Abstract
Ho endonuclease of Saccharomyces cerevisiae is a homing endonuclease that makes a site-specific double-strand break in the MAT gene in late G1. Here we show that Ho is rapidly degraded via the ubiquitin-26S proteasome system through two ubiquitin-conjugating enzymes UBC2Rad6 and UBC3Cdc34. UBC2Rad6 is complexed with the ring finger DNA-binding protein Rad18, and we find that Ho is stabilized in rad18 mutants. We show that the Ho degradation pathway involving UBC3Cdc34 goes through the Skp1/Cdc53/F-box (SCF) ubiquitin ligase complex and identify a F-box protein, Yml088w, that is required for Ho degradation. Components of a defined pathway of the DNA damage response, MEC1, RAD9, and CHK1, are also necessary for Ho degradation, whereas functions of the RAD24 epistasis group and the downstream effector RAD53 have no role in degradation of Ho. Our results indicate a link between the endonuclease function of Ho and its destruction.
Homing endonucleases are encoded by mobile genetic elements and introduce a double-strand break (DSB) into a long (12–40 bp) DNA cognate sequence. Repair by gene conversion leads to insertion of a copy of the endonuclease-encoding ORF into the middle of the recognition site, and this disruption of the cognate sequence protects the cells from further DNA cleavage (1). Ho endonuclease of Saccharomyces cerevisiae belongs to the LAGLIDADG family of homing endonucleases and introduces a DSB into a 24-bp sequence in the mating type (MAT) locus in late G1. Repair of the Ho-induced DSB uses one of the silent mating type (HM) cassettes as template and results in regeneration of a MAT gene of the opposite mating type. This new MAT allele is a substrate for Ho endonuclease, and therefore Ho activity must be stringently regulated. Tight transcriptional regulation ensures that HO is transcribed briefly in late G1 in haploid mother cells only (2, 3). Here we show that the temporal regulation of Ho activity also involves its rapid degradation and that this is done through the ubiquitin-26S proteasome system.
The ubiquitin-26S proteasome system has a major role in degradation of many regulatory proteins, particularly those of the cell cycle (4, 5). Proteins are ubiquitinated by a cascade of enzymes: ubiquitin is activated in a thioester linkage to a ubiquitin-activating enzyme, E1/Uba, transferred to a ubiquitin-conjugating enzyme, E2/Ubc, that together with a ubiquitin ligase complex, E3, covalently links ubiquitin to the target. Additional ubiquitin molecules subsequently are linked through their C-terminal Gly moieties to Lys-48 of the preceding ubiquitin, leading to assembly of polyubiquitin chains on the substrate protein that then is degraded by the 26S proteasome (6, 7).
F-box proteins are an expanding family of eukaryotic proteins that specifically recruit substrates for ubiquitination (8). They are a subunit of the Skpl/Cdc53/F-box (SCF) ubiquitin ligase complex that acts with the E2 UBC3Cdc34 (9). At the core of the SCF is a heterodimer of Cdc53 and Roc1; an adapter protein, Skp1, binds the SCF scaffold via the N terminus of Cdc53 and the F-box receptor through its F-box motif (8, 10–12). There are 17 ORFs in S. cerevisiae with an F-box motif (11); substrates recruited for ubiquitination by the F-box receptors, Cdc4, Met30, and Grr1 have been identified (10, 13–15). Recently, 26 human F-box proteins were identified, 25 are novel genes (16). Substrates targeted for degradation through F-box proteins typically are phosphorylated, indicating that the ultimate regulatory element that determines the protein half-life is a protein kinase (8).
DNA damage leads to transient arrest of the cell cycle through the action of highly conserved proteins (17–19). Mec1 is the central regulator of DNA damage response (DDR) and controls both cell cycle arrest and transcription of DNA repair genes through activating the protein kinases Rad53 and Dun1 (20–22). Mec1 belongs to a large phosphoinositide kinase family that includes the mammalian ATM (mutated in ataxia telangiectasia) and ATR genes and the catalytic subunit of the DNA-dependent protein kinase, DNA-PK (23). Other genes involved in the DNA damage checkpoint include RAD9 and members of the RAD24 epistasis group (RAD17, RAD24, MEC3, and DDC1) (24).
