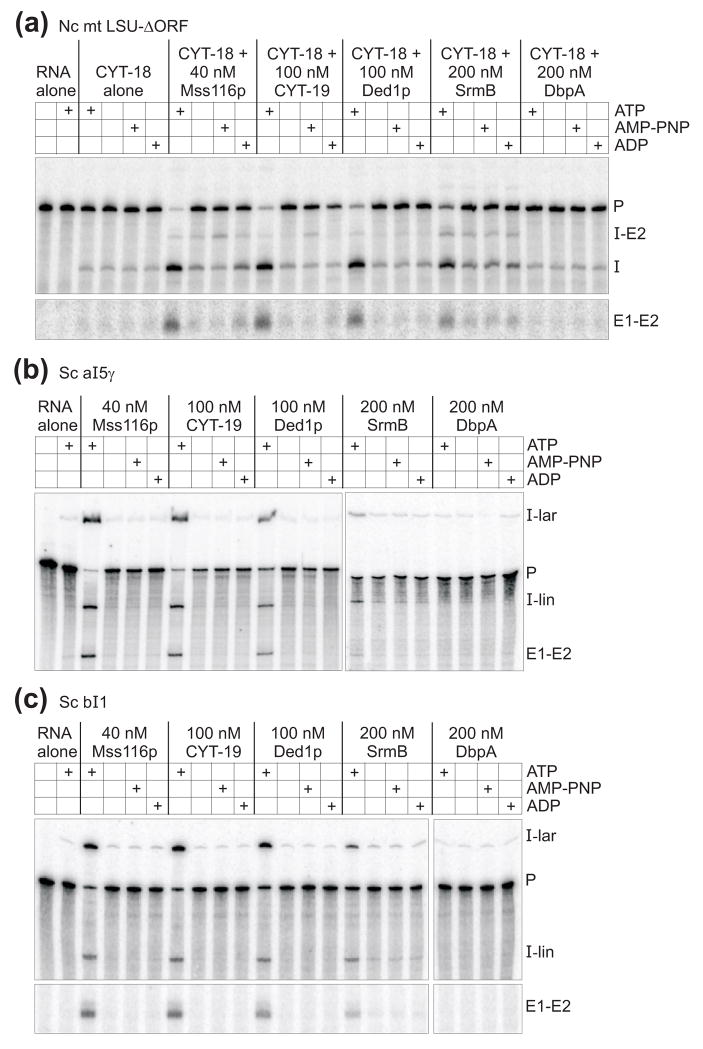
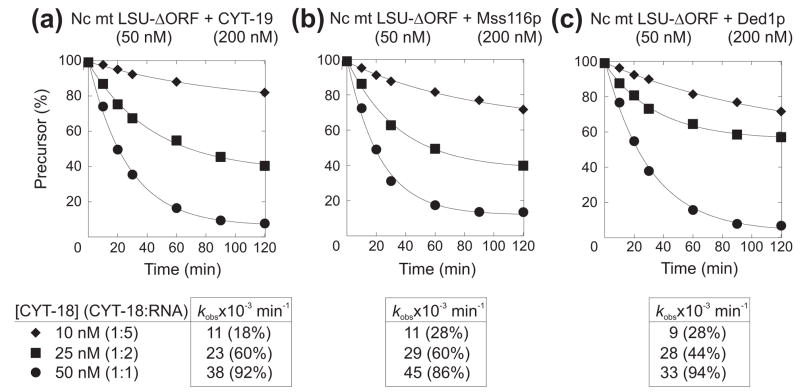
To analyze the effects of the different DExH/D-box proteins on RNA splicing in detail, we compared their ability to stimulate the splicing of group I and group II introns at different protein concentrations in vitro. For these experiments, we purified each protein by an established method and confirmed that the protein preparation has the expected RNA-dependent ATPase and RNA-duplex unwinding activities (see below). First, we analyzed the effect of DExH/D-box proteins on CYT-18-dependent splicing of the N. crassa mt LSU-ΔORF group I intron. Figure 3(a) shows splicing reactions in which 10 nM 32P-labeled precursor RNA was incubated with 40 nM CYT-18 and saturating concentrations of DEAD-box proteins (see Figure 4 and Table 2 below) for 120 min at 25°C. The reactions were done in the presence of 1 mM GTP and either ATP, ADP, or the non-hydrolyzable ATP analog AMP-PNP. Because high concentrations of NPH-II inhibited splicing and caused RNA degradation after 120-min incubation, we do not show end-point splicing reactions for this protein in Figure 3, but instead show time courses done at different protein concentrations in Supplementary Figure 1. We also tried to assay splicing in the presence of ADP-BeFx, which favors a protein conformation that promotes RNA unwinding without ATP-hydrolysis.14 We found, however, that BeCl2, which is added to form the non-covalent complex, by itself strongly inhibits both group I and II intron splicing, possibly by competing out essential Mg2+ ions within the intron RNA (not shown).
Figure 3.
Ability of different DEAD-box proteins to stimulate the in vitro splicing of group I and group II introns. (a) Group I intron splicing. Splicing reactions with the N. crassa mt LSU-ΔORF intron were done with 10 nM 32P-labeled precursor RNA and 40 nM CYT-18 dimer in reaction medium containing 100 mM KCl, 5 mM MgCl2, and 1 mM GTP for 120 min at 25°C. (b) and (c) Group II intron splicing. Splicing reactions with the S. cerevisiae group II introns aI5γ and bI1 were done with 10 nM 32P-labeled precursor RNA in reaction medium containing 100 mM KCl and 8 mM MgCl2 for 120 min at 30°C. DEAD-box proteins Mss116p, CYT-19, Ded1p, SrmB, and DbpA were added at the concentrations indicated, and ATP, ADP, and AMP-PNP were added at 1 mM. Products were analyzed by electrophoresis in a denaturing 4% polyacrylamide gel, which was dried and scanned with a phosphorimager. The first lane of each gel shows the precursor RNA immediately before it was added to initiate splicing reactions. Images in panels (a) and (c) are parts of the same gel, while those in panel (b) are from two different gels.
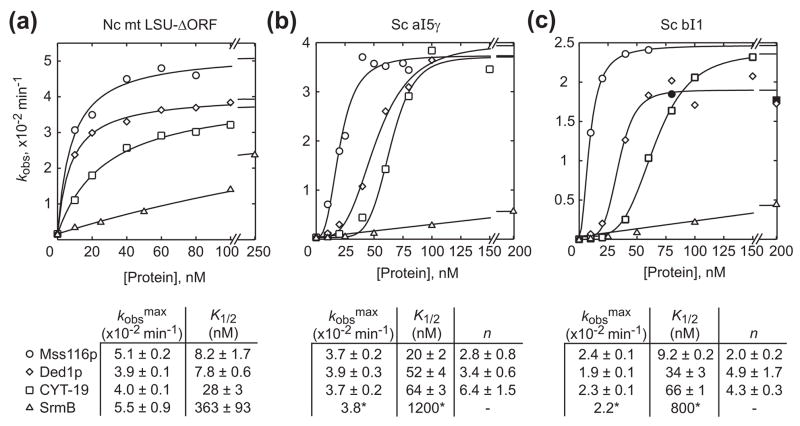
Figure 4.
Dependence of group I and II intron splicing rate constants on DEAD-box protein concentration. The plots show the observed rate constant (kobs) for (a) N. crassa mt LSU-ΔORF, (b) S. cerevisiae aI5γ, or (c) S. cerevisiae bI1 precursor RNA disappearance as a function of Mss116p, CYT-19, Ded1p, and SrmB concentration. The rate constants were calculated from the initial rates in splicing time courses (Supplementary Figure 2), with reactions done in the presence of 1 mM ATP under the same conditions as in Figure 3. The data for the N. crassa mt LSU-ΔORF intron were best fit by a hyperbolic function for all proteins tested. The data for S. cerevisiae aI5γ and bI1 were best fit by a sigmoidal function for all proteins, except SrmB, for which a hyperbolic fit was used. The maximum observed rate constants (kobsmax), protein concentrations giving half-maximal activation (K1/2), and Hill coefficients (n) along with standard errors of the fits are summarized beneath the plots. For SrmB stimulation of aI5γ and bI1 splicing, K1/2 values indicated with a * were estimated by fixing kobsmax at the mean of those for the other proteins. Filled symbols represent data not used in curve fitting because of splicing inhibition at higher protein concentrations. The data for Mss116p were taken from Mohr et al.25 and replotted with some newer data. Each data point is the mean of at least two independent experiments.
Table 2.
Equilibrium binding of DEAD-box proteins to group I and II intron RNAs.
| Protein | Nucleotide |
Kd (nM) |
|||
|---|---|---|---|---|---|
| Nc mt LSU-ΔORF | Tt LSU-ΔP5abc | Sc aI5γ | Sc bI1 | ||
| Mss116p | -NTP | 8 ± 2 | 14 ± 3 | 8 ± 2 | 11 ± 3 |
| AMP-PNP | 7 ± 1 | 11 ± 3 | 2 ± 1 | 3 ± 1 | |
| ATP | 13 ± 4 | 21 ± 6 | 4 ± 1 | 9 ± 2 | |
| ADP | 10 ± 2 | 16 ± 3 | 5 ± 1 | 9 ± 3 | |
| CYT-19 | -NTP | 52 ± 9 | 48 ± 16 | 113 ± 19 | 63 ± 9 |
| AMP-PNP | 12 ± 2 | 9 ± 2 | 8 ± 1 | 16 ± 2 | |
| ATP | 35 ± 4 | 27 ± 6 | 83 ± 17 | 75 ± 25 | |
| ADP | 51 ± 11 | 28 ± 5 | 102 ± 19 | 120 ± 27 | |
| Ded1p | -NTP | 24 ± 2 | 51 ± 14 | 87 ± 19 | 206 ± 51 |
| AMP-PNP | 36 ± 10 | 33 ± 12 | 51 ± 13 | 72 ± 14 | |
| ATP | 30 ± 10 | 32 ± 14 | 98 ± 21 | 111 ± 24 | |
| ADP | 43 ± 11 | 62 ± 24 | 95 ± 17 | 169 ± 44 | |
| SrmB | -NTP | 10 ± 1 | 17 ± 5 | 39 ± 6 | 83 ± 28 |
| AMP-PNP | 9 ± 2 | 15 ± 6 | 30 ± 6 | 33 ± 8 | |
| ATP | 7 ± 1 | 13 ± 5 | 40 ± 10 | 52 ± 11 | |
| ADP | 10 ± 1 | 13 ± 5 | 120 ± 29 | 84 ± 21 | |
| DbpA | -NTP | 359 ± 91 | 139 ± 14 | 327 ± 75 | 520 ± 286 |
| AMP-PNP | 85 ± 27 | 214 ± 39 | 178 ± 51 | 412 ± 124 | |
| ATP | 172 ± 22 | 132 ± 14 | 147 ± 40 | 612 ± 219 | |
| ADP | 256 ± 52 | 142 ± 42 | 171 ± 50 | 917 ± 353 | |
32P-labeled RNAs (5 pM) were incubated with increasing concentrations of protein in splicing reaction medium containing 100 mM KCl and 5 or 8 mM MgCl2 for group I and group II introns, respectively, for 60 min at 30°C with or without the indicated nucleotides and then filtered through nitrocellulose backed by nylon (see Materials and Methods). The percent bound RNA was plotted as a function of the protein concentration. The data were best fit by a hyperbolic function using Kaleidagraph (Synergy Software) to obtain Kd ± the standard error of the fit. The Kd values are from a single experiment in which all proteins and conditions were done in parallel using RNAs from a single transcription. In each case, essentially identical binding curves were obtained with incubation times of 15 min (SrmB and DbpA) or 15 and 90 min (Mss116p, CYT-19, Ded1p; data not shown). Representative binding curves are shown in Supplementary Figure 4.
