Abstract
Herpesviruses must cross the inner nuclear membrane and underlying lamina to exit the nucleus. HSV-1 US3 and PKC can phosphorylate lamins and induce their dispersion but do not elicit all of the phosphorylated lamin species produced during infection. UL13 is a serine threonine protein kinase conserved among many herpesviruses. HSV-1 UL13 phosphorylates US3 and thereby controls UL31 and UL34 nuclear rim localization, indicating a role in nuclear egress. Here, we report that HSV-2 UL13 alone induced conformational changes in lamins A and C and redistributed lamin B1 from the nuclear rim to intranuclear granular structures. HSV-2 UL13 directly phosphorylated lamins A, C, and B1 in vitro, and the lamin A1 tail domain. HSV-2 infection recapitulated the lamin alterations seen upon expression of UL13 alone, and other alterations were also observed, indicating that additional viral and/or cellular proteins cooperate with UL13 to alter lamins during HSV-2 infection to allow nuclear egress.
Keywords: herpes simplex virus, HSV-2, UL13, lamin, phosphorylation, nuclear, egress
INTRODUCTION
The nuclear envelope, comprised of the inner and outer nuclear membranes (INM and ONM, respectively), define the nuclear rim and separate the nucleus from the cytoplasm. The nuclear lamina is comprised primarily of nuclear lamins, which form a network that underlies and supports the nucleoplasmic face of the INM (Likhacheva and Bogachev, 2001;Gruenbaum et al., 2005). Lamins A and C are A-type lamins and alternative splice products of the same gene (Lin and Worman, 1993). B-type lamins, subdivided into lamins B1 and B2, are products of individual genes (Likhacheva and Bogachev, 2001). Nuclear lamins have three domains: 1) a central alpha-helical rod domain, 2) a globular head domain at the amino-terminus, and 3) a globular tail domain at the carboxy-terminus. The nuclear lamina is a dynamic structure that is disassembled during cell division. Reversible phosphorylation at conserved serines within lamin head and tail domains regulates the integrity of the nuclear lamina. Cdc2, PKC, and MAP kinases phosphorylate lamins (Peter et al., 1992;Fields and Thompson, 1995), and phosphorylation by Cdc2 of serine residues in the head and tail domains results in lamin depolymerization during mitosis (Heald and McKeon, 1990).
Herpesviruses are large, double-stranded DNA viruses that replicate and assemble progeny virions in the nuclei of infected cells, and hence must exit the nucleus to escape the cell. Because of their large size, progeny virions cannot exit through nuclear pores and therefore must cross the nuclear lamina to bud through the INM. Complete dismantling of the nuclear envelope does not occur during herpesvirus infection; however, infection appears to induce partial lamin disassembly or reorganization to allow virions to reach the INM (Scott and O'Hare, 2001;Mettenleiter, 2002;Reynolds et al., 2004).
The alphaherpesvirus, herpes simplex virus 1 (HSV-1), the betaherpesviruses, murine cytomegalovirus (MCMV) and human cytomegalovirus (HCMV), and the gammaherpesvirus Epstein-Barr virus (EBV) all induce nuclear lamin alterations (Scott and O'Hare, 2001;Reynolds et al., 2004;Muranyi et al., 2002;Marschall et al., 2005;Simpson-Holley et al., 2004;Lee et al., 2008). The MCMV proteins M50/p35 and M53/p38, and the HCMV proteins pUL50 and pUL53, form complexes that localize and recruit PKC to the nuclear rim (Muranyi et al., 2002;Milbradt et al., 2007). This recruitment leads to phosphorylation of lamins A, C, and B (Muranyi et al., 2002). The M50/p53 and M50/p38 homologues HSV-1 UL31 and UL34, respectively, also form complexes that localize to the nuclear rim (Reynolds et al., 2001). HSV-1 infection does not result in gross loss of lamins A and C. Rather, HSV-1 infection is thought to induce conformational changes within the head and tail domains of lamins A and C, leading to masking of the epitopes recognized by selected lamin A/C antibodies (Reynolds et al., 2004). HSV-1 UL31 and UL34 are necessary but not sufficient to cause the alterations to lamins A and C that occur during HSV-1 infection, indicating a role for additional viral protein(s) in this process (Reynolds et al., 2004). PKC recruitment to the nuclear rim during HSV-1 infection requires UL31 and UL34, and PKC also mediates phosphorylation of lamin B during infection (Park and Baines, 2006). However, lamin B phosphorylation is only partially reduced by PKC inhibitors and direct phosphorylation of lamin B by PKC has not been shown (Park and Baines, 2006). These observations indicate a role for additional viral protein kinases in lamin B phosphorylation during HSV infection.
HSV-1 and HSV-2 encode two serine/threonine protein kinases, US3 and UL13, which may phosphorylate lamins during nuclear egress. US3 is packaged into the virion and is conserved only among the alpha herpesviruses (Purves et al., 1987;Smith and Smith, 1989). Proper localization of UL34, UL31, and PKC at the nuclear rim during HSV-1 egress requires US3 (Kato et al., 2006), and phosphorylation of UL31 by US3 controls capsid egress from the nucleus (Mou et al., 2009). HSV-1 US3 also can phosphorylate lamins A and C in vitro at multiple sites (Mou et al., 2007). However, phosphorylation by US3 does not account for all of the phosphospecies of lamins A and C seen in HSV-1 infected cells (Mou et al., 2007). UL13 and its homologs are components of the viral tegument, are synthesized as late gene products, and are conserved among alpha, beta and gamma herpesviruses (Overton et al., 1992;Kawaguchi and Kato, 2003). This extensive conservation suggests an important role for UL13 in the herpesvirus replication cycle. The HCMV UL13 homolog UL97 may play a role in nuclear egress because it can directly phosphorylate lamins A, C, and B in vitro and induces lamin alterations in transfected and infected cells (Marschall et al., 2005). UL13 may be involved in tegument dissociation (Morrison et al., 1998), viral gene expression (Purves and Roizman, 1992;Purves et al., 1993;Tanaka et al., 2005), and cell cycle regulation (Advani et al., 2000). HSV-1 UL13 can phosphorylate US3, and proper localization of UL31 and UL34 also depends on UL13 (Kato et al., 2006), indicating an additional role for UL13 in HSV nuclear egress.
We previously noted alterations in nuclear morphology in cells transfected with UL13, but not with an enzymatically-inactive UL13 (K176A) (Geiss et al., 2004). These alterations were not due to apoptosis. Therefore, we tested the possibility that UL13 kinase activity alters the integrity of the nuclear lamina by targeting the nuclear lamins.
MATERIALS AND METHODS
Cells and viruses
Vero cells were maintained in Dulbecco’s modified Eagle medium (DMEM) supplemented with 3% fetal calf serum, 3% newborn calf serum, and 1% penicillin-streptomycin. HeLa cells were maintained in DMEM supplemented with 10% bovine growth serum and 1% penicillin-streptomycin. The wild-type HSV-2 strain 333, kindly provided by Jim Smiley, was propagated in Vero cells. To create HSV-2 333 UL13-HA virus, the HSV-2 UL13 ORF was amplified by PCR from HSV-2 333 genomic DNA in two fragments. For the first fragment, forward primer 5’-TTCTAACCGCACACCGACGGTACC and reverse primer 5’-AGCGTAGTCTGGGACGTCGTATGGGTACGACAGAGGATCCCTTCCGCCCTCGAGC were used to amplify a region containing UL13 and 145bp of upstream UL14 sequence and to insert an HA tag (underlined) at the C-terminus of UL13. For the second fragment, the forward primer 5’-TACCCATACGACGTCCCAGACTACGCTTGAGGTCCCTTCCGCCCTCGAGC and the reverse primer 5’-GGAATTCCGTACTTGGCTCGGCACTTAAC amplified a region which included the remainder of the UL13 ORF and 1106bp of downstream UL12 sequence. The forward primer from the first reaction and reverse primer from the second reaction were then used to amplify full-length UL13 with a 3’ HA tag and flanking sequences. The product was cloned into pRsetB (Invitrogen). To generate the 333-UL13-HA virus, pRsetB-UL13-HA was co-transfected into Vero cells with full-length 333 DNA using the Amaxa nucleofection system. DNA prepared from plaque isolates was subjected to PCR using primers flanking the HA tag, and the PCR products were screened by restriction enzyme digestion with AatII for a site present within the HA tag. Positive plaque isolates from two independent transfections underwent three rounds of plaque purification. The genotype of the 333-UL13-HA viruses was confirmed by Southern blot performed as previously described (Korom et al., 2008).
