Abstract
We provide a thought experiment in advanced molecular engineering leading to the construction of a molecular robot acting as a DNA-based closed-loop device for the management of diabetes mellitus. The key components of this robot are nucleic acid enzymes regulated allosterically by glucose. In the presence of glucose above a threshold level the robot moves, releasing insulin from a lawn covered with insulin, but stops in its tracks whenever glucose decreases to a level below threshold; thus, the robot could provide an insulin release response to spikes in glucose concentration in real time.
Keywords: allosteric regulation, deoxyribozymes, glucose sensor, insulin release, molecular robots
Deoxyribonucleic acid molecules are best known for their central biological role as carriers of genetic information, yet they have a rich alternative life in the field of molecular engineering.1 Here DNA can self-assemble into complex two (2D)- and three-dimensional (3D) patterned structures,2,3 can autonomously compute through self-assembly,4 can play games with perfect strategy,5 or can be used in the construction of primitive remote-controlled molecular robotic components.6 The question that now arises with these modern advances in DNA manipulation is: could any of these discoveries lead to therapeutic applications7 or are they destined to remain just scientific oddities, albeit impressive ones? This commentary starts as a thought experiment in advanced molecular engineering, developing further into a proposal for a stepwise approach to the construction of DNA-based closed-loop devices of increasing complexity for the management of diabetes mellitus. Such devices would either act in stand-alone fashion or provide support for other therapies.
Let us start by picturing a “robot” of molecular-scale dimensions (Figure 1). The robot resides in the human body on the surface of a biocompatible matrix (e.g., 2D surface of a bead or 3D gel) coated with a lawn of insulin. Instead of nuts and bolts, this robot consists of molecular sensors for glucose and insulin and a catalyst for releasing insulin. On sensing glucose above a threshold level the molecular robot is triggered to start a relentless roll over the lawn, releasing the insulin from the surface as it progresses. Because the robot senses glucose in real time, it can stop in its tracks whenever glucose decreases to a level below threshold, thereby suspending the release of insulin until glucose levels rise again. The insulin sensor of the robot helps it reposition in areas with unreleased insulin, while it is in an inactive phase (i.e., glucose levels below threshold); thus, the robot could provide an insulin release response to many spikes in glucose concentration. The most immediate (and quite reasonable) reaction to this proposal is that such a molecular device might be better discussed in the context of a science fiction novel, even at the level of a test-tube demonstration. Simply put, molecular behavior of this complexity, using unnatural components, has not been demonstrated before.
Figure 1.
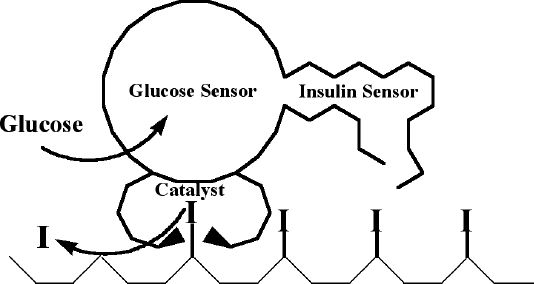
A schematic representation of a molecular “robot” that continuously releases insulin (I) when the level of glucose is high. The robot consists of a glucose sensor, insulin sensor, and catalysts releasing insulin.
We now argue that a thoughtful combination of already demonstrated molecular behaviors can lead to the eventual construction of a similar molecular device, with the development of less complex, but still potentially useful, devices as the intermediate steps. Let us now discuss the basic components that a molecular system for diabetes management would require and how these components can be combined into functional devices, culminating in a full-blown molecular robot capable of controlling glucose levels by insulin release without the side effects of hyper- or hypoglycemia.