As found previously, both Mss116p and CYT-19 strongly stimulated the CYT-18-dependent splicing of the N. crassa mt LSU-ΔORF intron in an ATP-dependent manner (Figure 3(a)). Among the other proteins, Ded1p and SrmB also stimulated splicing of this intron, whereas DbpA did not detectably stimulate splicing and NPH-II at best stimulated minimally at any protein concentration tested (Figure 3(a) and Supplementary Figure 1(a); DbpA tested at concentrations up to 1 μM; data not shown). Unlike DEAD-box proteins, NPH-II can hydrolyze GTP,30 which is added as the group I intron splicing cofactor. To exclude the possibility that this GTPase activity obscures group I intron splicing stimulation by NPH-II, initial experiments with GTP (not shown) were supplemented by splicing time courses with 1 mM GMP (Supplementary Figure 1(a)). For Mss116p, CYT-19, Ded1p, and SrmB, we observed maximal group I intron splicing stimulation in the presence of ATP, but all four proteins gave a small degree of stimulation in the absence of nucleotide or the presence of AMP-PNP or ADP. We previously described ATP-independent stimulation of CYT-18-dependent mt LSU-ΔORF splicing by Mss116p, which appeared to be greatest in the presence of ADP.21 For SrmB, however, the ATP-independent stimulation appeared to be the same whether or not AMP-PNP or ADP was present (Figure 3(a)). As discussed previously,21 such ATP-independent effects could reflect a low level of RNA chaperoning by cycles of single-strand capture and dissociation, a mechanism invoked to account for RNA chaperoning by non-specific RNA-binding proteins, or a contribution from other DEAD-box protein activities, such as non-specific RNA binding or strand annealing.
To compare quantitatively the ability of Mss116p, CYT-19, Ded1p, and SrmB to stimulate CYT-18-dependent group I intron splicing, we carried out a series of splicing time courses at different protein concentrations for each DEAD-box protein (Supplementary Figure 2) and plotted the observed rate constant (kobs) for precursor RNA disappearance as a function of protein concentration (Figure 4(a)). For each protein, the data from these single-turnover reactions were best fit by a hyperbolic function, which defines the apparent second-order rate constant for substrate binding and conversion to product (kobsmax/K1/2). The latter equals kcat/Km measured under multiple turnover conditions so long as the reaction favors product formation. Comparison of the kcat/Km values estimated in this way showed that Mss116p is the most efficient stimulator of mt LSU-ΔORF splicing, followed by Ded1p, CYT-19 and SrmB (kcat/Km values = 6.2 × 106, 5.0 ×106, 1.4 ×106, and 1.5×105 M−1 min−1, respectively). The differences in efficiency appear to be due largely to differences in K1/2, as the maximum observed rates (kobsmax) are similar. We were unable to achieve saturation with SrmB because the protein inhibited splicing at concentrations greater than 250 nM (data not shown).
Ability of different DExH/D-box proteins to stimulate group II intron splicing in vitro
Figures 3(b) and (c) show 120-min splicing reactions for the S. cerevisiae group II introns aI5γ and bI1 done by incubating 10 nM 32P-labeled precursor RNA with saturating concentrations of DEAD-box proteins at 30°C. As reported previously,21 CYT-19, Mss116p, and Ded1p stimulated the splicing of both aI5γ and bI1 in an ATP-dependent manner. Notably, SrmB + ATP also stimulated the splicing of both of these introns despite failing to do so in vivo, and SrmB was the only DEAD-box protein that showed some albeit barely detectable ATP-independent stimulation. Again, DbpA gave no detectable splicing stimulation and NPH-II at best stimulated minimally at any protein concentration tested (Figure 3(b) and (c) and Supplementary Figure 1(b) and (c); DbpA tested at concentrations up to 1 μM; data not shown). The ATP-independent effects of SrmB on group II intron splicing may reflect the same processes discussed above for ATP-independent effects in group I intron splicing.
Figures 4(b) and (c) show plots of kobs versus protein concentration from splicing time courses at different protein concentrations for the same two group II introns. By contrast to the hyperbolic curves for group I intron splicing, the plots for both aI5γ and bI1 show sigmoidal dependences on Mss116p, Ded1p, and CYT-19 concentration (Figure 4(b) and (c)). Although we previously used linear or hyperbolic fits for Mss116p splicing stimulation data for these introns,22 sigmoidal behavior is apparent from more measurements. The Hill coefficients for the curve fits range from 2 to 6, indicating that stimulation of group II intron splicing by Mss116p, Ded1p, and CYT-19 requires the cooperative participation of two or more molecules of DEAD-box protein per intron. For all three proteins the kobsmax values are similar (~4 ×10−2 min−1 for aI5γ and ~2 ×10−2 min−1 for bI1) with the magnitude of the K1/2 values indicating the same order of efficiency as for group I intron splicing (Mss116p > Ded1p > CYT-19 > SrmB). We could not achieve saturation with SrmB (as concentrations above 200 nM caused splicing inhibition), and so could not determine if SrmB displayed sigmoidal behavior in group II intron splicing. Because the other proteins reach similar kobsmax values on mt LSU-ΔORF, aI5γ, and bI1, and because SrmB likely reaches the same kobsmax on the N. crassa mt LSU-ΔORF intron, we fit the data for SrmB splicing of aI5γ and bI1 with a hyperbola with a kobsmax fixed at the mean for the other proteins in order to obtain a rough estimate for K1/2. Based on these estimates, SrmB appears about 20- and 12-fold less efficient than the next least active DEAD-box protein CYT-19 in stimulating aI5γ and bI1 splicing, respectively (similar to the ~13-fold difference between the two for the N. crassa mt LSU-ΔORF group I intron). The above findings supplant previous results with different protein preparations in which Ded1p stimulated bI1 splicing less efficiently than CYT-19 and SrmB stimulation of group I or II intron splicing was only barely detectable.21
Together, these findings show that Mss116p, CYT-19, Ded1p, and SrmB stimulate the splicing of both group I and II introns to different degrees in vitro, while DbpA and NPH-II are unable to stimulate splicing of these introns appreciably. The finding that the stimulation of aI5γ and bI1 group II intron splicing shows a sigmoidal dependence on DEAD-box protein concentration, while the stimulation of N. crassa mt LSU-ΔORF group I intron splicing shows a hyperbolic dependence could reflect that the required RNA structural transitions are more difficult in the group II introns than in the group I intron, requiring the action of multiple promoters acting together or sequentially.
DEAD-box proteins do not dissociate CYT-18 bound to group I introns
The CYT-18-protein binds tightly and stoichiometrically to group I intron RNAs and remains tightly bound to the excised intron RNA after splicing has occurred, so that splicing is effectively limited to one round of catalysis.31 Given that some DExH/D-box proteins have RNPase activity,32,33 it was possible that the observed stimulation of group I intron splicing simply reflects that they stimulate turnover of CYT-18 from the excised intron RNA. We reported previously that this was not the case for CYT-19,15 but it remained possible for the other DEAD-box proteins. To address this issue, we carried out splicing reactions with 50 nM N. crassa mt LSU-ΔORF RNA, stoichiometric or substoichiometric concentrations of CYT-18 dimer, and excess CYT-19, Mss116p, or Ded1p, which give the greatest splicing stimulation for this intron. Figure 5 shows that in all cases, the amount of precursor RNA spliced was equal to the amount of CYT-18-dimer within experimental error, indicating that none of these DEAD-box proteins stimulates group I intron splicing by enabling rapid CYT-18 turnover from the excised intron RNA. For unknown reasons, perhaps related to the presence of saturating concentrations of DEAD-box proteins, the rate of splicing also increased with increasing CYT-18 concentration, in contrast to similar experiments in the absence of a DEAD-box protein.31
Figure 5.
DEAD-box proteins do not dissociate CYT-18 from a group I intron RNA. Splicing reactions with the N. crassa mt LSU-ΔORF intron were carried out with 50 nM 32P-labeled precursor RNA, indicated amounts of CYT-18 dimer, and 200 nM CYT-19 (a), Mss116p (b), or Ded1p (c) in reaction medium containing 100 mM KCl, 5 mM MgCl2, plus 1 mM GTP and ATP at 25°C. The plots show disappearance of precursor RNA as a function of time with the data fit to a single exponential equation. Rate constants (kobs) and amplitudes (% precursor RNA spliced at 120 min) are summarized below the plots. The amplitudes are equal to molar ratio of CYT-18 dimer to precursor RNA (20, 50 or 100%) within error. In this experiment, splicing was initiated by adding CYT-18.
Comparison of DExH/D-box protein ATPase activities
RNA-stimulated ATPase activity provides a measure of whether a given RNA is bound productively by an RNA helicase. Table 1 shows ATP hydrolysis rate constants for the different DEAD-box proteins in the presence of group I and II intron RNAs or a 172-nt fragment of E. coli 23S rRNA (nts 2454–2625) containing the DbpA target helix. The assays were done under in vitro splicing conditions at saturating ATP and RNA concentrations. Although the ATPase activities of all of these proteins have been measured previously under a variety of different reaction conditions, for comparative purposes here, it was important to measure them in parallel under in vitro splicing conditions.