Plasmids and transfections
UL13 constructs were derived from pSG5-UL13 (Geiss et al., 2004), which contains the HSV-2 UL13 gene downstream of the SV40 early promoter in pSG5 (Stratagene). pSG5-UL13-K176A contains an alanine mutation at lysine 176, resulting in an enzymatically inactive UL13. The UL13 plasmids contain an HA tag fused to the carboxy-terminus of UL13. pcDNA-333VP16 (Korom et al., 2008) contains HSV-2 VP16 downstream of a CMV promoter in pcDNA3.1 (Stratagene). Transfections were performed using Lipofectamine Plus (Invitrogen) according to the manufacturer’s recommendations.
Infections
For immunofluorescence studies, HeLa cells were infected with HSV-2 333 or 333-UL13-HA at MOI of 5. For growth curves, Vero and HeLa cells were infected with 333 or 333-UL13-HA at low (0.01) MOI. Cells were harvested at various time points and virus titers were determined by plaque assay. Statistical significance of difference in titers at individual time points was determined by t test.
Antibodies
Mouse anti-human lamin A/C monoclonal antibody 636 and goat anti-human lamin B1 polyclonal antibody C-20 were obtained from Santa Cruz Biotechnology. Rabbit anti-human lamin A/C polyclonal antibody 2032 was obtained from Cell Signaling Technologies. Mouse anti-VP16 monoclonal antibody 1–21 was obtained from Santa Cruz Biotechnology. The rat anti-HA antibody 3F10 was obtained from Roche Diagnostics. All antibodies were used according to the manufacturer’s recommendations. Secondary antibodies conjugated with Alexa Fluors were obtained from Molecular Probes, and FITC-conjugated anti-rat antibody was obtained from Jackson Immunoresearch.
Indirect immunofluorescence and confocal microscopy
HeLa cells were seeded onto glass coverslips in 12-well plates and transfected with 2µg of plasmid DNA, or were infected at a multiplicity of infection (MOI) of 5. At 2 hr intervals from 12 to 24 hr post-infection or post-transfection cells were washed three times with PBS and fixed with ice-cold methanol at −20ºC for 20 min. Cells were washed with PBS and incubated for 1 hr at room temperature in PBS containing 1% BSA. Cells were then probed with primary and secondary antibodies, washed with PBS, and mounted onto glass slides using SlowFade containing DAPI (Molecular Probes). Fluorescence was visualized using a 60x oil objective on a BioRad 1024 confocal scanning microscope. Images were captured with Image J and formatted using Adobe Illustrator.
Quantitation of immunofluorescence images
Ten random fields of cells were examined in masked fashion for differences in lamin staining intensity (high, medium, or undetectable): High intensity was sharp and intense staining; medium intensity was intermediate staining; and not detectable was nearly complete absence of lamin staining. At least 100 cells were counted per sample in each experiment by a blinded observer, and each experiment was carried out in triplicate. The percentage of cells in each category was determined by dividing the number of cells staining high, medium, or undetectable by the total number of cells counted. The percentages from three independent experiments were used to ascertain the mean and standard deviation. An unpaired t-test was used to determine statistical significance of differences.
Mixed-bed immunoprecipitation
UL13-HA and UL13-K176A-HA were synthesized in vitro using the TNT® T7 coupled transcription/translation reticulocyte lysate system (Promega) according to the manufacturer’s recommendations. The translation products were pre-cleared twice for 30 min each with protein A/G agarose beads (Oncogene Research) at 4°C in immunoprecipitation (IP) buffer (50 mM Tris HCl [pH 8.0], 150 mM NaCl, 1% NP-40) supplemented with 40 mM sodium fluoride, 1mM sodium orthovanadate, 10mM β-glycerophosphate, 2µg/ml aprotinin, 3µg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride, then were bound by incubation with 3F-10 for 2 hr. Lamins A, C, and B1 were derived from HeLa cells and HSV-2 VP16 was derived from VP16-transfected HeLa cells. To isolate these proteins, HeLa cells were scraped into the medium and centrifuged at 1,200 × g for 15 minutes at 4°C. The supernatant was removed and the pellet was resuspended in IP buffer supplemented with complete EDTA-free protease inhibitor cocktail (Roche). Cells were lysed on ice for 20 min with periodic mechanical disruption through a 25-g needle. The lysate was centrifuged at 12,000 × g for 15 min at 4°C and the supernatant was retained for IP. Supernatants were pre-cleared twice for 30 min each with protein A/G agarose beads at 4°C in IP buffer and then were incubated with anti-VP16, anti-lamin A/C or anti-lamin B1 antibodies for 2 hr. Enzyme-antibody and VP16-antibody or lamin-antibody mixtures were combined and incubated with protein A/G agarose beads for 2 hr at 4°C. The beads were washed three times with cold IP buffer, and used in an in vitro kinase assay.
Full length lamin A with the farnesylation site was cloned by PCR amplification of cDNA for human lamin A/C obtained from Open Biosystems and insertion into pCDNA3.1 using primers forward 5’-CACGAATTCCTGCCGGCCATCGAGACCCCG-3’ and reverse 3’-CTGGTCACCCGCTCCTACTAACTCGAGCACGACCAGTGGGCGAGGATGATTGAGCTCGTGGTGCTCGAGTTAGTAGGAGCGGGTGACCAG-5’. The lamin A tail domain was amplified using the same reverse primer and forward primer 5’-CACGAATTCGCCGCCATGGAGAGGCTACGC-3’. Full-length and tail domains of lamin A were expressed using the TNT® T7 coupled transcription/translation system in the absence or presence, respectively, of 35S-methionine and incubated with anti-lamin antibody 636. These were individually mixed with UL13-antibody complexes and mixed bed immunoprecipitations were performed as described above.
Mixed-bed in vitro kinase assay
Kinase assays were performed with UL13 and lamins or VP16 immobilized on protein A/G beads in a total volume of 25µl kinase assay buffer (25 mM Tris-HCl [pH7.5], 20µM EGTA) supplemented with Magnesium/ATP cocktail (Upstate) and 10 µCi [γ-32P]ATP (Perkin Elmer; 3000Ci/mmol) at 30°C for 30 min. The reaction was then centrifuged to separate supernatant from proteins bound to the beads.
Western blot analysis
Proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes (PVDF; Millipore). Target proteins were probed with the appropriate antibody and detected with an alkaline phosphatase-conjugated secondary antibody and BCIP/NBT (Promega). To detect in vitro phosphorylated proteins, the blots were dried and subjected to autoradiography and phosphorimager analysis. Two layers of film were used when the substrate was identified by 35S labeling, the first to detect both 35S and 32P signals and the second the 32P signal.