DNA-Based Molecular Sensors for Glucose
The first step in the construction of molecular devices for glucose-sensitive insulin release is the engineering of a glucose sensor into the device. For a DNA-based device, this means we first have to isolate a glucose-recognition region based on oligonucleotides (Figure 2). Many oligonucleotide-based recognition regions for small molecules, named aptamers, are known. These are isolated through a process of in vitro selection and amplification (also known as SELEX). In a multistep process, which starts from a large library of some 1014 members of random oligonucleotide sequences, one can select (through repeated polymerase chain reaction amplifications) the members of the library that bind with great specificity to an affinity material displaying the targeted molecule.8 The diagnostic potential of aptamers in the context of managing diabetes has been discussed elsewhere.9
Figure 2.
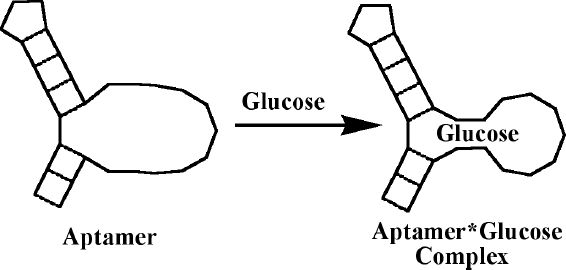
A schematic representation of a hypothetical aptamer binding glucose. Free aptamer has a poorly defined binding site, which forms a tightly organized structure in the presence of glucose (i.e., adaptive binding).
While no aptamers to glucose have been reported as of now, binding motifs to several sugars, including cellobiose (a glucose dimer),10 and glucose-6-phosphate11 are known. Despite the small size of glucose and the lack of any charged or hydrophobic groups, there is no reason why an oligonucleotide fold binding glucose could not be isolated under carefully chosen selection conditions. For example, proteins bind glucose with micromolar dissociation constants12,13 and all glucose-binding protein functionalities are also naturally available to nucleic acids. Alternatively, it would be relatively easy to engraft bisboronic acid functionalities,14 which show natural preference for complexing glucose, onto oligonucleotide folds, prior to the selection process.
In order to be useful, the DNA-based recognition regions for glucose should be sensitive to glucose in the low millimolar range, preferably with a response threshold just above fasting glucose levels. These requirements are well within the reach of DNA-based sensors. For example, with a micromolar sensor as a starting point, it is easy to introduce point mutations and obtain desired millimolar binders. Furthermore, two aptameric-binding pockets can be combined into a single aptamer with multiple, connected binding sites,15 with a potential for cooperative binding, and a more threshold-like response.
Glucose-Triggered Catalytic Release of Insulin
Once we have glucose-binding recognition regions, our next task is to integrate it into a catalytic system for the release of insulin (Figure 3). This article does not mention numerous options of using noncatalytic molecular systems (e.g., two-state switches binding exclusively either insulin or glucose); such a system might be effective in releasing insulin, but is unlikely to be very effective over extended lengths of time.
Figure 3.
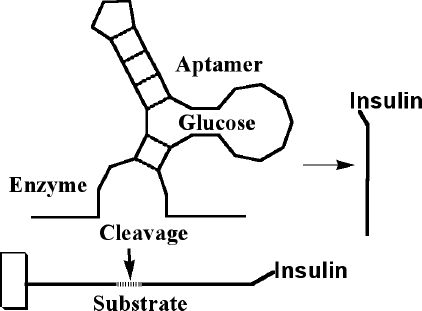
The aptamer can be combined with a nucleic acid enzyme (e.g. deoxyribozyme) in such a way that glucose controls cleavage of the substrate oligonucleotide. If the substrate oligonucleotide is conjugated with insulin on one end, and attached to matrix on other, this system can be used to release insulin from beads.
Aptameric recognition regions can be used to regulate activities of nucleic acid enzymes (called ribozymes if made of RNA or deoxyribozymes if they are DNA analogues) by using some of the already published approaches.16 Advantages of nucleic acid enzymes go beyond their ability to interface with aptamers; as shown later, they have strong residual binding to products (which is usually considered a drawback for turnover, but in our case will be helpful to ensure processivity), and their kinetic and binding properties can be optimized easily. This can eventually lead to emulating complex multiphase insulin release exhibited by the natural system.