Table 1.
RNA-dependent ATPase activity of DEAD-box proteins.
| [Mg2+] | RNA |
kcat (min−1) |
||||
|---|---|---|---|---|---|---|
| Mss116p | CYT-19 | Ded1p | SrmB | DbpA | ||
| 5 or 8 | No RNA | n.d.a | n.d. | n.d. | n.d. | n.d. |
| 5 | Nc mt LSU-ΔORF | 180 ± 24 | 148 ± 50 | 199 ± 30 | 30 ± 5 | n.d. |
| 5 | Tt LSU-ΔORF | 119 ± 17 | 107 ± 52 | 144 ± 46 | 64 ± 27 | n.d. |
| 8 | Sc aI5γ | 168 ± 10 | 142 ± 35 | 154 ± 10 | 33 ± 12 | n.d. |
| 8 | Sc bI1 | 172 ± 29 | 109 ± 27 | 116 ± 9 | 25 ± 5 | n.d. |
| 5 | Ec 23S rRNA (2454–2625)b | 132 ± 12 | 125 ± 64 | 160 ± 10 | 21 ± 11 | 245 ± 47 |
ATPase activity was assayed in splicing reaction medium containing 100 mM KCl, 5 or 8 mM MgCl2 as indicated, 10 mM Tris-HCl, pH 7.5, and 10% glycerol at 30 C with 25 nM Mss116p, CYT-19, or Ded1p, and 25 or 50 nM SrmB or DbpA in the presence of saturating unlabeled ATP (3 mM) and RNA (300 nM). kcat values were determined by measuring the initial rate (r0), corresponding to < 1% of final product in a 90-min time course at saturating ATP and RNA concentrations. All reactions were linear for ≥10 min. kcat values are the mean ± the standard deviation for at least three measurements. kcat values for Mss116p with the mt LSU-ΔORF, aI5γ, and bI1 intron RNAs were reported previously.22,25
n.d., no detectable ATPase activity, kcat < 1 min−1.
172-nt segment of E. coli 23S rRNA (residues 2454-2625), containing the DbpA target helix.54
The ATPase activities of Mss116p, CYT-19, Ded1p, and SrmB were stimulated by all group I and II intron RNAs tested, as well as by the DbpA target helix, with each protein showing no significant difference in activity with the different RNAs. The kcat values for Mss116p, CYT-19, and Ded1p were similar to each other within error (107–199 min−1) and comparable to values determined previously for Mss116p (~100 min−1),21 CYT-19 (~55 min−1; using a different method for protein preparation),15 and Ded1p (120–600 min−1).9 The kcat values for SrmB’s ATPase activity were several fold lower than those for the other DEAD-box proteins (21–64 min−1), also in agreement with published values (20–40 min−1).9
DbpA is a targeted DEAD-box protein that acts on a specific helix in E. coli 23S rRNA.27,28,34 As expected from previous work,28,34 its ATPase activity was not stimulated by group I or group II intron RNAs, but was stimulated by the E. coli 23S rRNA segment containing its target helix (Table 1).
Finally, NPH-II is a processive RNA helicase with a high basal ATPase activity.30 Because it was only available in smaller amounts than the other proteins, we assayed its ATPase activity by carrying out time courses using 1 nM protein and 200 nM RNA in splicing reaction medium at 30°C. Under these conditions, the kobs calculated from the initial rate was 5.1 × 103 min−1 in the absence of RNA, and it increased two- to three-fold in the presence of 200 nM group I or group II intron RNAs (kobs = 1.7 × 104 min-1, 1.8 × 104 min−1, and 1.2 × 104 min−1 with the N. crassa mt LSU-ΔORF, aI5γ, and bI1 introns, respectively; Supplemental Figure 3). These results indicate that NPH-II can interact productively with group I and II intron RNAs in a manner that activates its ATPase and presumably RNA-unwinding activities.
Together, the above findings show that the ATPase activity of the DEAD-box proteins Mss116p, CYT-19, Ded1p, and SrmB is stimulated by the binding of all group I and II intron RNAs tested with no apparent specificity, in agreement with previous findings of non-specific stimulation of their ATPase activity by other RNAs,9,15,21 while DbpA differs in that its ATPase activity is stimulated only by an RNA containing its target helix and not by group I or group II intron RNAs. The ATPase activity of NPH-II is also stimulated by group I and group II intron RNAs, suggesting that NPH-II can unwind helices in these RNAs, even though it does not promote their splicing. The findings for NPH-II also demonstrate that RNA-stimulated ATPase activity by itself does not necessarily correlate with RNA splicing activity.
Comparison of DEAD-box protein RNA-binding affinity
To assay directly the binding of group I and II intron RNAs by the DEAD-box proteins, we carried out equilibrium binding assays under in vitro splicing conditions in the presence or absence of ATP, ADP, and AMP-PNP (Table 2). Although binding assays had been done previously for some of these proteins, it was again important to compare them in parallel assays under in vitro splicing conditions. We note that the dissociation constants (Kds) for these large RNAs are expected to be a composite for interactions involving the helicase core and other protein regions, such as the C-terminal α-helical region and basic tail in CYT-19 and Mss116p.
As we found previously for CYT-19 and Mss116p,19,21 the binding curves for all the DEAD-box proteins with all the group I and II intron RNAs tested were equivalently fit to a hyperbolic function or to the Hill equation with n close to 1, indicating that binding is not cooperative (Supplementary Figure 4 and data not shown). Among the four DEAD-box proteins that promote group I and group II intron splicing, Mss116p binds group I and II intron RNAs the tightest (Kds = 7 to 21 nM for group I intron RNAs and 2 to 11 nM for group II intron RNAs) and shows only a small decrease in Kd in the presence of AMP-PNP, in agreement with previous results.21 CYT-19 binds more weakly in the absence of nucleotides or presence of ATP or ADP (Kds = 27 to 52 nM for group I intron RNAs and 63 to 120 nM for group II intron RNAs), but shows a more pronounced decrease in Kd in the presence of AMP-PNP to Kds in the range 8 to 12 nM, comparable to those for Mss116p, in agreement with previous results.15,21,25 These differences between CYT-19 and Mss116p could reflect that CYT-19 undergoes a larger conformational change in the presence of AMP-PNP under the conditions of the assay or that Mss116p relies to a greater extent on regions other than the helicase core for tight binding of group I and group II intron RNAs. Notably, Mss116p and CYT-19 show little or no preference for binding their natural RNA substrate (Mss116p for aI5γ or bI1 or CYT-19 for the N. crassa mt LSU-ΔORF) compared to other intron RNAs.
Ded1p and SrmB, which also stimulate group I and II intron splicing, bind to group I and II intron RNAs with Kds in the same ranges as Mss116p and CYT-19, with little or no decrease in the presence of AMP-PNP in most cases. SrmB appears to show some preference for binding group I rather than group II introns RNAs, with Kds for group I introns ranging from 7 to 17 nM, comparable to binding group I intron RNAs by Mss116p. DbpA, which does not stimulate RNA splicing, binds the group I and group II intron RNAs with Kds ranging from 85 to 917 nM, substantially higher than those for the other DEAD-box proteins in most cases.
Importantly, under in vitro splicing conditions, Mss116p, CYT-19, and Ded1p bind to group I and group II intron RNAs with Kd values that are similar to their K1/2 values for promoting splicing of these RNAs (see Figure 4). Thus, the K1/2 values for these proteins may reflect their functional equilibrium binding to the intron RNAs. By contrast, SrmB binds group I and group II intron RNAs with Kd values that are much lower than the K1/2 values for promoting splicing (~36 fold for group I introns and between 10- and 40-fold for group II introns). The latter finding shows that binding affinity is not correlated with splicing activity and suggests that many binding events by SrmB are non-productive, so that a higher concentration of the protein is required to promote splicing. As discussed below, this inefficient splicing stimulation by bound SrmB appears to be due to its relatively low RNA-unwinding activity. The finding that the binding curves are hyperbolic presumably reflects that association of a single protomer is sufficient to cause the RNA to be retained on the filter, regardless of whether multiple promoters act cooperatively in RNA splicing, as appears to be the case for group II introns (see above).
RNA unwinding by DExH/D-box proteins under splicing conditions
To quantitatively correlate the RNA-unwinding activities of the different DExH/D-box proteins with their in vitro splicing activities, we measured duplex unwinding by the different proteins under in vitro splicing conditions using a model substrate containing a 10-bp duplex and 25-nt unpaired RNA 3′ to the duplex region. This substrate was chosen because it allowed us to determine and thus compare unwinding rate constants for all proteins. Unwinding time courses were measured over a range of protein concentrations, and the observed unwinding rate contents were calculated and plotted as a function of protein concentration.35
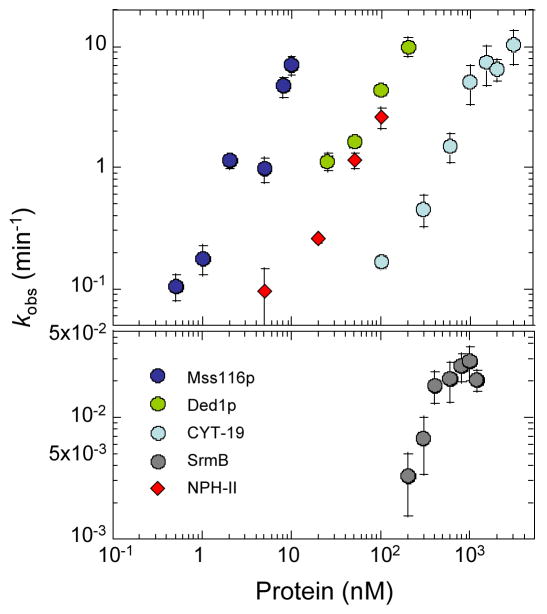
The DExH/D-box proteins unwound the substrate under in vitro splicing conditions at significantly different concentrations (Figure 6). For the DEAD-box proteins tested, comparable unwinding rate constants of kobs ~ 1 min−1 were measured for Mss116p at ~ 3 nM, for Ded1p at ~25 nM, and for CYT-19 at ~ 500 nM. SrmB showed lower unwinding activity, consistent with previous results.26 At saturating protein concentration, kobsmax for SrmB was 0.026 min−1, whereas the unwinding rate constants at similar or lower concentrations of the other DEAD-box proteins exceeded kobs > 10 min−1, the limit for reliable measurements by the methods used here. The DExH-box protein NPH-II unwound the substrate under these reaction conditions with a concentration dependence similar to that of Ded1p, while DbpA showed no ATP-dependent unwinding activity with this model duplex (not shown), as expected since it does not match its normal target site.