RESULTS
HSV-2 UL13 induces conformational changes in lamins A and C
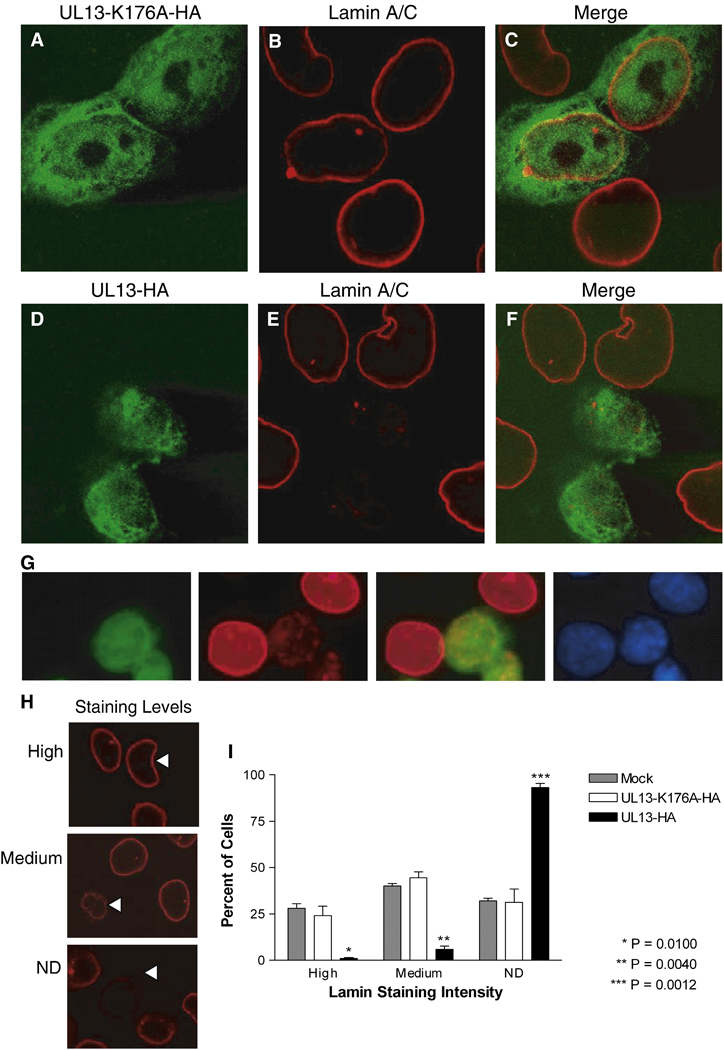
To investigate the effects of UL13 kinase activity on lamins A and C, cells were transfected with plasmids expressing UL13-HA or the enzymatically inactive UL13-K176A-HA, and lamins A and C were detected using a monoclonal antibody previously found to recognize epitopes in the lamin tail domain (Reynolds et al., 2004) (Fig. 1). In UL13-K176A-HA-expressing cells, UL13-K176A-HA was both nuclear and cytoplasmic (Fig. 1A) and lamins A and C were tightly confined to the nuclear rim (Fig. 1B), resembling the staining seen in untransfected cells (Fig. 1B arrowhead). The majority of UL13-K176A-HA did not colocalize with lamins A and C (Fig. 1C). In contrast, in UL13-HA-expressing cells, UL13-HA was found almost exclusively in the nucleus (Fig. 1D) and staining with the lamin A/C monoclonal antibody was decreased at or absent from the nuclear rim (Fig. 1E). In many UL13-HA-expressing cells displaying undetectable lamin staining at the nuclear rim, some lamins remained in the nuclear interior (Fig. 1G). To quantify the change in lamin staining intensity, the intensity in UL13-HA and UL13-K176A-HA expressing cells was analyzed in multiple fields and ranked as high, medium, or undetectable (Fig. 1H). The results revealed a statistically significant difference in lamin staining between UL13-HA- and UL13-K176A-HA- expressing cells at all intensity levels (Fig. 1I), with the greatest difference occurring in undetectable staining. The images shown are cells 18 hrs post-transfection; however, the same phenotype was observed at all two-hour increments from 12 to 24 hrs post-transfection (data not shown). Therefore, the enzymatic activity of UL13 induces changes in nuclear lamins A and/or C such that a decrease in lamin staining intensity is observed at the nuclear rim with a lamin A/C monoclonal antibody recognizing an epitope in the tail domain.
Figure 1. The intensity of lamin A and C staining with a monoclonal antibody differs in HeLa cells expressing UL13-HA or UL13-K176A-HA.
HeLa cells were transfected with UL13-K176A-HA (A–C) or UL13-HA (D–F) expression constructs. At 18 h post-transfection, cells were fixed, stained with anti-HA antibody (green) and monoclonal lamin A/C antibody (red), and examined by confocal microscopy. Images showing both transfected and untransfected cells were merged (C and F). All images were obtained at the same microscope settings and representative images are shown. (G) Confocal images showing cells with lamins remaining in the nuclear interior. Cells were fixed, stained with anti-HA antibody (green), monoclonal lamin A/C antibody (red), and DAPI (blue). (H) Ten random fields were analyzed and lamin staining in each cell was ranked as high or medium intensity or not detectable (ND) by a masked observer. Examples are denoted by arrowheads. (I) Graph shows the mean ± SD from three experiments.
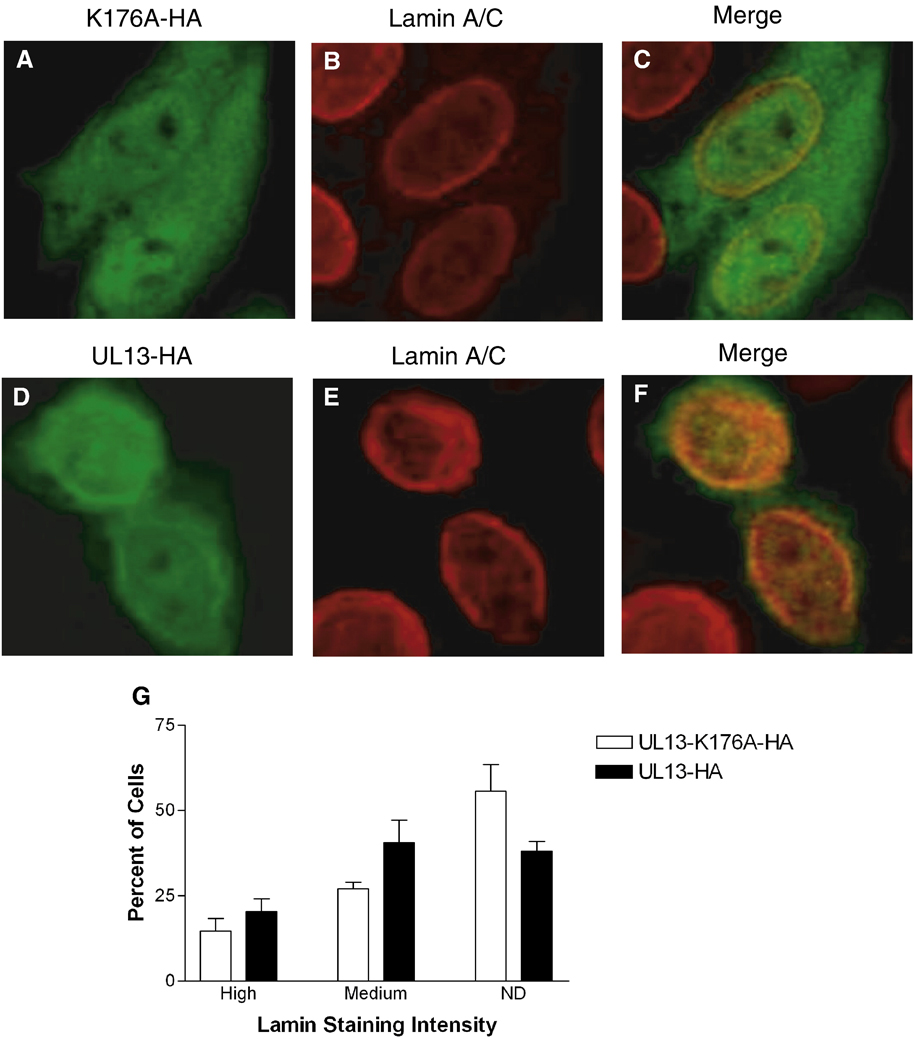
Differential detection of lamins A/C in HSV-1-infected cells led to the conclusion that HSV-1 infection causes conformational changes to lamins A and C that mask the epitope for the lamin A/C monoclonal antibody in the tail domain (Reynolds et al., 2004). To investigate the possibility that the reduction in lamin staining induced by HSV-2 UL13 was also due to altered lamin conformation rather than loss of lamins A/C, we utilized a polyclonal lamin A/C antibody recognizing epitopes in the rod domain of lamins A and C to observe the staining intensity of lamins A and C in UL13-HA and UL13-K176A-HA transfected cells. In mock transfected cells, the polyclonal lamin A/C antibody detected more intranuclear lamins (Figs. 2 B & E) than had the lamin A/C monoclonal antibody (Figs. 1 B & E), as previously observed (Reynolds et al., 2004). Importantly, there was no statistically significant difference in lamin staining intensity between UL13-HA and UL13-K176A-HA expressing cells (Fig. 2G). UL13-HA colocalized with intranuclear lamins, whereas UL13-K176A-HA did not (Fig. 2C & 2F). However, staining with the polyclonal antibody against the rod domain revealed that both UL13-HA and UL13-K176A-HA colocalized more frequently with the lamins at the nuclear rim (Figs. 2C & 2F). These results indicate that there was no large-scale loss of lamins A and C in UL13-HA transfected cells.