MEC1 and RAD9 participate in two parallel pathways of checkpoint arrest: one pathway involves the RAD24 epistasis group and the protein kinase Rad53; the other involves Chk1 (22). Both Chk1 and Rad53 have a role in restraint of anaphase and mitotic exit after DNA damage. The main effector of the Chk1 branch is Pds1; phosphorylation of Pds1 blocks the metaphase-to-anaphase transition (22, 25). Rad53 controls the DNA damage checkpoint through polo kinase, Cdc5 (22). Rad53 has an additional role in arrest of S phase in the presence of unrepaired lesions in DNA by inhibiting firing of late origins of replication (26, 27).
Cells respond to DNA damage by transient arrest of the cell cycle and induction of genes involved in repair (28). A DSB introduced into the genome late in G1 is expected to evoke the G1/S checkpoint response, enabling repair of the DSB before replication (29). Indeed after Ho-induced mating-type switching both progeny cells have the new mating type, indicating that repair precedes replication of MAT (30, 31). We find that checkpoint functions of the DDR pathway, MEC1, RAD9, and CHK1, are necessary for regulated proteolysis of Ho. In this study of regulation of the half-life of an endonuclease by the ubiquitin system, we have found a link between the endonuclease function of Ho and its destruction.
Materials and Methods
Determination of the Half-Life of Ho Endonuclease in Yeast.
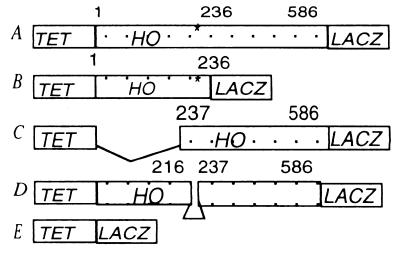
We fused the whole HO ORF, or the N- and C-terminal halves separately, to LACZ in plasmid pCM190 where expression from the Tet promoter is tightly regulated (32). Subsequently the N-terminal HO fragment was truncated by 60 bp to delete the PEST (proline/glutamate/serine/threonine/aspartic acid) residues. This N-terminal fragment subsequently was fused to the C-terminal fragment to give a HO ORF lacking the 20 PEST residues (Fig. 1). Fusion of LACZ to HO results in loss of endonucleolytic activity.
Figure 1.
pCM190-HO-LACZ fusions. (A) HO ORF encoding 586 aa. *, PEST residues. (B) N-terminal half to residue 236. (C) C-terminal half from residue 237. (D) HO ORF without PEST residues 216–236. (E) LACZ for expression from Tet promoter.
The plasmids were transformed into yeast by using the lithium acetate method (33). Yeast transformants were selected on SD plates lacking uracil with 2 μg/ml doxycycline to prevent expression of HO. In a typical experiment overnight 10-ml liquid cultures were grown to saturation in the presence of doxycycline, diluted to OD600 0.3, and grown for 2–3 h to obtain logarithmic cells. The cells were washed three times and resuspended in medium lacking doxycycline for induction. After 30 min they were pulsed with 35S-Redivue (Amersham Pharmacia) for 5 min, and aliquots were taken at different times for immunoprecipitation as described in ref. 34 using anti-β-galactosidase from ICN.
o-Nitrophenyl β-d-Galactoside (ONPG) Assay for Measuring the Steady-State Level of Ho-LacZ in Mutant Yeast and Their Congenic Wild Types.
Transformants were treated as above for induction of HO and LACZ except that the cells were induced for 18 h to reach steady state. The activity of Ho-LacZ was compared with that of LacZ by itself (33), and the ratio between the two was calculated for each mutant strain and its congenic wild type. Each assay was performed in triplicate, and the results are an average of at least two separate experiments.
Cell Cycle Arrest.
An overnight cell culture was diluted 10-fold into minimal selective medium with doxycycline and containing 2 μg/ml α factor, 0.1 M hydroxyurea (HU), or 15 μg/ml nocodazole. The arrest was followed visually, and usually all of the cells were arrested within 2–3 h. At that point the cells were washed three times in medium without doxycycline but with the same cell cycle inhibitor, for induction of HO. After 30 min they were pulsed with 35S-Redivue, and aliquots were collected for immunoprecipitation as described above.