Figure 6.
Unwinding activity of DExH/D-box proteins. Unwinding reactions for Mss116p, Ded1p, CYT-19, SrmB, and NPH-II were done with a substrate containing a 10-bp duplex and a 25-nt single-stranded overhang 3′ to the duplex region in reaction medium containing 100 mM KCl and 5 mM Mg2+ at 30°C (see Materials and Methods). Observed unwinding rate constants (kobs) in the presence of Mss116p (blue spheres), Ded1p (green spheres), CYT-19 (light blue spheres), SrmB (grey spheres, lower panel) and NPH-II (red diamonds) were plotted as a function of protein concentrations, using a double-logarithmic scale.
Taken together, the biochemical assays show that the efficiency of the DEAD-box proteins Mss116p, CYT-19, Ded1p, and SrmB in stimulating either group I or group II intron splicing correlates with their RNA-unwinding activity. The failure of the targeted DEAD-box protein DbpA to stimulate group I and II intron splicing reflects that its RNA-unwinding activity is activated only by binding of its specific RNA substrate. Importantly, the results for NPH-II show that RNA-unwinding activity per se is not sufficient to stimulate group I or II intron splicing unless it has mechanistic features specific to the DEAD-box protein family (i.e., unwinding by local strand separation). The finding that the unwinding rate constants for a single, isolated duplex are higher than those for stimulation of group I or II intron RNA splicing is not surprising, as the latter likely requires the unwinding of multiple duplexes, some of which may have inherently higher stability than the model duplex or be stabilized by tertiary structure in the context of the large RNA.
CYT-19 and Ded1p function similarly to rescue RNA-folding mutations
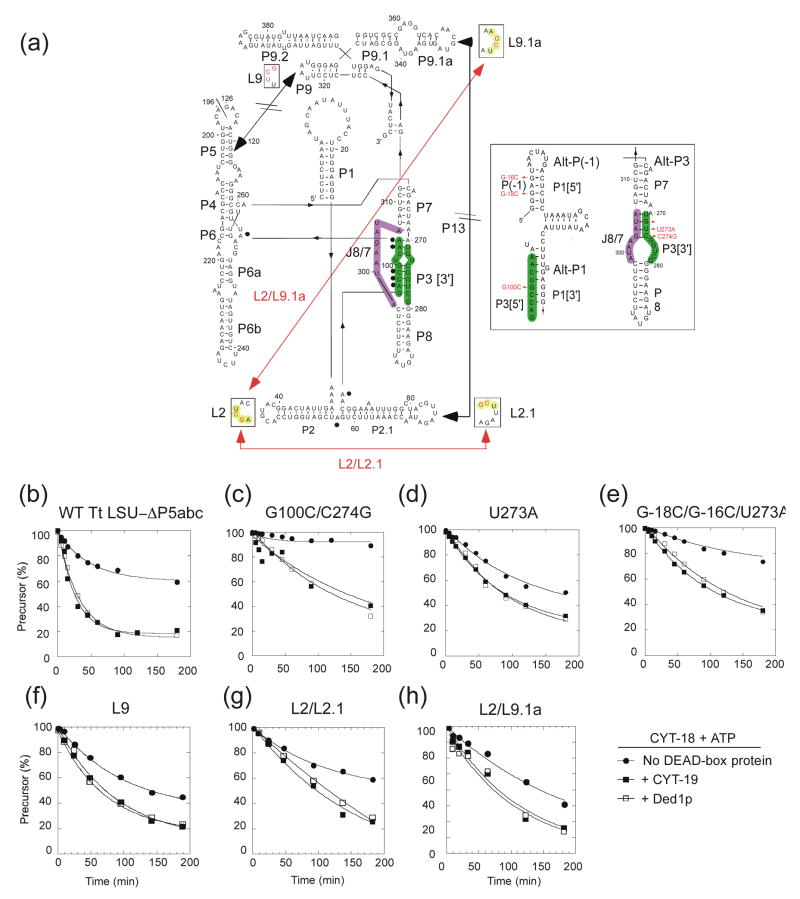
DEAD-box proteins that function as general RNA chaperones should be able to resolve diverse kinetic traps. To test this prediction, we used the Tetrahymena thermophila LSU-ΔP5abc intron, whose splicing at low Mg2+ concentrations is dependent upon CYT-18 for structural stabilization (Figure 7).36 The T. thermophila LSU-ΔP5abc intron has a propensity to form a non-native secondary structure in which sequences from P3 form alternate pairings with P1[3′] and J8/7 (Alt-P(−1) and Alt-P3, respectively; Figure 7(a), inset right), which bias the intron toward a long-lived misfolded state.37–40 We showed previously that CYT-19 + ATP stimulates the splicing of T. thermophila LSU-ΔP5abc by disrupting this non-native structure, enabling the intron RNA to fold into the active structure for RNA splicing.15
Figure 7.
Ability of CYT-19 and Ded1p to resolve kinetic traps in the T. thermophila LSU-ΔP5abc intron. (a) Predicted secondary structure of the T. thermophila LSU-ΔP5abc intron. The inset shows the predicted alternative pairings Alt-P(−1) and Alt-P3. Mutations that influence the formation of alternative secondary structures or perturb tertiary interactions are indicated in red, with red lines indicating potential non-native tertiary interactions. Sequences involved in the P3 pairing are highlighted in green; sequences involved in J8/7 are highlighted in purple; and sequences involved in loop-loop pairings in mutants are highlighted in yellow. (b) RNA splicing assays. Reactions were done with 20 nM 32P-labeled precursor RNA and 30 nM CYT-18 in the presence or absence of 200 nM CYT-19 or Ded1p at 30°C and were initiated by adding CYT-18 protein. Reactions media contained 1 mM ATP and GTP, 100 mM KCl, and 5 mM (b) to (e) or 10 mM (f) to (h) Mg2+. The products were analyzed by electrophoresis in a denaturing 6% polyacrylamide gel, which was dried and scanned with a phosphorimager. The plots show disappearance of precursor RNA as a function of time, with the data fit to a single exponential. kobs values and amplitudes (percent precurosr RNA spliced at 180 min) were: (b) CYT-18 alone, 0.02 min−1, 38%; + CYT-19, 0.04 min−1, 80%; + Ded1p, 0.03 min−1, 80%; (c) CYT-18 alone, 0.003 min−1, 10%; + CYT-19, 0.007 min−1, 60%; + Ded1p, 0.005 min−1, 70%; (d) CYT-18 alone, 0.008 min−1, 50%; + CYT-19, 0.01 min−1, 70%; + Ded1p, 0.01 min−1, 70%; (e) CYT-18 alone, 0.007 min−1, 20%; + CYT-19, 0.01 min−1, 65%; + Ded1p, 0.007 min−1, 65%; ((f) CYT-18 alone, 0.016 min−1, 52%; + CYT-19, 0.018 min−1, 80%; + Ded1p, 0.022 min−1, 80%; (g) CYT-18 alone, 0.013 min−1, 40%; + CYT-19, 0.009 min−1, 75%; + Ded1p, 0.008 min−1, 75%; (h) CYT-18 alone, 0.006 min−1, 60%; + CYT-19, 0.018 min−1, 75%; + Ded1p, 0.014 min−1, 80%.
First, we compared the ability of CYT-19 and Ded1p to resolve the long-lived non-native structure by assaying CYT-18-dependent splicing of the wild-type T. thermophila LSU-ΔP5abc group I intron. The splicing reactions were done with 20 nM precursor RNA, 30 nM CYT-18 and saturating (200 nM) CYT-19 or Ded1p in reaction medium containing 100 mM KCl and 5–10 mM Mg2+ plus 1 mM GTP and 1 mM ATP. (We could not test Mss116p because it inhibits splicing of the T. thermophila LSU-ΔP5abc intron.)21 Figure 7(b) shows that CYT-19 and Ded1p were equally efficient in stimulating CYT-18-dependent splicing of the T. thermophila LSU-ΔP5abc intron, indicating equivalent ability to resolve the inactive structure at saturating protein concentration.
Next, we tested a series of mutant T. thermophila LSU-ΔP5abc introns patterned after previously characterized RNA-folding mutants of the full-length intron with mutations that strengthen or destabilize native or non-native secondary structures.37,38 These mutations lead to different rate-limiting steps and/or different relative stabilities of the native and non-native states, resulting in different rates and amplitudes of CYT-18-dependent splicing (Figure 7). The mutation G100C, which weakens P3 by introducing a G-G mismatch, abolished splicing by CYT-18 alone or by CYT-18 in the presence of CYT-19 or Ded1p (data not shown), consistent with its drastic effect on folding of the full-length Tetrahymena LSU intron.37 The mutant G100C/C274G, in which G100C is rescued by introducing a compensatory C-G pair in P3 while simultaneously weakening Alt-P3, showed weak splicing in the presence of CYT-18 that was stimulated equally by CYT-19 or Ded1p + ATP (Figure 7(c)). Mutants containing U273A, which stabilizes P3, alone or together with C-18C/G-16C, which are expected to destabilize Alt-P(−1), are likewise equally suppressed by CYT-19 or Ded1p (Figure 7(d) and (e)). These findings show that CYT-19 and Ded1p function similarly to resolve non-native structures that have a range of different stabilities relative to the wild-type structure.
Finally, we also tested three mutations that perturb long-range tertiary interactions.41 The mutant L9 has three changes in L9 (A324U, A325C, U326G) that eliminate the tetraloop-receptor interaction between P5 and L9, resulting in more rapid CYT-18-dependent splicing, presumably because the less stable intron tertiary structure decreases the energy barrier for the transition from the non-native to the native state.39,42 CYT-19 and Ded1p plus ATP equally stimulate the CYT-18-dependent splicing of this mutant RNA (Figure 7(f)). The remaining two mutants L2/L2.1 and L2/L9.1a have modifications that disrupt the native P13 pairing between L2.1 and L9.1a and enable alternate loop-loop interactions between L2 and L2.1 (L2: G44C, C45G and L2.1: G77C, C78G) or between L2 and L9.1. (L2: G44C, C45G and L9.1a: G349C, C350G). These mutations potentially require that the DEAD-box protein disrupt non-native tertiary interactions. Again, the CYT-18-dependent splicing of both mutants is equally stimulated by either CYT-19 or Ded1p (Figure 7(g) and (h)). For each of the secondary or tertiary structure mutants, we confirmed that stimulation by the DEAD-box protein requires ATP and is not observed in the presence of AMP-PNP or hexokinase-treated ATP (not shown).