Figure 2. Lamin A and C staining with a polyclonal antibody does not differ in HeLa cells expressing UL13-HA or UL13-K176A-HA.
HeLa cells were transfected with UL13-K176A-HA (A–C) or UL13-HA (D–F) expression constructs. At 18 h post-transfection, cells were fixed, stained with anti-HA antibody (green) and a polyclonal lamin A/C antibody (red), and examined by confocal microscopy. Images showing both transfected and untransfected cells were merged (C and F). All images were obtained at the same microscope settings and representative images are shown. Ten random fields were analyzed and lamin staining in each cell was ranked as high or medium intensity or not detectable (ND) by a masked observer. (G) Graph shows the mean ± SD from three experiments. No significant differences between UL13-K176A-HA and UL13-HA were observed (P = 0.2482 to 0.4685).
HSV-2 UL13 induces lamin B1 alterations
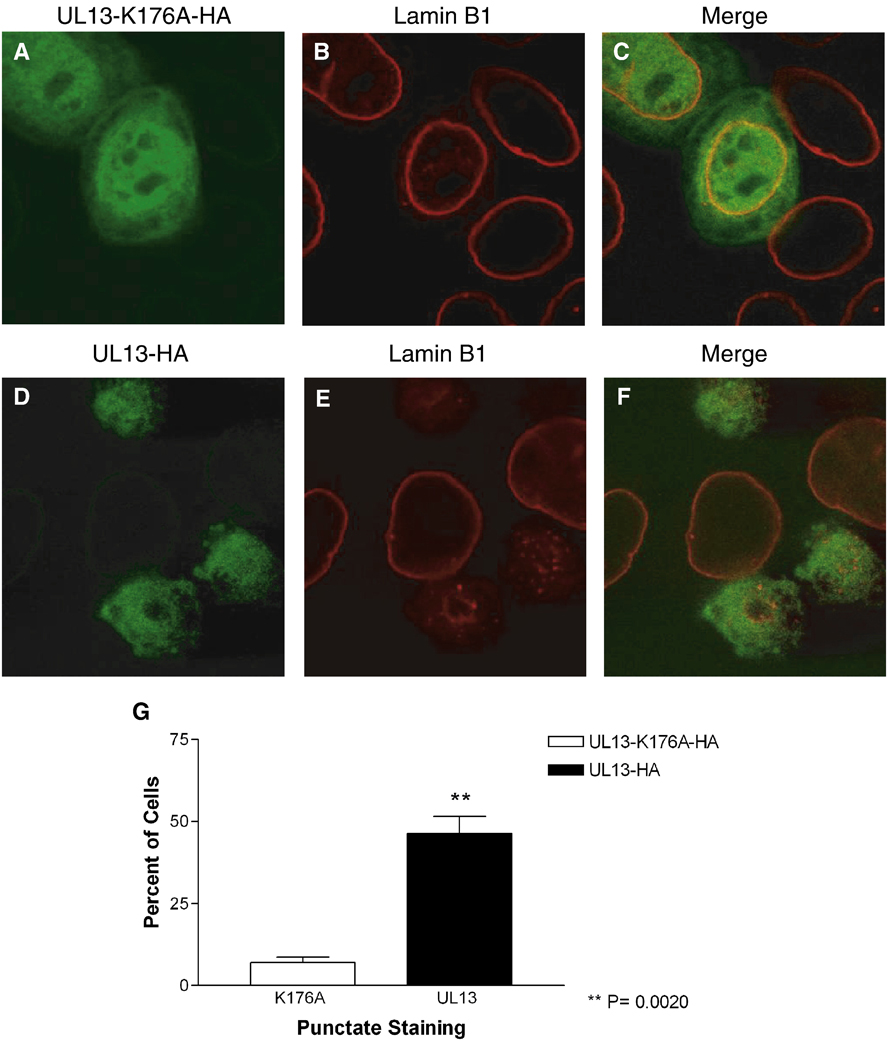
To determine the effects of HSV-2 UL13 expression on lamin B1, HeLa cells were transfected with plasmids expressing UL13-HA or enzymatically inactive UL13-K176A-HA, and lamin B1 was detected using a polyclonal antibody. The distribution of UL13-K176-HA and UL13-HA after transfection was consistent with earlier observations: UL13-K176A-HA localized to the nucleus and cytoplasm and UL13-HA to the nucleus (Figs. 3A and 3D). In UL13-K176A-HA expressing cells, lamin B1 staining was confined to the nuclear rim (Fig. 3B) and resembled that of untransfected cells (Fig. 3B, arrowhead). UL13-HA expression caused redistribution of lamin B1 from the nuclear rim to an intra-nuclear punctuate pattern (Fig. 3E). This pattern was observed from 12 through 24 hr post-transfection (data not shown). UL13-K176A-HA colocalized with lamin B1 at the nuclear rim (Fig. 3C) and UL13-HA colocalized with lamin B1 at some of the intranuclear puntae (Fig. 3F). The difference in redistribution of lamin B1 from the nuclear rim to intranuclear puncta in UL13-HA- compared with UL13-K176A-HA-expressing cells was statistically significant (Fig. 3G). Therefore, enzymatically active UL13 can induce lamin B1 redistribution.
Figure 3. The pattern of lamin B1 staining differs in HeLa cells expressing UL13-HA or UL13-K176A-HA.
HeLa cells were transfected with UL13-K176A-HA (A–C) or UL13-HA (D–F) expression constructs. At 18 h post-transfection, cells were fixed, stained with anti-HA antibody (green) and a polyclonal lamin B1 antibody (red), and examined by confocal microscopy. Images showing both transfected and untransfected cells were merged (C and F). All images were obtained at the same microscope settings and representative images are shown. Ten random fields were analyzed for punctate, intranuclear lamin staining by a masked observer. (G) Graph shows the mean ± SD from three experiments.