As it stands now, most nucleic acid enzymes cleave other oligonucleotides; therefore, the fastest way to adapt insulin to being released by deoxyribozymes is to attach insulin to a bead or matrix using a deoxyribozyme substrate as a tether. For example, the LysB29 of insulin is a possible point of attachment for a polyethylene glycol linker that does not perturb biological activity and, in fact, actually increases the stability of the preparations17; thus, it could be used for conjugation of an oligonucleotide tether as well. The simplest molecular device that can take advantage of such a conjugate is a bead cocoated with a glucose-sensitive enzyme surrounded with insulin–substrate conjugates. Upon sensing glucose the enzyme will cleave some of the proximal substrates. In principle, such beads could be used for more than one increase in glucose concentrations, as long as we accept that the response to the second challenge will be slower and that glucose concentrations will reach higher values.
Oligonucleotide–insulin conjugates are unlikely to be any more immunogenic than synthetic insulin or to have any toxicity (save for some idiosyncratic reason, which could be avoided by changing the oligonucleotide conjugated to insulin). The drawback of such devices is an increased price, at least initially, while improved synthetic schemes are developed.
Moving Molecular Devices
We have discussed an approach that allows us to release insulin in a glucose-dependent manner using catalytic cleavage dependent on glucose. However, are there any known physical phenomena that could be used to construct a moving molecular “robot” from our catalytic parts, as described in the introduction? As a reminder, we stress that the robot has to reposition itself in areas that are rich with unreleased insulin in order to be equally effective after each glucose spike. As a starting point, we can picture an enzyme that processively grazes substrates tethered to a bead, behaving as a self-avoiding random walker (i.e., it avoids already visited areas), and that is always moving in the direction of new substrates. There are several problems with such an enzyme, for example, how to achieve any prolonged processivity (i.e., migration toward new substrates without leaving the matrix) and how to stop an enzyme from diffusing away from the matrix, upon grazing itself into a space without substrates, or becoming inactive in a low glucose environment. We now propose to solve these problems by attaching multiple nucleic acid enzymes sensitive to glucose as “legs” to an inert body in a design we have named “spider” molecules18 (Figure 4).
Figure 4.
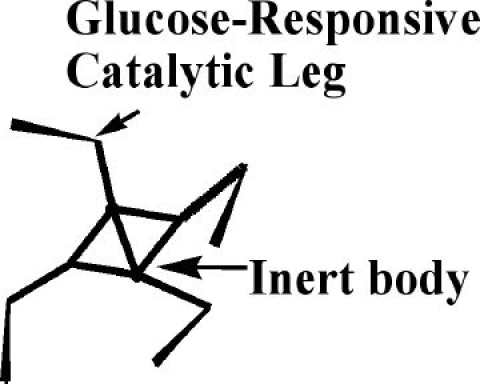
A schematic representation of a four-legged spider molecule that continuously releases insulin when the glucose level is high. The spider consists of glucose-sensitive nucleic acid catalysts as legs, connected to an inert body. Catalytic legs can release insulin by cleaving from the surface insulin–oligonucleotide conjugates.
Let us consider what happens if a spider molecule, containing several catalytic “legs” sensitive to glucose, attaches to a matrix displaying substrates with insulin attached (Figure 5). With properly adjusted parameters, such as leg span, intersubstrate distance, and various catalytic constants, individual legs will start to randomly cleave substrates upon addition of glucose. Upon cleavage, individual legs will dissociate from complexes with products (releasing product conjugated to insulin into solution) and associate with new substrates, displaying a net migration in the direction of new substrates. The processivity can be increased by adjusting binding parameters to substrates and products, as well as the number of legs. Importantly, if captured in an area covered with only matrix-bound products, the spider molecule will not immediately leave the matrix. The spider will proceed with the process of dissociation and association of individual legs from the matrix until it migrates to new substrate-covered areas, wherein binding of the spider to the matrix is the strongest.
Figure 5.