Together, the results above show that CYT-19 and Ded1p at saturating concentrations are equally proficient in rescuing different RNA-folding mutations in the T. thermophila LSU-ΔP5abc intron. The DEAD-box proteins could act directly to disrupt the aberrant secondary or tertiary structure and/or indirectly by destabilizing other secondary and tertiary structure interactions, thereby accelerating equilibrium between native and non-native states.
Discussion
Here, we compared the RNA chaperone activities of six DExH/D-box proteins as measured by their ability to stimulate group I and group II intron splicing. In addition to the mt DEAD-box proteins Mss116p and CYT-19, we find that the S. cerevisiae DEAD-box protein Ded1p and the E. coli DEAD-box protein SrmB stimulate both group I and II intron splicing, while the targeted E. coli DEAD-box protein DbpA, and the vaccinia virus DExH-box protein NPH-II, which unwinds duplexes by directional translocation, gave little or no stimulation of group I or II intron splicing. By correlating their ability to stimulate group I and II intron splicing with biochemical activities, we identify characteristics of DExH/D-box proteins that are critical for RNA chaperone function.
Ded1p and SrmB can function as general RNA chaperones
The S. cerevisiae DEAD-box protein Ded1p ordinarily functions in translation initiation in the cytoplasm and pre-mRNA splicing in the nucleus (reviewed in ref. 43), and the E. coli DEAD-box protein SrmB has been implicated in ribosome biogenesis.44,45 Here, we show that Ded1p efficiently complements the mt translation and group I and II intron splicing defects in mss116Δ strains, promotes group I as well as group II intron splicing in vitro, and functions indistinguishably from CYT-19 to resolve a spectrum of non-native structures that slow CYT-18-dependent splicing of the T. thermophila LSU-ΔP5abc intron. We also show that the E. coli DEAD-box protein SrmB promotes group I and II intron splicing in vitro, albeit with lower activity than the other DEAD-box proteins tested here. The group I and II introns whose splicing is stimulated by Mss116p, Ded1p, CYT-19 and SrmB have different secondary and tertiary structures, use different RNA-folding pathways with different rate-limiting steps, and require different protein factors to stabilize their active RNA structures. The ability of these four DEAD-box proteins to act on structurally diverse RNAs and RNPs indicates that they bind these substrates non-specifically and have an innate ability to function as general RNA chaperones. The non-specific binding of RNA substrates is central to the physiological roles of Mss116p and CYT-19 and could also be relevant to the physiological functions of Ded1p and SrmB.
DEAD-box proteins that function as general RNA chaperones interact non-specifically with RNP substrates
The CYT-18-dependent splicing of the N. crassa mt LSU-ΔORF intron provides a model system for studying how DEAD-box proteins act on RNP substrates as presumably occurs in vivo. CYT-18 binds tightly and specifically to group I intron RNAs to stabilize their catalytically active RNA structure, which in the case of non-self-splicing introns like N. crassa mt LSU-ΔORF does not correspond to the thermodynamically most stable RNA structure.46 A co-crystal structure of CYT-18 with a bound group I intron RNA shows that CYT-18 binds on one side of the RNA interacting extensively with the backbone of the P4–P6 stacked helices and making additional contacts with the P3–P9 domain to stabilize the two domains in the correct relative orientation to form the intron’s active site.47 We find here that DEAD-box proteins do not promote rapid dissociation of CYT-18 from the excised intron RNA after splicing, likely reflecting that CYT-18 remains anchored to the RNA by its extensive interactions with the P4–P6 stacked helices. If so, the DEAD-box proteins must bind the intron RNA primarily on the side opposite CYT-18, where they could disrupt misfolded structures involving the P3–P9 domain or the interaction of the P3–P9 domain with the P4–P6 stacked helices. The disruption of such inactive structures would enable the intron RNA to fold iteratively into the active structure, which is stabilized relative to other structures in the presence of CYT-18. It is also possible that the DEAD-box proteins partially disrupt the binding of CYT-18 providing greater access to other regions of the intron RNA or dissociate CYT-18 from misfolded intermediates to which it may be less stably bound, thereby further accelerating refolding. In principle, a DEAD-box protein RNA chaperone could also act on naked RNA to promote binding of a splicing factor, but previous results indicate that CYT-19 acts on the N. crassa mt LSU-ΔORF intron after CYT-18 binding15 and that Mss116p acts on the S. cerevisiae aI2 group II intron after binding of the intron-encoded maturase.16
Although we found previously that the cyt-19-1 mutation specifically inhibits the splicing of CYT-18-dependent group I introns, as noted at the time, this phenotype could reflect either targeting of CYT-19 to CYT-18-dependent introns or that the cyt-19-1 mutation results in only partial loss of function, so that only those introns that most stringently require an RNA chaperone for folding are affected.15 This issue is difficult to address by genetic analysis in N. crassa as it is an obligate aerobe in which disruption of the cyt-19 gene is ascospore lethal (S.M. and A.M.L., unpublished data). Our finding here that the non-cognate DEAD-box proteins Mss116p and Ded1p stimulate splicing of the CYT-18/group I intron RNP as well or better than the cognate DEAD-box protein CYT-19 suggests that the DEAD-box proteins interact with this RNP complex non-specifically, as they do with free RNAs, and that specific protein-protein interactions are not required. This conclusion is also supported by the finding that the presence or absence of CYT-18 does not affect the unwinding rate of the P1 duplex in a ribozyme derivative of the N. crassa mt LSU-ΔORF intron10 and by the failure to detect interactions between CYT-18 and CYT-19 by GST pull-down or yeast two-hybrid assays (S.M. and A.M.L. unpublished data). More generally, a lack of targeting via specific protein-protein interactions is indicated by the ability of the heterologous DEAD-box proteins CYT-19 and Ded1p to stimulate the in vivo splicing of diverse S. cerevisiae group I and group II intron RNAs, which are associated with a variety of different splicing factors (this work and ref. 16). Thus, in many if not all cases, DEAD-box proteins that function as general RNA chaperones likely interact with group I and II intron RNPs via non-specific binding to exposed RNA surfaces and possibly via non-specific protein-protein interactions.
Characteristics of DEAD-box proteins that have general RNA chaperone activity
Our study enables us to define several characteristics of DExH/D-box proteins that are required for general RNA chaperone function. First, we find that among the proteins tested only DEAD-box proteins have the ability to stimulate group I or group II intron splicing. The non-DEAD-box RNA helicase NPH-II, which unwinds duplexes by directional translocation, does not stimulate group I and group II intron splicing in vitro. Importantly, we show that RNA-unwinding activity of NPH-II on a model duplex is comparable to that of DEAD-box proteins that stimulate group I and II intron splicing and that NPH-II’s ATPase activity is stimulated by group I and II intron RNAs, implying a functional interaction. In a previous study, the hepatitis C virus DExH-box protein NS3, which, like NPH-II, unwinds duplexes by directional translocation, also failed to stimulate aI5γ splicing in vitro.20 The finding that only DEAD-box proteins promote group I and II intron splicing implies a requirement for a helicase mode based on local strand separation. The failure of translocating RNA helicases to stimulate group I and II intron splicing may reflect that their continued association with the RNA during unwinding impedes RNA refolding, that their action results in excessive RNA unfolding when ATP is present, or that their loading mechanism is not suitable for the disruption of critical duplexes in misfolded RNAs.
Second, we find that only those DEAD-box proteins that bind RNA substrates non-specifically in a manner that activates their ATPase and unwinding activities are capable of stimulating group I and II intron splicing. Mss116p, CYT-19, Ded1p, and SrmB bind group I and II intron RNAs with Kds ranging from 2 to 206 nM, and the binding of both types of RNAs equally stimulates their ATPase activity. By contrast, the targeted DEAD-box protein DbpA, which does not stimulate RNA splicing, binds group I and II intron RNAs non-specifically with Kds ~10-fold higher than those for the other DEAD-box proteins, but this binding does not activate ATPase activity. The latter requires specific binding of the E. coli 23S rRNA helix 92 flanked by a short non-specific single-strand region.34,48 Thus, the targeted DEAD-box protein DbpA fails to promote group I and II intron splicing because its remains inactive when bound to group I or II introns RNAs.
Finally, we find that the ability of non-specific DEAD-box proteins to stimulate group I and II intron splicing correlates with their RNA-unwinding activity, which for the protein preparations used here was highest for Mss116p, followed by Ded1p, CYT-19, and SrmB. This order of activity does not correlate with RNA-binding affinity or strand-annealing activity, which are highest for Mss116p and SrmB (Table 2, ref. 21, and H.J. and E.J., unpublished results). Importantly, the same order of activity holds for all group I and group II intron RNAs tested, implying that the DEAD-box proteins act by a fundamentally similar mechanism on both types of introns. A correlation between RNA splicing and RNA-unwinding activity was found previously for Mss116p motif III mutants that weaken ATP-dependent unwinding activity and correspondingly decrease the in vivo and in vitro splicing of both group I and II introns.22,49
Structural basis for DEAD-box protein RNA chaperone activity
Available evidence suggests that the most critical DEAD-box protein region for RNA chaperone activity is the helicase core, which binds RNAs non-specifically and unwinds duplexes by local strand separation rather than translocation. This unwinding mechanism enables the helicase core to act at multiple sites on large RNAs and refold localized RNA regions or structures without globally disrupting the RNA structure. Many DEAD-box proteins including CYT-19, Mss116p, Ded1p and SrmB that stimulate group I and II intron splicing have ancillary N- or C-terminal domains that are linked to the helicase core and may enhance or modulate its RNA chaperone function. In CYT-19 and Mss116p, the helicase core is linked to a C-terminal α-helical extension, and all four DEAD-box proteins that stimulate group I and II intron splicing end with a basic hydrophilic tail region.25 The C-terminal α-helical extension in Mss116p and CYT-19 appears to be required for structural stability of the helicase core and may contribute to non-specific RNA binding and RNA unwinding, but is not present in Ded1p or SrmB and thus cannot be essential for group I or II intron splicing activity or general RNA chaperone function.25 The basic tail is not required for ATPase or RNA-unwinding activity, but provides a second non-specific RNA binding site that likely enhances RNA chaperone activity by tethering the helicase core to large RNA substrates for multiple rounds of RNA-unwinding.24,25
Targeted DEAD-box proteins, like DbpA, differ from those that function as general RNA chaperones in that the helicase core must be activated by the binding of a specific RNA or RNP target. In general, the activating RNA or RNP could interact directly with the helicase core or activate it indirectly via a protein conformational change induced by binding to an ancillary domain. The latter appears to be the case for DbpA, whose C-terminal domain is responsible for the specific binding of helix 92.7 Notably, the binding of helix 92 is not sufficient to activate DbpA’s ATPase activity unless it is linked to a sufficiently long non-specific single-strand RNA.34 Thus, after the binding of the C-terminal domain to helix 92, the helicase core of DbpA may be activated by non-specific RNA binding and function non-specifically to unwind proximate RNAs and perform RNA chaperone functions in the same manner as the helicase cores of non- or less specifically targeted DEAD-box proteins.