HSV-2 UL13 can directly phosphorylate lamins A, C, and B1
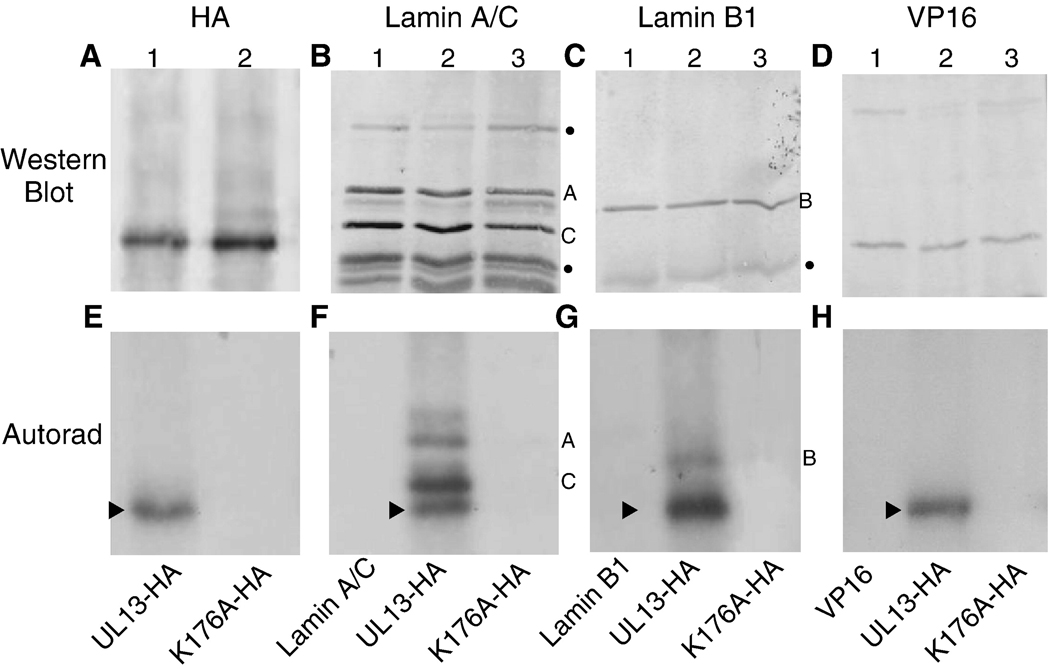
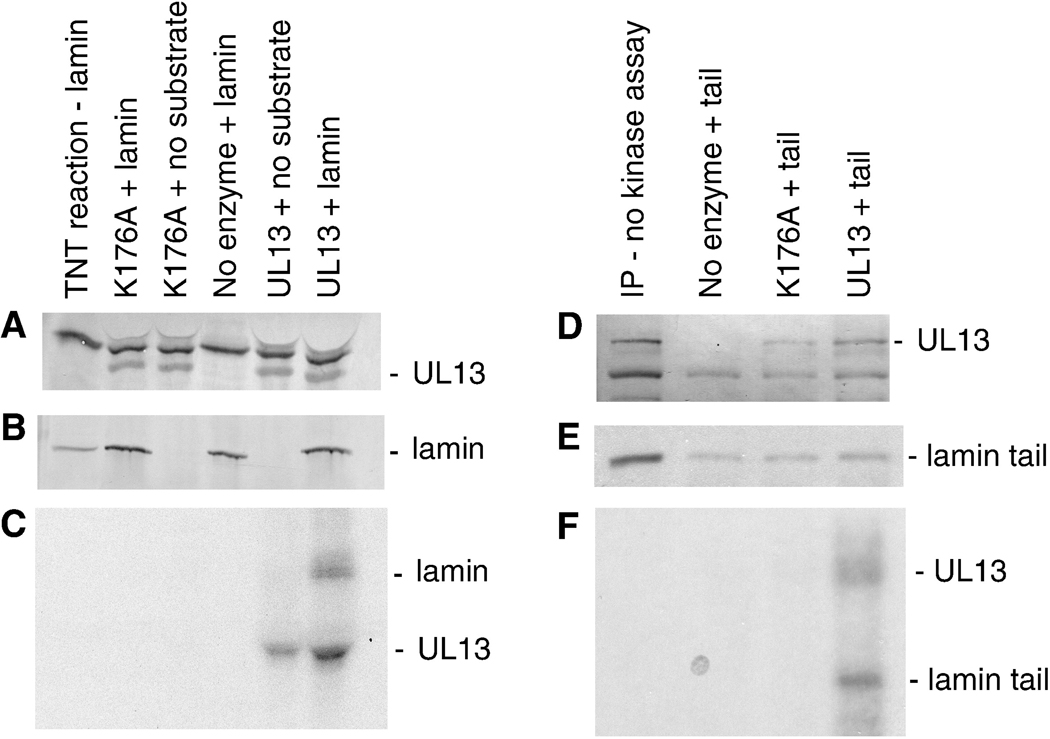
Phosphorylation is one mechanism by which HSV-2 UL13 could alter conformation or distribution of lamins A, C and B1. Thus, a mixed-bed in vitro kinase assay was performed to determine the capacity of UL13 to directly phosphorylate lamins A, C, and B1. The active enzyme, UL13-HA, and inactive enzyme, UL13-K176A-HA, were expressed by in vitro translation and then incubated with anti-HA antibody. The substrates, endogenous lamins A, C, B1 from HeLa cell lysates, were incubated with their respective anti-lamin antibodies. As a negative control substrate for the phosphorylation assay, HSV-2 VP16 was transfected into HeLa cells and the lysates were incubated with anti-VP16 antibody. The enzymes and substrates bound to their respective antibodies were co-immunoprecipitated using protein A/G agarose beads, then utilized in an in vitro kinase assay containing [γ32P] ATP. Following the in vitro kinase assay, the products were separated by SDS-PAGE and probed by western blot for HA-tagged UL13 or UL13-K176A, lamins A, C, and B1, and VP16 (Figs. 4 A–D, respectively). 32P-labeled products were observed by autoradiography of the western blots (Figs. 4E–H). As expected, UL13-HA and UL13-K176A-HA were expressed (Fig. 4A) but only UL13-HA autophosphorylated (Cano-Monreal et al., 2008) (Figs. 4E–H). Lamins A, C, and B1 were phosphorylated by UL13-HA but not by UL13-K176A-HA (Figs. 4F and 4G, respectively). Phosphorylation of lamins by UL13-HA was specific because VP16 was not phosphorylated (Figs. 4D and H). Thus, HSV-2 UL13 can phosphorylate lamins A, C, and B1 in vitro.
Figure 4. HSV-2 UL13-HA can phosphorylate lamins A, C, and B1 in vitro.
In vitro translated UL13-HA and UL13-K176A-HA, lamins A, C or B1 extracted from HeLa cells, and VP16 from transfected HeLa cells were isolated by mixed bed immunoprecipitation. The immunoprecipitates were subjected to an in vitro kinase assay, the products were resolved by SDS-PAGE and detected by western blot using A) anti-HA antibody, B) lamin A/C monoclonal antibody, C) lamin B1 polyclonal antibody, or D) VP16 monoclonal antibody. (E–H) 32P-labeled proteins were detected by autoradiography of the western blots. The UL13-HA and UL13-K176A-HA western blot shown in panel A is representative of the entire experiment. The arrowhead denotes UL13-HA autophosphorylation. Bullets beside western blots denote non-specific bands. Lamin A, B and C bands are denoted by their respective letters.
UL13 can directly phosphorylate the tail domain of lamin A
Lamins must be phosphorylated at conserved serines in their head and tail domains during cell division for disassembly to occur. The loss of monoclonal antibody binding to the lamin A tail domain that occurred in the presence of UL13 would be consistent with a phosphorylation-induced conformational change leading to disassembly. We therefore determined whether HSV-2 UL13 can phosphorylate the lamin tail domain. Full-length lamin A and the tail domain were cloned and used as substrates in an in vitro kinase assay with HSV-2 UL13. UL13-HA and UL13-K176A-HA, and full-length lamin A or the tail domain were expressed by in vitro transcription/translation reactions and then incubated with HA-specific or lamin-specific antibodies, respectively. These enzyme-antibody and substrate-antibody combinations were mixed and coimmunoprecipitated, and then subjected to an in vitro kinase assay. UL13-HA and UL13-K176A-HA were detected by western blot in immunoprecipitations alone or in coimmunoprecipitations with full-length lamin A (Fig. 5A). Full-length lamin A was readily detectable following translation in the reticulocyte lysate and after immunoprecipitation by western blot with anti-lamin antibody (Fig. 5B). Autoradiography revealed autophosphorylated UL13-HA, and a 32P-labeled band corresponding to the mobility of full-length lamin A in coimmunoprecipitates with UL13-HA but not UL13-K176A-HA (Fig. 5C). In a second kinase assay, 35S-labeled lamin A tail domain was coimmunoprecipitated with UL13-HA or UL13-K176A-HA. The western blot probed with anti-HA antibody showed UL13-HA and UL13-K176A-HA were present as expected (Fig. 5D). The first layer of film revealed a radiolabeled band of mobility corresponding to the 35S-labeled lamin A tail which was immunoprecipitated from the reticulocyte lysate and coimmunoprecipitated with UL13-HA and UL13-K176A-HA (Fig. 5E). The second layer of film revealed 32P-labeled bands corresponding to lamin A tail in the presence of UL13-HA but not UL13-K176A-HA, as well as autophosphorylated UL13-HA (Fig. 5F). Thus, UL13 can phosphorylate in vitro full-length lamin A and one or more sites in the tail domain.
Figure 5. HSV-2 UL13-HA can directly phosphorylate the tail domain of lamin A.
In vitro transcribed and translated UL13-HA and UL13-K176A-HA, and full-length or tail domain of lamin A were isolated by mixed bed immunoprecipitation and the immunoprecipitates were subjected to an in vitro kinase assay. The products were resolved by SDS-PAGE (8% gel for full-length lamin and 12% gel for the tail domain), and detected by western blot and autoradiography. A) UL13-HA and UL13-K176A-HA detected with anti-HA antibody, B) full-length lamin A detected by western blot with anti-lamin antibody, and C) 32P-labeled proteins detected by autoradiography; D) UL13-HA and UL13-K176A-HA detected by western blot, E) 35S-labeled and 32P-labeled proteins and F) 32P-labeled proteins detected by autoradiography. The lamin tail domain was not discernible by western blot because it has the same mobility as light chain and other cross reacting bands.