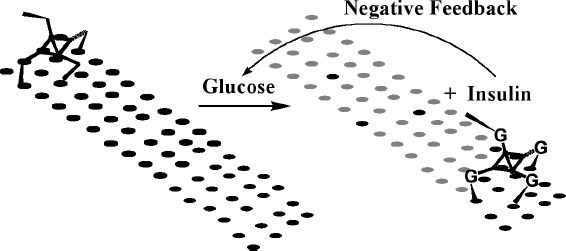
A schematic representation of a spider bound to a matrix (surface) covered with insulin–oligonucleotide conjugates (black ellipses). The spider is bound to the surface tightly through multivalent Watson–Crick-based pairing. Upon sensing glucose, legs become activated and the spider moves over the surface, releasing insulin (turning black ellipses into gray). Released insulin diminishes glucose concentration, slowing down the spider in a feedback loop.
We have demonstrated the behavior of the first molecular spiders with two to six deoxyribozyme legs, sensitive to the presence of Zn2+ ions, and releasing oligonucleotide products from a dextran matrix.18 Figure 6 shows the time-dependent release of oligonucleotide products from the surface using spider molecules to carry out the release. The rate of release is almost the same, regardless whether spiders cleave 10, 40, or 60% of the substrates available to them in the matrix. This is indicative of spiders migrating into new substrate-rich areas. Furthermore, the release of products by spiders can be fully stopped and restarted by the removal and addition of necessary cofactors (this process can be demonstrated easily with Zn2+/EDTA switching, but, in principle, oscillations in glucose concentration can do the same). The cumulative binding, as a result of its multiple numbers of legs, keeps the spider bound to the matrix, with or without movement, albeit the spider will always move to areas with substrates. We can also predict that with an increased number of glucose-sensitive legs, inactive legs will slow down the spiders, and this phenomenon can be used to engineer movement only above certain glucose concentrations. Mixed formulations of spiders with different glucose recognition regions (or completely without them) will offer opportunities to mimic complex natural release profiles.
Figure 6.
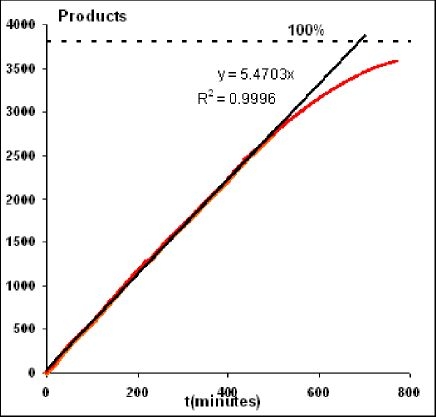
Spider with Zn2+-sensitive legs releasing oligonucleotide products from the three-dimensional dextran matrix (results obtained by surface plasmon resonance). Deposited at a ratio of 1:3800 to substrates, spiders release products with an almost constant rate until 70% of substrates are cleaved. With substrates attached to the matrix, this observation supports spider migration toward new substrates. Processivity (substrate cleavage without leaving the matrix) was estimated independently at approximately 800 substrates per spider. Cleavage can be stopped and restarted by switching between Zn2+ and EDTA solutions.
There are numerous hurdles to the implementation of spiders in vivo, but they seem mostly practical (technological) in nature rather than being fundamental objections. For example, the stability of oligonucleotides in vivo is a great concern, but it can be addressed by constructing enantiomeric forms of deoxyribozymes, which are completely inert in biological fluids.19 The enantiomeric approach should also eliminate potential immunogenicity of spiders, although the possibility of various idiosyncratic toxic events can never be completely dismissed without actually doing experiments. It is hoped that after optimization of their stability, and properties of biodegradable matrices, spiders could be operational after intramuscular injection for periods of several weeks at a time.
Conclusions
The goal of our exercise in synthetic molecular robotics was to provide insight into a new approach that is becoming more accepted among basic scientists, but has not yet “spilled over” into the therapeutic arena. While we cannot guarantee that our approach will ever actually become fully implemented, as we certainly hope, we are convinced that similar studies will play an important role in improving the health of diabetes patients in years to come.