Non-specific DEAD-box proteins must evolve to function optimally on a collection of RNA substrates
DEAD-box proteins that function non-specifically as general RNA chaperones must evolve to function optimally on a collection of RNA substrates that differs in each organism and may require a different balance of activities. Our comparative analysis shows that Mss116p binds RNAs more tightly than Ded1p and CYT-19, has the highest RNA-unwinding activity, and is the most efficient stimulator of group I and II intron splicing in vitro and in vivo. Ded1p and CYT-19 on the other hand have RNA-dependent ATPase activities comparable to that of Mss116p, but lower RNA-binding affinity, RNA-unwinding activity, and group I and group II intron splicing activity, suggesting less efficient coupling of ATP hydrolysis to RNA unwinding. SrmB, the least efficient RNA chaperone, binds RNAs tightly but has low RNA-dependent ATPase activity, RNA-unwinding, and group I and II intron splicing activities and thus appears to least efficiently couple RNA binding to ATP hydrolysis and RNA unwinding. The relatively high RNA-unwinding activity of Mss116 could reflect that it evolved to unwind duplexes with relatively high stability in its natural RNA substrates, while the low unwinding activity of SrmB may reflect that it normally unwinds duplexes with relatively low stability and/or that some of its functions require RNA binding in the absence of unwinding. It is also possible that the RNA-unwinding activity of SrmB is enhanced by binding to a specific partner protein or RNA in vivo.
Notably, too high concentrations of Mss116p and other DEAD-box proteins inhibit the splicing of some introns in vitro (this work and ref. 21), and this also appears to be the case for CYT-19 and Ded1p in vivo (Figure 2(c)). This inhibition may reflect excessive RNA unwinding or non-specific RNA binding and suggests that the expression levels of DEAD-box proteins that function as general RNA chaperones must evolve in concert with biochemical activities to avoid deleterious effects on some RNA substrates.21 It also suggests that DEAD-box proteins with very high RNA-unwinding activity, which may be needed to unwind very stable duplexes or promote difficult structural rearrangements, could be deleterious as general RNA chaperones and require targeting mechanisms that mask their RNA-unwinding activity until they bind the desired substrate, as appears to be the case for DpbA.
DEAD-box proteins that function as general RNA chaperones may be widespread and carry out multiple functions
Based on our current understanding of the DEAD-box protein unwinding mechanism, it is likely that all DEAD-box proteins have an inherent RNA chaperone activity by virtue of their distinctive RNA-unwinding mechanism. Some, like Mss116p, CYT-19, Ded1p, and SrmB, bind RNAs non-specifically, while others, like DbpA, are targeted but may act non-specifically after their unwinding activity is activated by substrate binding. Those DEAD-box proteins that function as non-specific RNA chaperones provide a means of resolving a wide variety of kinetic traps that may arise in the folding of different cellular RNAs or RNPs, without needing to target a specific DEAD-box protein to each kinetic trap.10 Like protein chaperones, non-specific RNA chaperones provide a buffer against deleterious effects of spontaneous mutations that impair RNA folding.50,51 As noted previously, general chaperones could not only resolve non-native structures, as found for the T. thermophila group I intron, but could also accelerate rearrangement of stable on-pathway intermediates, as may be the case for the group II intron aI5γ.21,22 Targeting may be needed in some cases to accelerate a specific structural transition or because excessive unregulated RNA-unwinding activity or non-specific RNA binding is deleterious. We also note that a primary function as an RNA chaperone does not preclude the acquisition of other functions involving RNA binding, protein displacement, structural stabilization, or strand annealing, endowing DEAD-box proteins with additional versatility for carrying out RNA and RNP structural transitions that differ in detail for each RNA.
Materials and Methods
Recombinant plasmids
Recombinant plasmids used for DExH/D-box protein expression were: pMAL-CYT-19 and pMAL-Mss116p, which express CYT-19 and Mss116p, respectively, with their C-termini fused to maltose-binding protein (MalE) via a TEV protease-cleavable linker;21,24 pET3a::srmB, which expresses N-terminal hexahistidine-tagged SrmB;7 pET3a::dbpA, which expresses N-terminal hexahistidine tagged DbpA;52 pET22b-His6-DED1, which expresses N-terminal hexahistidine tagged Ded1p;53 and pTM-His-NPH-II, which expresses N-terminal hexahistidine tagged NPH-II.54
pBlueLSU-ΔP5abc contains the T. thermophila LSU-ΔP5abc intron and flanking exons cloned behind the T7 promoter in the HindIII site of pBluescript KS (+) (Stratagene, La Jolla, CA). Mutant T. thermophila LSU-ΔP5abc introns were constructed via PCR with primers that introduce the mutations.
Analysis of DExH/D-box protein function in S. cerevisiae
S. cerevisiae strains have the nuclear background of 161-U7 (MATa ade1, lys1, ura3). Strains 161-U7 Δmss116::kanR and 161-U7 Δmss116::ura3 have most of the mss116 gene replaced by kanR and ura3 markers, respectively.16
The wild-type MSS116myc gene and the MSS116/cyt-19myc chimeric gene were described.16 Other Mss116/DExH/D-box protein chimeric genes are similar to Mss116/cyt-19myc and consist of the promoter, 5′ UTR, and codons 1–36 (mt targeting sequence) of MSS116 fused in frame via an NdeI site to the start codon of the DExH/D-box protein coding sequence, followed by a 0.34-kb segment containing the myc tag, tandem UAA stop codons, and the 3′ UTR of MSS116. The 3′ UTRs are identical in different constructs except for the restriction site immediately following the termination codons (SpeI for CYT-19 and Ded1p, and BamHI for DbpA, SrmB and NPH-II). The chimeric genes were cloned between the HindIII and SacII sites in the polylinker of the yeast centromere plasmid pRS416 (New England Biolabs, Ipswich, MA) or 2 μ-plasmid pRS42655 and were transformed into yeast, as described.56
The MSS116/cyt-19 knock-in (cyt-19-ki) was described previously.16 The MSS116/dbpA and Mss116/DED1 knock-ins were constructed by transforming the respective chimeric genes into the 161-U7 Δmss116::ura3 strain containing GII-5γ mtDNA and selecting against URA3 with 5-fluoro-orotic-acid.57 The Mss116/srmb and Mss116p/NPH-II knock-ins were made in the Δmss116::kanR strain by a two-step gene replacement with the yeast integrating vector pRS306 containing the chimeric genes and a URA3 marker.58,59
Strains containing different mtDNAs were constructed by cytoduction with previously constructed derivatives of MCC109 (MATα ade2-101 ura3-52 kar1-1) carrying one of the following mtDNAs: I0, lacking introns; GII-5γ containing group I introns bI4, bI5, aI3, and aI4, and the group II intron aI5γ; and mtDNAs containing only the group II intron aI5γ or bI1.16,49
For Northern hybridizations, cells were grown at 30°C in YNB (yeast nitrogen base) medium containing 2% raffinose, 1% casamino acids, and appropriate nutritional supplements until O.D.600 = 1.0–2.0. RNA was isolated as described,60 extracted twice with phenol-chloroform-isoamyl alchohol (phenol-CIA; 25:24:1), and ethanol precipitated. The RNA was then run in a 1% agarose gel containing 0.24% guanidinium thiocyanate and 1X TBE,61 blotted to a nylon membrane (GE Healthcare Biosciences, Piscataway, NJ), and hybridized with 5′-32P-labeled DNA oligonucleotide probes (COX1 exon 6, 5′-GAG TGT ACA GCT GGT GGA GA; COB exon 6, 5′-AAG GTA CTT CTA CAT GGC ATG CT; COX2, 5′-AGGTAATGATACTGCTTCGATC). After hybridization, the blots were washed, air-dried and scanned with a phosphorimager. Immunoblotting was as described.25
Protein purification
CYT-19 and Mss116p were expressed from pMAL-CYT-19 and pMAL-Mss116p, respectively, and purified as described.21,22,24
Ded1p was expressed from pET22b-His6-DED1 in E. coli Rosetta2(DE3);53 DbpA was expressed from pET3a::dbpA in E. coli BL21(DE3);52 and SrmB was expressed from pET3a::srmB in E. coli HMS174(DE3).7 Protein expression was induced either by adding IPTG (1 mM final) to mid-log phase cells or by growing cells in auto-induction medium (LB containing 0.2% lactose, 0.05% glucose, 0.5% glycerol, 24 mM (NH4)2SO4, 50 mM KH2PO4, 50 mM Na2HPO4).62 In both cases, induction was for ~20 h at 20–25 °C, after which cells were harvested by centrifugation, suspended in NiA buffer (20 mM Tris-HCl, pH 7.5, 0.5 M KCl, 10 mM imidazole), and frozen at −80 °C. To purify the proteins, the cell suspension was thawed, treated with lysozyme (1 mg/ml; Sigma-Aldrich, St. Louis, MO) for 15 min on ice, sonicated, and centrifuged for 40 min at 18,500 × g at 4 °C. Nucleic acids were precipitated by adding 10% PEI to a final concentration of 0.2% and centrifuging for 15 min at 15,000 × g at 4 °C. The resulting supernatant was applied to a 5-ml Hi-Trap Ni2+ column (GE Healthcare Biosciences), which was washed sequentially with 10 column volumes each of NiA buffer containing 0.5 M, 1.5 M, or 0.5 M KCl, and then eluted with a linear gradient of 10 to 600 mM imidazole. Protein fractions eluted with imidazole were pooled and purified further by gel-filtration through a Superdex 200 column (GE Healthcare Biosciences), which was pre-equilibrated and run in 20 mM Tris-HCl, pH 7.5, 0.5 M KCl, 1 mM EDTA, 1 mM DTT, and 10% glycerol. The peak fractions from the gel filtration column were pooled and dialyzed against a solution containing 20 mM Tris-HCl, pH 7.5, 0.5 M KCl, 1 mM EDTA, 1 mM DTT, and 50% glycerol for storage. The Ded1p used for RNA duplex unwinding assays was expressed in E. coli and purified as described.32 NPH-II was expressed from pTM-His-NPH-II in baculovirus-infected insect cells and purified as described.54
CYT-18 was expressed from pEX560 and purified as described63 with an additional final purification through a Superdex 200 column (GE Healthcare Biosciences) in 25 mM Tris-HCl, pH 7.5, 100 mM KCl followed by dialysis against a solution containing 20 mM Tris-HCl, pH 7.5, 0.5 M KCl, 1 mM EDTA, 2 mM DTT, and 50% glycerol.