HSV-2 infection changes the nuclear lamina
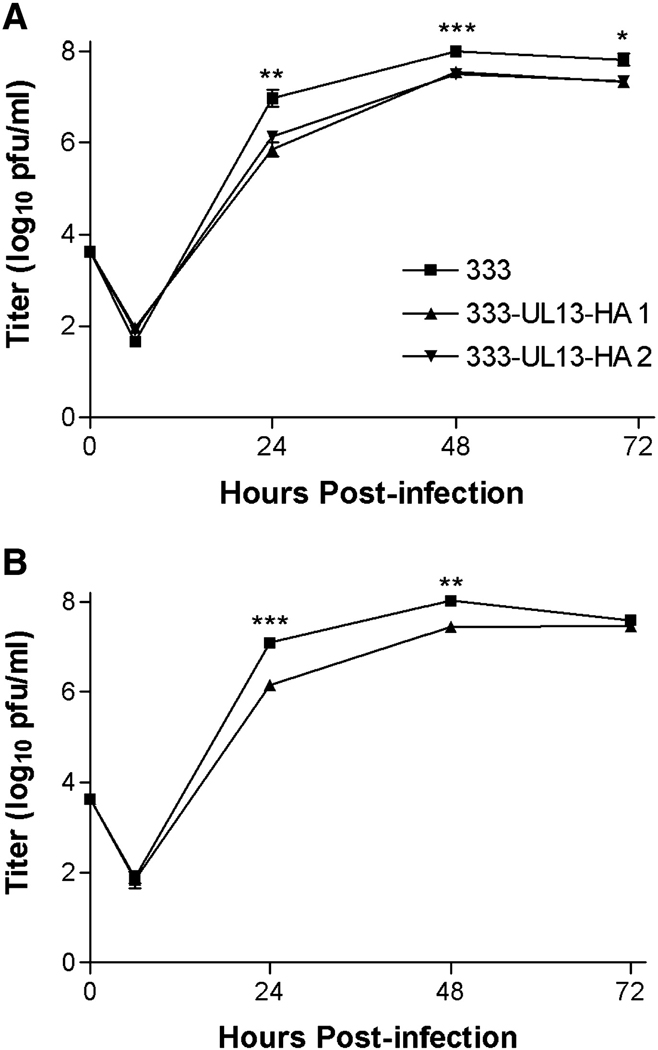
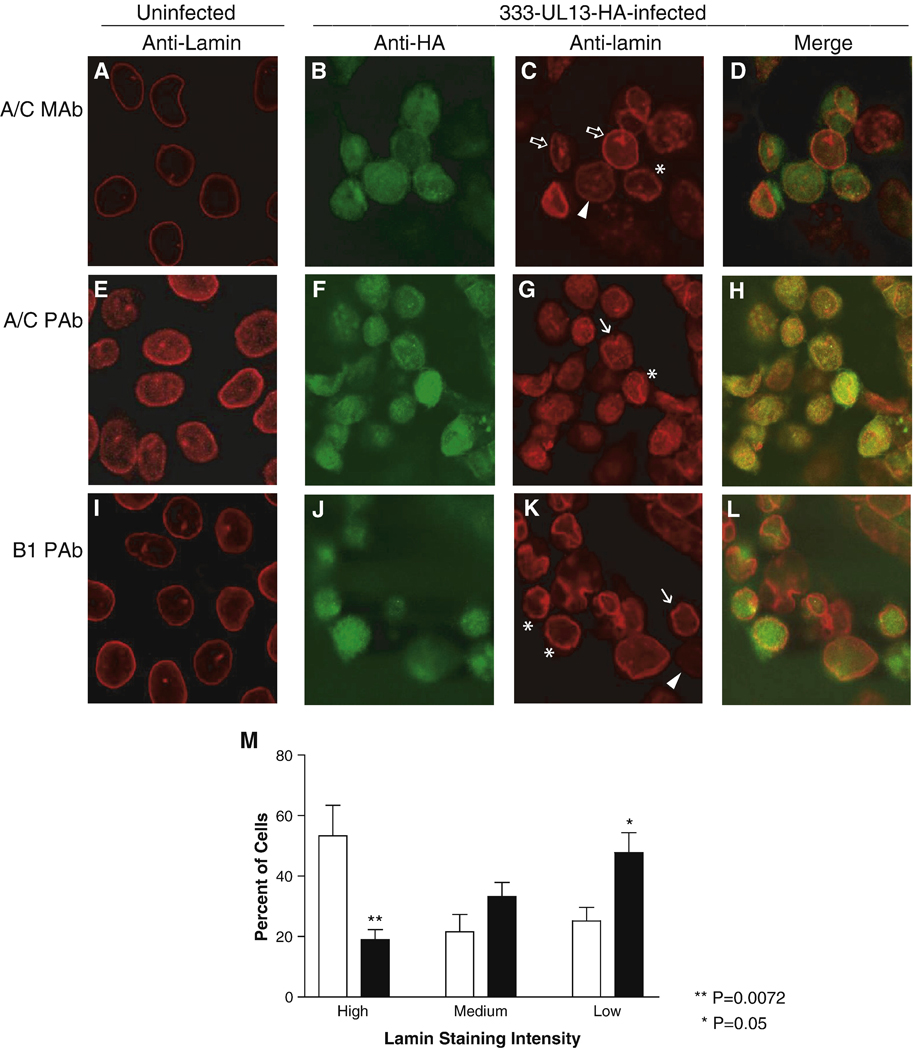
We next asked whether the alterations to lamins A, C and B1 observed in transfected cells expressing UL13-HA occur during infection with HSV-2. Because antibodies to HSV-2 UL13 have been difficult to raise, we constructed HSV-2 recombinants of the parental strain 333 encoding an HA-tagged UL13 (HSV-2 333-UL13-HA). Two independent isolates replicated nearly as well as wild-type 333 in Vero and HeLa cells after infection at low moi (Fig. 6), though the minor differences at each time point were statistically significant. We used HSV-2 333-UL13-HA to observe UL13-HA and the distribution of lamins A, C, and B1 in the context of HSV-2 infection. HeLa cells were left uninfected (Figs. 7A, E, I) or were infected with 333-UL13-HA at MOI of 5 for 18 hr. Cells were stained with anti-HA and anti-lamin monoclonal and polyclonal antibodies used in the transfection studies to determine whether differential staining with the two also occurs during infection, and were observed by confocal microscopy. As in the UL13-HA transfection experiments, UL13 staining was predominantly nuclear during infection (Figs. 7B, F, J). Also as seen in UL13-HA-transfected cells, some infected cells showed a decrease in lamin staining intensity with the monoclonal lamin A/C antibody (Fig. 7C, arrowhead). Decreased lamin A/C staining occurred in more cells infected with HSV-2 333-UL13-HA, and high levels of lamin staining were observed more often in mock-infected cells (Fig. 7M). Additional abnormalities in lamins A/C were observed in infected cells, including discontinuous nuclear rim staining (Fig. 7C, asterisk), and increased intranuclear lamin staining (Fig. 7C, open block arrows). Staining intensity with the polyclonal lamin A/C antibody did not decrease, resembling what had been observed in UL13-HA and UL13-K176A-HA-transfected cells. There were, however, abnormalities in the lamin staining pattern observed with the polyclonal lamin A/C antibody, including invagination (Fig. 7G, arrow) and discontinuous staining at the nuclear rim (Fig. 7G, asterisk). Staining for lamin B1 in infected cells did not reveal the redistribution seen in UL13-HA-transfected cells. Instead, decreased lamin B1 staining was observed (Fig. 6K, arrowhead), as well as invagination (Fig. 7K, arrows), and discontinuous nuclear rim staining (Fig. 7K, asterisks). These lamin alterations were observed in cells stained 12 through 24 hr post-transfection and in HeLa cells infected with wild-type HSV-2 (data not shown). As was seen in transfected cells, the majority of UL13-HA in infected cells did not appear to colocalize with lamins which had been stained with the monoclonal lamin A/C or polyclonal lamin B1 antibodies. However, a high degree of colocalization was observed when the polyclonal lamin A/C antibody was used. Altogether, the variety of phenotypes seen in infected cells was greater than that seen in UL13-HA-transfected cells. Therefore, UL13 is sufficient to cause some but not all of the lamin alterations observed in infected cells.