Yeast strains used were: ubc2 and ubc3 (34); ubc4, ubc5, ubc6, and ubc7 (35); ubc9 (36); mec3 and ddc1 (24); all other DDR mutants (37); chk1 (22); rpt2 (38); hrr25 (39); cak1 (40); cdc7 (41); pho85 and cdc53 (42); and skp1 (43). For F-box mutants, Research Genetics BY4730 is wild type, and #482 is Δyml088w; Euroscarf #10000 M is wild type, and Δynl311c is #10293A; Δyjl149w is #10429A; and Δyor080w is #Y11856. All F-box mutants are in the S288c strain background. ubr1, ubc1, ubc4, ubc5, ubc6, ubc7, ubc12, ubc13, and their double mutants all are in the same W303 strain background (35), and mec1, rad9, rad17, rad24, and rad53 also were made in the same parental strain (37).
Results
Determination of the Half-Life of Ho Endonuclease.
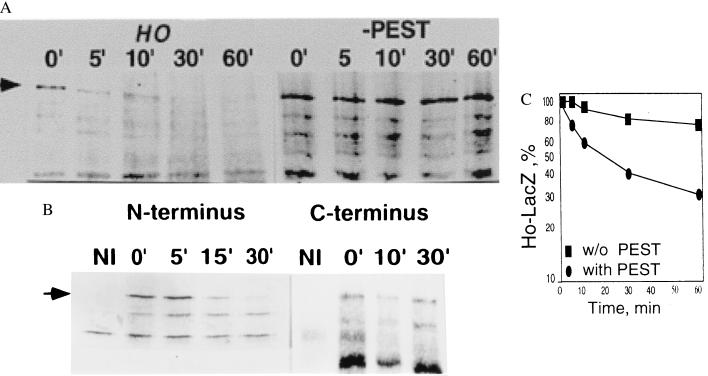
The pCM190-HO-LACZ constructs were transformed into W303 wild-type yeast. Logarithmic cultures were induced for 30 min by removal of doxycycline and then pulsed with S35-Redivue for 5 min. The cells were washed three times, and the amount of protein remaining was determined at each time point by immunoprecipitation with anti-β-galactosidase (Fig. 2A Left). Our results indicate that Ho undergoes rapid degradation in wild-type yeast with a half-life of ca. 10 min.
Figure 2.
(A) Pulse–chase experiment showing that the half-life of the native Ho protein fused to LacZ is ca. 10 min compared with Ho without PEST residues 216–236 fused to LacZ that is stable over 60 min. Arrow points to Ho-LacZ. (B) Pulse–chase experiment of the N-terminal (Fig. 1B) and C-terminal (Fig. 1C) halves of Ho fused separately to LacZ. The N-terminal Ho-fusion protein that has the PEST residues is degraded with a half-life of ca. 10 min in wild-type yeast (Left) whereas the C-terminal Ho-LacZ fusion protein is stable over 30 min (Right). NI, noninduced; arrow points to Ho-LacZ.
We identified a sequence rich in PEST residues in Ho between residues 216 and 236 by using the program pestfind (44). An engineered HO gene from which this potential PEST sequence was deleted is stabilized (Fig. 2A Right). When the N-terminal (residues 1–236 including the PEST residues, Fig. 1B) and C-terminal (residues 237–586, Fig. 1C) halves of Ho were fused separately to β-galactosidase and their half-life was measured by pulse–chase and immunoprecipitation we found that the N-terminal half has a half-life similar to that of the whole Ho protein, whereas the C-terminal half is far more stable with 70% of the protein remaining after 30 min (Fig. 2B). These results indicate that the sequence between residues 216 and 236 constitutes a major degradation signal for Ho.
Involvement of the Ubiquitin-26S Proteasome System in Degradation of Ho.
To see whether degradation of Ho involves the ubiquitin system we compared the steady-state level of Ho in yeast overexpressing a mutant form of ubiquitin (Ubi K48R, G76A) from the CUP1 promoter with that in cells overexpressing normal ubiquitin. Mutation of Lys-48 prevents formation of polyubiquitin conjugates and gives a dominant negative phenotype (45).