In vitro transcription of RNA substrates
Group I and II intron RNAs were transcribed using T3 or T7 Megascript kits (Ambion, Austin, TX) from recombinant plasmids digested with restriction enzymes as follows: N. crassa mt LSU-ΔORF intron, pBD5a/BanI;64 T. thermophila LSU-ΔP5abc intron and mutants thereof, pBlueΔP5abc/XhoI; S. cerevisiae aI5γ intron, pJD20/HindIII;65 S. cerevisiae bI1 intron, pHrH154#4/EcoRI.16 The E. coli 23S rRNA segment 2454–2625 was transcribed from pUC19::rrnB 2454–2625 digested with BamHI.52 Transcription was according to the manufacturer’s instructions for 4 h at 37 °C. 32P-labled RNAs were synthesized by adding [α-32P]-UTP (3,000 Ci/mmol; Perkin Elmer, Wellesley, MA). After digesting the DNA template with Turbo DNase I (5 min, 37°C), RNAs were extracted with phenol-CIA and purified by two cycles of gel filtration through Sephadex G-50 (Sigma-Aldrich) spin columns. Intron RNAs were stored in Milli-Q-grade H2O and either used the same day or kept at 4 °C. Immediately prior to use, RNAs were denatured at 92 °C for 2 min and then incubated at reaction temperature for at least 5 min before adding to the reaction mixture.
RNA splicing assays
Group I intron splicing reactions were done with either 10 or 20 nM 32P-labeled precursor RNA and amounts of CYT-18 protein dimer and DEAD-box proteins indicated for individual experiments in 100 μl of reaction medium containing 100 mM KCl, 5 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 10% glycerol, 1 mM GTP, and 1 mM ATP, AMP-PNP, or ADP. Group II splicing reactions were done with 10 nM 32P-labeled precursor RNA and the indicated amounts of a DEAD-box protein in 100 μl of reaction medium containing 100 mM KCl, 8 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 10% glycerol, and 1 mM ATP, AMP-PNP, or ADP. All nucleotides were added from stock solutions containing equimolar MgCl2. 10 mM stocks of AMP-PNP-Mg2+ and ADP-Mg2+ were treated with hexokinase (1.5 units/ml; 60 min, 30°C; Roche Diagnostics, Indianapolis, IN) plus 25 mM glucose to eliminate any residual ATP. MgCl2 used in splicing experiments was purchased as a 1 M stock solution (Molecular Biology Grade; RNase-free; Sigma-Aldrich). For both group I and group II introns, RNAs were added last to initiate splicing. To terminate the reactions, portions (10 μl) were removed at different times into tubes containing 20 μl of 75 mM EDTA, 0.1% SDS, and 1 mg/ml proteinase K (Ambion) and extracted with phenol-CIA. Products were analyzed by electrophoresis in a denaturing 4% polyacrylamide gel, which was dried and quantified with a phosphorimager (Typhoon Trio), using ImageQuant TL software (GE Healthcare Biosciences). The data for disappearance of precursor RNA from splicing time courses were fit to single exponential equations, using KaleidaGraph (Synergy Software, Reading, PA).
ATPase assays
ATPase assays were as described15 at 30°C in 20μl of splicing reaction medium containing 100 mM KCl, 5 or 8 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 10% glycerol, 25 or 50 nM DEAD-box protein, 10 μCi [γ-32P]ATP (10 Ci/mmol; Perkin Elmer), and saturating unlabeled ATP (3 mM). At each time point, a 1-μl portion was removed and applied to a PEI-cellulose thin-layer chromatography plate (20 × 20 cm; Scientific Adsorbents Inc., Atlanta, GA), which was developed with 0.75 M LiCl and 1 M formic acid, then dried, and quantified with a phosphorimager.
RNA-binding assays
Equilibrium-binding assays were done by incubating 5 pM 32P-labeled RNA with increasing concentrations of DEAD-box protein at 30°C in reaction medium containing 100 mM KCl, 5 or 8 mM MgCl2, 10 mM Tris-HCl, pH 7.5 and 0.1 mg/ml bovine serum albumin to stabilize proteins at low concentrations. The solutions were then filtered through nitrocellulose (BioRad, Hercules, CA) backed by nylon (Hybond-N; GE Healthcare Biosciences), and the radioactivity retained on the filters was quantified with a phosphorimager.
Duplex-unwinding reactions
The RNA substrate containing a 10-bp duplex and a 25-nt single stranded overhang 3′ to the duplex region was prepared by mixing oligonucleotides R10C (5′-AGCACCGUAA) and R35-3 (5′-UUACGGUGCUUAAAACAAAACAAAACAAAACAAAA) as described.29 Unwinding reactions were done at 30°C in 30 μl of group I intron splicing medium (100 mM KCl, 5 mM MgCl2, 25 mM Tris-HCl, pH 7.5), supplemented with 10% glycerol, 0.7 U/μl RNasin (Promega, Madison, WI), 2 mM DTT, and 0.01% NP-40. DExH/D-box proteins at concentrations specified in the legend of Figure 6 were pre-incubated with 0.1 nM [32P]-labeled RNA duplex in the absence of ATP for 5 min, and reactions were initiated by adding ATP to a final concentration of 2 mM. Aliquots (3 μl) were removed at the indicated times, and the reactions were stopped by adding 3 μl of a solution containing 0.1% (w/v) SDS, 50 mM EDTA, 0.01% (w/v) xylene cyanol blue, 0.01% (w/v) bromophenol blue, and 20% (v/v) glycerol. The samples were run at 4°C in a 15% non-denaturing polyacrylamide gel at ~15 V/cm. The gels were then dried and exposed to a phosphorimager screen. Single-stranded and duplex RNA species were quantified with a phosphorimager and ImageQuant 5.2 software (GE Healthcare Biosciences), as described.11
Supplementary Material
Figure S1. Ability of NPH-II to promote group I and group II intron splicing in vitro. (a) Splicing reactions with the N. crassa mt LSU-ΔORF group I intron done with 1 nM 32P-labeled precursor RNA and 2 nM CYT-18 dimer in reaction medium containing 100 mM KCl, 5 mM MgCl2, 1 mM GMP, and 1 mM ATP for 120 min at 25°C. (b) and (c) Splicing reactions with the S. cerevisiae aI5γ and bI1 group II introns, respectively, done with 1 nM 32P-labeled precursor RNA in reaction medium containing 100 mM KCl, 8 mM MgCl2, and 1 mM ATP for 120 min at 30°C. NPH-II was added in the amounts indicated. Lane P shows precursor RNA immediately before addition to reactions. Each time course consists of samples taken at 7, 15, 25, 45, 98, and 120 min. P, precursor RNA; I-E2, intron-3′-exon; I, intron; “?”, unidentified product; E1–E2, ligated exons.
Figure S2. Time courses of precursor RNA disappearance at different DEAD-box protein concentrations. (a) Splicing reactions for the N. crassa mt LSU-ΔORF intron group I intron were done with 10 nM 32P-labeled precursor RNA, 20 nM CYT-18 dimer, and the indicated amount of DEAD-box protein in reaction medium containing 100 mM KCl, 5 mM MgCl2, 1 mM GTP, and 1 mM ATP at 25°C. (b) and (c) Splicing reactions with the S. cerevisiae aI5γ and bI1 group II introns, respectively, were done with 10 nM 32P-labeled precursor RNA and the indicated amount of DEAD-box protein in reaction medium containing 100 mM KCl, 8 mM MgCl2, and 1 mM ATP at 30°C. For simplicity, the data in each plot were fit by single exponential equations with end-points of zero, assuming RNA splicing could go to completion. In a small number of cases, apparent leveling off of the reaction prior to completion could be due to time-dependent loss of DEAD-box protein activity or ADP inhibition. The curve fits to zero end-points minimize the influence of such effects on the rate-constants by giving greater weight to the initial time points.
Figure S3. The ATPase activity of NPH-II is stimulated by group I and group II intron RNAs. Reactions were done at 30°C with 1 nM NPH-II, 200 nM RNA, and 1 mM ATP in splicing reaction medium containing 100 mM KCl and 5 or 8 mM Mg2+ for group I and II intron RNAs, respectively. kobs values were calculated from the initial rate.
Figure S4. Equilibrium binding of DEAD-box proteins to the S. cerevisiae aI5γ group II intron. 32P-labeled RNA (5 pM) was incubated with increasing concentrations of different DEAD-box proteins in reaction medium containing 100 mM KCl and 8 mM MgCl2 for 60 min at 30°C with or without the indicated nucleotides and then filtered through nitrocellulose backed by nylon. In all cases, the data for percent RNA bound as a function of protein concentration were fit by a hyperbolic equation. The data were plotted using a log scale for protein concentration so that binding curves measuring different Kds over a wide range of protein concentrations could be compared readily on the same plots.
Acknowledgments
We thank Dr. Quansheng Yang for initial RNA unwinding assays, Ken Johnson for helpful discussions, and Rick Russell for comments on the manuscript. This work was supported by NIH grants GM037951 to A.M.L and GM067700 to E.J. Mark Del Campo was the recipient of NIH postdoctoral fellowship F32 GM076961.