Figure 6. HSV-2 333-UL13-HA replicates similarly to wild-type HSV-2 333.
A) Vero cells or B) HeLa cells were infected in duplicate at MOI 0.01 with HSV-2 333 or with independent isolates of HSV-2 333-UL13-HA. Cultures were harvested at the indicated times and virus titers were determined by plaque assay. Data points are the geometric mean ± SEM of duplicate wells from three independent experiments. *, P = 0.0274; **, P = 0.0066 to 0.0098; ***, P <0.0001 to 0.0007 for 333 compared with 333-UL13-HA 1.
Figure 7. Infection with HSV-2 encoding HA-tagged UL13 alters the nuclear lamin network.
HeLa cells were left uninfected (A, E, I) or were infected with HSV-2 333-UL13-HA at an MOI of 5. At 18 hr post-infection, cells were fixed, stained with anti-HA antibody (green) and anti-lamin antibodies (red). Antibodies used were (A–D) monoclonal lamin A/C antibody, (E–H) polyclonal lamin A/C antibody, or (I–L) polyclonal lamin B1 antibody. Cells were analyzed by confocal microscopy and the anti-HA and anti-lamin images were merged. All images were obtained at the same microscope settings, and representative images are shown. Abnormalities in the lamin staining pattern include decreased lamin staining, arrowhead; invagination, arrow; increased intranuclear lamin staining, open block arrow; and discontinuous staining at the nuclear rim, asterisk. (M) Fifteen random fields of infected cells and three random fields of mock-infected cells were analyzed and lamin staining intensity was ranked as high, medium or low by a masked observer. Graph shows the mean ± SD.
DISCUSSION
Herpesviruses replicate in the host cell nucleus and must disrupt the nuclear lamina to continue the process of virion assembly and egress from the cell. We have found that the HSV-2 UL13 protein kinase alters the nuclear lamins. Extensive colocalization is observed between nuclear lamins and UL13-HA or UL13-K176A-HA, especially at the nuclear rim. HSV-2 UL13-HA but not the enzymatically-inactive UL13-K176A-HA induces changes in lamins A and C that result in loss of monoclonal but not polyclonal antibody binding, presumably due to masking of the monoclonal antibody epitope in the tail domain. UL13-HA but not UL13-K176A-HA also causes redistribution of lamin B1 to the nuclear interior. HSV-2 UL13 can directly phosphorylate lamins A, C, and B1 in an in vitro kinase assay, suggesting that lamin alterations induced by HSV-2 UL13 in cells could result from direct phosphorylation by UL13. Examination of lamins in HSV-2 infected cells revealed multiple alterations, not all of which were seen in cells expressing UL13 alone, indicating that multiple viral proteins play a role in altering lamins during infection.
Differential staining with antibodies to lamins A and C detected a change in the lamin tail domain in the presence of UL13-HA. Decreased staining of lamins A and C at the nuclear rim of UL13-HA-expressing cells compared with untransfected cells was observed when a monoclonal lamin A/C antibody that recognizes epitopes in the lamin tail domain was used. Lamins A and C that had relocated to the nuclear interior in UL13-HA expressing cells were detected only at low levels, and the remaining lamin staining did not substantially colocalize with UL13-HA. This decrease in lamin A/C staining was not observed, however, with a polyclonal lamin A/C antibody recognizing epitopes in the lamin rod domain. Therefore UL13 appears to induce conformational changes in the tail domain of lamins A and C, and only residual lamins not associated with and modified by UL13 remain detectable with the monoclonal antibody. Alternatively, decreased staining of lamins A and C could represent preferential degradation of a subset of lamins A and C when associated with UL13, a subset recognized by the monoclonal antibody. Such loss of lamins A and C would not be detected if the subset recognized by the monoclonal antibody was only a minor fraction of those lamins recognized by the polyclonal antibody, whose staining intensity was maintained.
Lamins contain conserved serines in their head and tail domains that must be phosphorylated for lamin disassembly to occur. This normally occurs during cell division though the action of cellular kinases. The Cdc2-kinase phosphorylates lamin A at sequences with the motif S/TPxS/R that are functionally important in lamin disassembly (Haas and Jost, 1993). We have previously shown that the amino acid pair SP can be a minimal recognition sequence for HSV-2 UL13 phosphorylation (Cano-Monreal et al., 2008). Lamins A and C contain one SP sequence in the head domain and three SP sequences in the mature tail domain, but no SP sequences in the intervening rod domain. Here, we have shown that HSV-2 UL13 can phosphorylate lamins A and C in vitro. UL13 also can phosphorylate the tail domain of lamin A in vitro, suggesting that the decreased reactivity seen with the monoclonal antibody recognizing epitopes in the tail domain of lamins A and C could be due to a conformational change in this region induced by HSV-2 UL13 phosphorylation. Alternatively, phosphorylation of the tail domain by UL13 may permit the association of lamins A and C with another cellular protein, leading to epitope masking.
The capacity of HSV-2 UL13 to phosphorylate the lamin A tail domain implies that UL13 phosphorylates the lamin tail in infected cells, leading to a conformational change that facilitates their dissolution. Disruption of lamin A polymers by UL13 would likely facilitate virion egress through the nuclear envelope (Mou et al., 2008). Cdc2 causes disassembly of lamin A/C polymers by phosphorylation of serine 22 in the head domain and 390 and 392 in the tail domain (Heald and McKeon, 1990). The UL13 homolog in Epstein-Barr virus, BGLF4 kinase, can phosphorylate lamin A head and tail domains in vitro, and serine22, serine390 and serine392 are important in EBV-induced redistribution of nuclear lamin A (Lee et al., 2008). The HCMV homolog of UL13, UL97, also mimics Cdc2 activity by phosphorylating serine22 and serine 390 in vitro. In addition, phosphorylation of serine22 and serine 392 in HCMV-infected cells and disruption of the nuclear lamina depends upon UL97 (Hamirally et al., 2009). Although HSV-2 UL13 does not recognize substrates by using the Cdc2 motif, its capacities to utilize SP as a minimal recognition sequence (Cano-Monreal et al., 2008), to disrupt the nuclear lamina, and to phosphorylate the SP-containing tail domain of lamin A in vitro suggest that HSV-2 UL13 and its HCMV and EBV counterparts mediate lamin disassembly through similar mechanisms. An investigation of the site(s) phosphorylated on lamin A by HSV-2 UL13 is underway to address this question.
HSV-2 UL13-HA but not the enzymatically inactive UL13-K176A-HA redistributed lamin B1 from the nuclear rim to the intranuclear space, but a decrease in lamin B1 staining was not observed (Fig. 3). Lamin B associates with the lamin B receptor (LBR), an integral membrane protein that anchors lamin B to the INM. Upon lamin phosphorylation during mitosis, lamin B remains bound to the LBR and remnants of the nuclear envelope (Gerace and Blobel, 1980). HSV-1 infection of COS-1 cells results in redistribution of a LBR-GFP fusion protein to the cytoplasm and lamin B1 to the cytoplasm and nuclei (Scott and O'Hare, 2001). Lamin B relocalization to the cytoplasm also occurs during HSV-1 infection of HEp-2 and HeLa cells (Park and Baines, 2006). The HCMV UL13 homologue UL97 induces redistribution of LBR from the nuclear rim to intranuclear granular structures, and this event is dependent on UL97 enzymatic activity. We did not examine the effect of HSV-2 UL13-HA on LBR; however we did observe a similar relocalization of lamin B1 to the intranuclear space. Therefore, the protein kinase activities of UL13 and UL97 may redistribute lamin B and LBR to similar intranuclear structures.