Abbreviations
- 2D
two dimensional
- 3D
three dimensional
References
- 1.Seeman NC. Nanotechnology and the double helix. Sci Am. 2004 Jun;290(6):64–69. 72–75. doi: 10.1038/scientificamerican0604-64. [DOI] [PubMed] [Google Scholar]
- 2.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006 Mar 16;440(7082):297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 3.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004 Feb 12;427(6975):618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 4.Rothemund PW, Papadakis N, Winfree E. Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2004 Dec;2(12):e424. doi: 10.1371/journal.pbio.0020424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stojanovic MN, Stefanovic D. Deoxyribozyme-based automaton. Nat Biotechnol. 2003 Sep;21(9):1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 6.Ding B, Seeman N. Operation of a DNA robot arm inserted into a 2D DNA crystalline substrate. Science. 2006 Dec 8;314(5805):1583–1585. doi: 10.1126/science.1131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margolin AA, Stojanovic MN. Boolean calculations made easy (for ribozymes) Nat. Biotechnol. 2005 Nov;23(11):1374–1376. doi: 10.1038/nbt1105-1374. [DOI] [PubMed] [Google Scholar]
- 8.Wilson D, Szostack JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 9.Yan A, Ellington AD. Aptamers as potential diagnostic reagents for diabetes. Diabetes Technol Ther. 2002;4(3):339–346. doi: 10.1089/152091502760098483. [DOI] [PubMed] [Google Scholar]
- 10.Yang Q, Irwin J, Mei H-Y, Engelke DR. DNA ligands that bind tightly and selectively to cellobiose. Proc Natl Acad USA. 1998 May 12;95(10):5462–5467. doi: 10.1073/pnas.95.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein DJ, Ferre-D'Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006 Sep 22;313(5794):1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 12.Li T, Lee HB, Park K. Comparative stereochemical analysis of glucose-binding proteins for rational design of glucose-specific agents. J Biomater Sci Polym Ed. 1998;9(4):327–344. doi: 10.1080/09205063.1998.9753059. [DOI] [PubMed] [Google Scholar]
- 13.Loris R, Imberty A, Beeckmans S, Van Driessche E, Read JS, Bouckaert J, De Greve H, Buts L, Wyns L. Crystal structure of Pterocarpus angolensis lectin in complex with glucose, sucrose, and turanose. J Biol Chem. 2003 May 2;278(18):16297–16303. doi: 10.1074/jbc.M211148200. [DOI] [PubMed] [Google Scholar]
- 14.Alexeev VL, Das S, Finegold DN, Asher SA. Photonic crystal glucose-sensing material for noninvasive monitoring of glucose in tear fluid. Clin Chem. 2004 Dec;50(12):2353–2360. doi: 10.1373/clinchem.2004.039701. [DOI] [PubMed] [Google Scholar]
- 15.Stojanovic MN, Kolpashchikov DM. Modular allosteric sensors. J Am Chem Soc. 2004 Aug 4;126(30):9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 16.Emilsson GM, Breaker RR. Deoxyribozymes: new activities and new applications. Cell Mol Life Sci. 2002 Apr;59(4):596–607. doi: 10.1007/s00018-002-8452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinds KD, Kim SW. Effects of PEG conjugation on insulin properties. Adv Drug Deliv Rev. 2002 Jun 17;54(4):505–530. doi: 10.1016/s0169-409x(02)00025-x. [DOI] [PubMed] [Google Scholar]
- 18.Pei R, Taylor S, Rudchenko S, Stefanovic D, Mitchell TE, Stojanovic MN. Behavior of polycatalytic assemblies in a substrate-displaying matrix. J Am Chem Soc. 2006 Oct 4;128(39):12693–12699. doi: 10.1021/ja058394n. [DOI] [PubMed] [Google Scholar]
- 19.Eulberg D, Klussmann S. Spiegelmers: biostable aptamers. Chembiochem. 2003 Oct 6;4(10):979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]