Abbreviations
- AMP-PNP
adenylyl-imidodiphosphate
- CEN
centromere-containing plasmid
- COB
cytochrome b gene
- COX1
cytochrome oxidase subunit I gene
- COX2
cytochrome oxidase subunit II gene
- COX3
cytochrome oxidase subunit III gene
- Gly
glycerol
- ki
knock-in
- LSU
large subunit rRNA
- mt
mitochondrial
- phenol-CIA
phenol-chloroform-isoamyl alcohol 25:24:1
- UTR
untranslated region
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 2.Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 3.Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol Cell. 2007;27:339–352. doi: 10.1016/j.molcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Jankowsky E, Fairman ME. RNA helicases--one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Guthrie C. PRP16, a DEAH-box RNA helicase, is recruited to the spliceosome primarily via its nonconserved N-terminal domain. RNA. 1998;4:1216–1229. [PMC free article] [PubMed] [Google Scholar]
- 6.Schneider S, Schwer B. Functional domains of the yeast splicing factor Prp22p. J Biol Chem. 2001;276:21184–21191. doi: 10.1074/jbc.M101964200. [DOI] [PubMed] [Google Scholar]
- 7.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExD/H protein YxiN is sufficient to confer specificity for 23 S rRNA. J Mol Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 8.Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol. 2004;24:10101–10110. doi: 10.1128/MCB.24.23.10101-10110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 10.Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci USA. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 12.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, Potratz JP, Tijerina P, Del Campo M, Lambowitz AM, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci USA. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 16.Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Séraphin B, Simon M, Boulet A, Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989;337:84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- 18.Lambowitz AM, Caprara MG, Zimmerly S, Perlman PS. Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In: Gesteland RF, Cech T, Atkins JF, editors. The RNA World. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 1999. pp. 451–485. [Google Scholar]
- 19.Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci USA. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solem A, Zingler N, Pyle AM. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 21.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and II intron splicing. Biochemical characterization of MSS116p, ATP-hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, Russell R, Lambowitz AM. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grohman JK, Del Campo M, Bhaskaran H, Tijerina P, Lambowitz AM, Russell R. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: The C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohr G, Del Campo M, Mohr S, Yang Q, Jia H, Jankowsky E, Lambowitz AM. Function of the C-terminal domain of the DEAD-box protein Mss116p analyzed in vivo and in vitro. J Mol Biol. 2008;375:1344–1364. doi: 10.1016/j.jmb.2007.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 27.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23S rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsu CA, Uhlenbeck OC. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry. 1998;37:16989–16996. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 29.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 30.Shuman S. Vaccinia virus RNA helicase: an essential enzyme related to the DE-H family of RNA-dependent NTPases. Proc Natl Acad Sci USA. 1992;89:10935–10939. doi: 10.1073/pnas.89.22.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saldanha RJ, Patel SS, Surendran R, Lee JC, Lambowitz AM. Involvement of Neurospora mitochondrial tyrosyl-tRNA synthetase in RNA splicing. A new method for purifying the protein and characterization of physical and enzymatic properties pertinent to splicing. Biochemistry. 1995;34:1275–1287. doi: 10.1021/bi00004a022. [DOI] [PubMed] [Google Scholar]
- 32.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 33.Bowers HA, Maroney PA, Fairman ME, Kastner B, Lührmann R, Nilsen TW, Jankowsky E. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12:903–912. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 36.Mohr G, Caprara MG, Guo Q, Lambowitz AM. A tyrosyl-tRNA synthetase can function similarly to an RNA structure in the Tetrahymena ribozyme. Nature. 1994;370:147–150. doi: 10.1038/370147a0. [DOI] [PubMed] [Google Scholar]
- 37.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 38.Pan J, Deras ML, Woodson SA. Fast folding of a ribozyme by stabilizing core interactions: evidence for multiple folding pathways in RNA. J Mol Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 39.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 40.Ohki Y, Ikawa Y, Shiraishi H, Inoue T. Mispaired P3 region in the hierarchical folding pathway of the Tetrahymena ribozyme. Genes to Cells. 2002;7:851–860. doi: 10.1046/j.1365-2443.2002.00567.x. [DOI] [PubMed] [Google Scholar]
- 41.Pan J, Woodson SA. The effect of long-range loop-loop interactions on folding of the Tetrahymena self-splicing RNA. J Mol Biol. 1999;294:955–965. doi: 10.1006/jmbi.1999.3298. [DOI] [PubMed] [Google Scholar]
- 42.Rook MS, Treiber DK, Williamson JR. Fast folding mutants of the Tetrahymena group I ribozyme reveal a rugged folding energy landscape. J Mol Biol. 1998;281:609–620. doi: 10.1006/jmbi.1998.1960. [DOI] [PubMed] [Google Scholar]
- 43.Linder P. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol Cell. 2003;95:157–167. doi: 10.1016/s0248-4900(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 44.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 45.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caprara MG, Mohr G, Lambowitz AM. A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J Mol Biol. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- 47.Paukstelis PJ, Chen JH, Chase E, Lambowitz AM, Golden BL. Structure of a tyrosyl-tRNA synthetase splicing factor bound to a group I intron RNA. Nature. 2008;451:94–97. doi: 10.1038/nature06413. [DOI] [PubMed] [Google Scholar]
- 48.Henn A, Cao W, Hackney DD, De La Cruz EM. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J Mol Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang HR. PhD Thesis. University of Texas Southwestern Medical Center; Dallas, TX: 2004. Functional studies of intron- and nuclear-encoded splicing factors in the mitochondria of Saccharomyces cerevisiae. [Google Scholar]
- 50.Sangster TA, Lindquist S, Queitsch C. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. Bioessays. 2004;26:348–362. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 51.Sangster TA, Salathia N, Undurraga S, Milo R, Schellenberg K, Lindquist S, Queitsch C. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc Natl Acad Sci USA. 2008;105:2963–2968. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karginov FV, Uhlenbeck OC. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res. 2004;32:3028–3032. doi: 10.1093/nar/gkh640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 54.Gross CH, Shuman S. Mutational analysis of vaccinia virus nucleoside triphosphate phosphohydrolase II, a DExH box RNA helicase. J Virol. 1995;69:4727–4736. doi: 10.1128/jvi.69.8.4727-4736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 56.Agatep R, Kirkpatrick RD, Parchaliuk DL, Woods RA, Gietz RD. Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol protocol. Technical Tips Online. 1998;3:133–137. [Google Scholar]
- 57.Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 58.Sherer S, Davis RW. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goda SK, Minton NP. A simple procedure for gel electrophoresis and northern blotting of RNA. Nucleic Acids Res. 1995;23:3357–3358. doi: 10.1093/nar/23.16.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 63.Paukstelis PJ, Coon R, Madabusi L, Nowakowski J, Monzingo A, Robertus J, Lambowitz AM. A tyrosyl-tRNA synthetase adapted to function in group I intron splicing by acquiring a new RNA binding surface. Mol Cell. 2005;17:417–428. doi: 10.1016/j.molcel.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 64.Guo QB, Akins RA, Garriga G, Lambowitz AM. Structural analysis of the Neurospora mitochondrial large rRNA intron and construction of a mini-intron that shows protein-dependent splicing. J Biol Chem. 1991;266:1809–1819. [PubMed] [Google Scholar]
- 65.Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS. Group II intron self-splicing. Alternative reaction conditions yield novel products. J Biol Chem. 1988;263:3432–3439. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Ability of NPH-II to promote group I and group II intron splicing in vitro. (a) Splicing reactions with the N. crassa mt LSU-ΔORF group I intron done with 1 nM 32P-labeled precursor RNA and 2 nM CYT-18 dimer in reaction medium containing 100 mM KCl, 5 mM MgCl2, 1 mM GMP, and 1 mM ATP for 120 min at 25°C. (b) and (c) Splicing reactions with the S. cerevisiae aI5γ and bI1 group II introns, respectively, done with 1 nM 32P-labeled precursor RNA in reaction medium containing 100 mM KCl, 8 mM MgCl2, and 1 mM ATP for 120 min at 30°C. NPH-II was added in the amounts indicated. Lane P shows precursor RNA immediately before addition to reactions. Each time course consists of samples taken at 7, 15, 25, 45, 98, and 120 min. P, precursor RNA; I-E2, intron-3′-exon; I, intron; “?”, unidentified product; E1–E2, ligated exons.
Figure S2. Time courses of precursor RNA disappearance at different DEAD-box protein concentrations. (a) Splicing reactions for the N. crassa mt LSU-ΔORF intron group I intron were done with 10 nM 32P-labeled precursor RNA, 20 nM CYT-18 dimer, and the indicated amount of DEAD-box protein in reaction medium containing 100 mM KCl, 5 mM MgCl2, 1 mM GTP, and 1 mM ATP at 25°C. (b) and (c) Splicing reactions with the S. cerevisiae aI5γ and bI1 group II introns, respectively, were done with 10 nM 32P-labeled precursor RNA and the indicated amount of DEAD-box protein in reaction medium containing 100 mM KCl, 8 mM MgCl2, and 1 mM ATP at 30°C. For simplicity, the data in each plot were fit by single exponential equations with end-points of zero, assuming RNA splicing could go to completion. In a small number of cases, apparent leveling off of the reaction prior to completion could be due to time-dependent loss of DEAD-box protein activity or ADP inhibition. The curve fits to zero end-points minimize the influence of such effects on the rate-constants by giving greater weight to the initial time points.
Figure S3. The ATPase activity of NPH-II is stimulated by group I and group II intron RNAs. Reactions were done at 30°C with 1 nM NPH-II, 200 nM RNA, and 1 mM ATP in splicing reaction medium containing 100 mM KCl and 5 or 8 mM Mg2+ for group I and II intron RNAs, respectively. kobs values were calculated from the initial rate.
Figure S4. Equilibrium binding of DEAD-box proteins to the S. cerevisiae aI5γ group II intron. 32P-labeled RNA (5 pM) was incubated with increasing concentrations of different DEAD-box proteins in reaction medium containing 100 mM KCl and 8 mM MgCl2 for 60 min at 30°C with or without the indicated nucleotides and then filtered through nitrocellulose backed by nylon. In all cases, the data for percent RNA bound as a function of protein concentration were fit by a hyperbolic equation. The data were plotted using a log scale for protein concentration so that binding curves measuring different Kds over a wide range of protein concentrations could be compared readily on the same plots.