Infection of cells with HSV-2 revealed additional alterations not observed when UL13-HA was expressed alone, although UL13-HA was found almost exclusively within the nucleus and nuclear rim in both transfected and infected cells. Previous transfection and infection studies have implicated the HSV-1 proteins UL31, UL34, and US3 (Reynolds et al., 2004;Mou et al., 2007) and the cellular PKCs (Park and Baines, 2006) in altering lamins. However, none of these proteins fully account for the lamin alterations seen during infection (Park and Baines, 2006), indicating additional viral or cellular proteins are necessary. During HSV-1 infection, UL13 phosphorylates US3 and participates in determining the proper localization of UL31 and UL34 at the nuclear rim (Kato et al., 2006). We found that HSV-2 UL13, when expressed alone, induces alterations in lamins A, C, and B1 and can directly phosphorylate lamins A, C, and B1 in vitro. The influence of HSV-1 UL13 on other proteins that regulate integrity of the nuclear lamina raises the possibility that HSV-2 UL13 could phosphorylate lamins as well as other viral protein(s) to cooperatively achieve sufficient lamin alteration and disassembly for virion egress through the nuclear membrane to occur.
Finally, it is interesting to note that UL13-HA expressed by transfection and UL13-HA in infected cells localizes almost exclusively to the nucleus, but UL13-K176A-HA is distributed throughout the cytoplasm as well as in the nucleus. Possibly HSV-2 UL13 becomes trapped in the nucleus due to interaction with substrates or binding partners. Protein kinase activity either plays a role in retention or the mutant UL13-K176A protein does not adopt the proper conformation for stable interaction with critical binding partner(s). Further work will be needed to elucidate the mechanism underlying nuclear retention of HSV-2 UL13.
ACKNOWLEDGEMENTS
We thank Hong Wang for assistance in isolating HSV-2 333-UL13-HA. We thank Maria Korom, Jane Schrimpf, and the Blight, Leib, Diamond, Pekosz, Stuart, Wang, and Yu labs for helpful advice and discussions. This work was supported by PHS AI061845 to G. C.-M. and PHS AI057573 and AI059050 to L. A. M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Advani SJ, Brandimarti R, Weichselbaum RR, Roizman B. The disappearance of cyclins A and B and the increase in activity of the G(2)/M-phase cellular kinase cdc2 in herpes simplex virus 1-infected cells require expression of the alpha22/U(S)1.5 and U(L)13 viral genes. J. Virol. 2000;74:8–15. [PMC free article] [PubMed] [Google Scholar]
- Cano-Monreal GL, Tavis JE, Morrison LA. Substrate specificity of the herpes simplex virus type 2 UL13 protein kinase. Virology. 2008;374:1–10. doi: 10.1016/j.virol.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields AP, Thompson LJ. The regulation of mitotic nuclear envelope breakdown: a role for multiple lamin kinases. Prog. Cell Cycle Res. 1995;1:271–286. doi: 10.1007/978-1-4615-1809-9_22. [DOI] [PubMed] [Google Scholar]
- Geiss BJ, Cano GL, Tavis JE, Morrison LA. Herpes simplex virus 2 VP22 phosphorylation induced by cellular and viral kinases does not influence intracellular localization. Virology. 2004;330:74–81. doi: 10.1016/j.virol.2004.08.034. [DOI] [PubMed] [Google Scholar]
- Gerace L, Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat.Rev.Mol.Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Haas M, Jost E. Functional analysis of phosphorylation sites in human lamin A controlling lamin disassembly, nuclear transport and assembly. Eur.J.Cell Biol. 1993;62:237–247. [PubMed] [Google Scholar]
- Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, Golan DE, Marto JA, Coen DM. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS.Pathog. 2009;5:e1000275. doi: 10.1371/journal.ppat.1000275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 1990;61:579–589. doi: 10.1016/0092-8674(90)90470-y. [DOI] [PubMed] [Google Scholar]
- Kato A, Yamamoto M, Ohno T, Tanaka M, Sata T, Nishiyama Y, Kawaguchi Y. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 2006;80:1476–1486. doi: 10.1128/JVI.80.3.1476-1486.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kato K. Protein kinases conserved in herpesviruses potentially share a function mimicking the cellular protein kinase cdc2. Rev.Med.Virol. 2003;13:331–340. doi: 10.1002/rmv.402. [DOI] [PubMed] [Google Scholar]
- Korom M, Wylie KM, Morrison LA. Selective ablation of virion host shutoff protein RNase activity attenuates herpes simplex virus 2 in mice. J. Virol. 2008;82:3642–3653. doi: 10.1128/JVI.02409-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Huang YH, Lin SF, Chang Y, Chang YH, Takada K, Chen MR. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J. Virol. 2008;82:11913–11926. doi: 10.1128/JVI.01100-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likhacheva EV, Bogachev SS. Lamins and their functions in cell cycle. Membr.Cell Biol. 2001;14:565–577. [PubMed] [Google Scholar]
- Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol.Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- Marschall M, Marzi A, aus dem SP, Jochmann R, Kalmer M, Auerochs S, Lischka P, Leis M, Stamminger T. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol.Chem. 2005;280:33357–33367. doi: 10.1074/jbc.M502672200. [DOI] [PubMed] [Google Scholar]
- Mettenleiter TC. Herpesvirus assembly and egress. J. Virol. 2002;76:1537–1547. doi: 10.1128/JVI.76.4.1537-1547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milbradt J, Auerochs S, Marschall M. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen.Virol. 2007;88:2642–2650. doi: 10.1099/vir.0.82924-0. [DOI] [PubMed] [Google Scholar]
- Morrison EE, Wang YF, Meredith DM. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 1998;72:7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou F, Forest T, Baines JD. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 2007;81:6459–6470. doi: 10.1128/JVI.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou F, Wills EG, Park R, Baines JD. Effects of lamin A/C, lamin B1, and viral US3 kinase activity on viral infectivity, virion egress, and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane. J. Virol. 2008;82:8094–8104. doi: 10.1128/JVI.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou F, Wills E, Baines JD. Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 2009;83:5181–5191. doi: 10.1128/JVI.00090-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranyi W, Haas J, Wagner M, Krohne G, Koszinowski UH. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science. 2002;297:854–857. doi: 10.1126/science.1071506. [DOI] [PubMed] [Google Scholar]
- Overton HA, McMillan DJ, Klavinskis LS, Hope L, Ritchie AJ, Wong-kai-in P. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology. 1992;190:184–192. doi: 10.1016/0042-6822(92)91204-8. [DOI] [PubMed] [Google Scholar]
- Park R, Baines JD. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 2006;80:494–504. doi: 10.1128/JVI.80.1.494-504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Sanghera JS, Pelech SL, Nigg EA. Mitogen-activated protein kinases phosphorylate nuclear lamins and display sequence specificity overlapping that of mitotic protein kinase p34cdc2. Eur.J.Biochem. 1992;205:287–294. doi: 10.1111/j.1432-1033.1992.tb16779.x. [DOI] [PubMed] [Google Scholar]
- Purves FC, Longnecker RM, Leader DP, Roizman B. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 1987;61:2896–2901. doi: 10.1128/jvi.61.9.2896-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves FC, Roizman B. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein alpha 22. Proc.Natl.Acad.Sci.U.S.A. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves FC, Ogle WO, Roizman B. Processing of the herpes simplex virus regulatory protein alpha 22 mediated by the UL13 protein kinase determines the accumulation of a subset of alpha and gamma mRNAs and proteins in infected cells. Proc.Natl.Acad.Sci.U.S.A. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 2001;75:8803–8817. doi: 10.1128/JVI.75.18.8803-8817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AE, Liang L, Baines JD. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 2004;78:5564–5575. doi: 10.1128/JVI.78.11.5564-5575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott ES, O'Hare P. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 2001;75:8818–8830. doi: 10.1128/JVI.75.18.8818-8830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Holley M, Baines J, Roller R, Knipe DM. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 2004;78:5591–5600. doi: 10.1128/JVI.78.11.5591-5600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RF, Smith TF. Identification of new protein kinase-related genes in three herpesviruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J. Virol. 1989;63:450–455. doi: 10.1128/jvi.63.1.450-455.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Nishiyama Y, Sata T, Kawaguchi Y. The role of protein kinase activity expressed by the UL13 gene of herpes simplex virus 1: the activity is not essential for optimal expression of UL41 and ICP0. Virology. 2005;341:301–312. doi: 10.1016/j.virol.2005.07.010. [DOI] [PubMed] [Google Scholar]