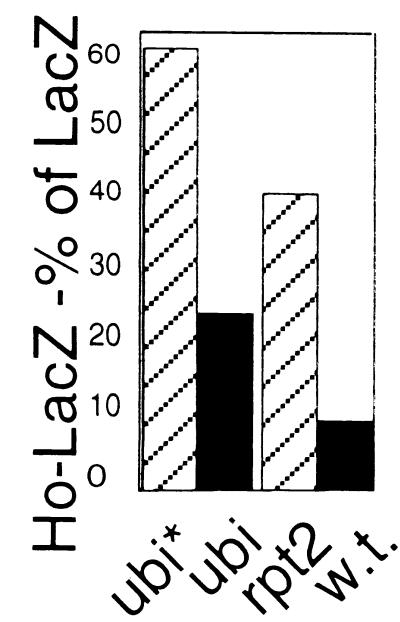
We found that overexpression of mutant ubiquitin leads to a steady-state level of Ho-LacZ, expressed as a percent of LacZ induced under the same conditions, that is 4-fold that obtained under conditions in which normal ubiquitin was overexpressed. This indicates that the ubiquitin system is involved in degradation of Ho (Fig. 3). In addition, we examined the steady-state level of Ho in cells mutant for the Rpt2 ATPase subunit of the 19S subunit of the proteasome (38) by the ONPG assay. Ho accumulates in rpt2 mutants, providing proof for the involvement of the 26S proteasome in its proteolysis (Fig. 3).
Figure 3.
The steady-state level of Ho-LacZ activity expressed as a percent of LacZ activity in cells in which mutant (K48R,G76A) ubiquitin (ubi*) is overexpressed is compared with that in cells overexpressing normal ubiquitin (ubi). The same experiment is shown for a mutant of the Rpt2 subunit of the 19S lid of the proteasome compared with its congenic wild type. Ho is stabilized by mutant ubiquitin overexpression and also in the rpt2 mutant.
Functions of the Ubiquitin-26S Proteasome System that Are Important for Ho Degradation.
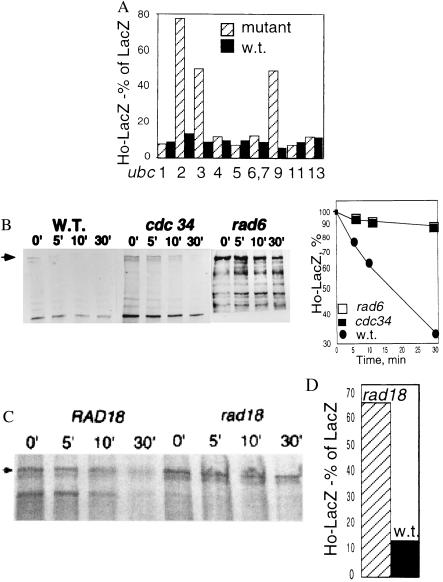
To characterize the pathway of Ho degradation we determined the steady-state level of Ho in yeast mutant for different ubiquitin-conjugating enzymes by the ONPG assay (Fig. 4A). The half-life of Ho then was determined by pulse–chase in mutants that tested positive in the ONPG assay (Fig. 4B). These experiments identified UBC2Rad6 and UBC3Cdc34 as two ubiquitin-conjugating enzymes that are involved in Ho degradation. In addition, Ho is stabilized in ubc9 mutants.
Figure 4.
(A) The steady-state level of Ho-LacZ activity is compared with that of LacZ in a series of ubc mutants and their congenic wild types. Ho is stabilized in ubc2rad6, ubc3cdc34, and ubc9 mutants. (B) The degradation of Ho was followed by pulse–chase and immunoprecipitation in ubc2rad6 and ubc3cdc34 mutants and their isogenic wild types. Graph shows the amount of protein remaining at each time point. (C) Degradation of Ho-LacZ in a rad18 mutant and its congenic wild type. (D) Steady-state level of Ho-LacZ compared with that of LacZ in these strains after 18 h induction. Ho is stabilized in the rad18 mutant.
Degradation of Ho via UBC2Rad6.
Ho is stabilized in UBC2Rad6 mutants, indicating a role for this ubiquitin-conjugating enzyme. The E3 Ubr1 acts with UBC2Rad6 when degradation proceeds via the N-end rule (46); however, there was no stabilization of Ho in a ubr1 mutant (not shown). UBC2Rad6 forms a complex with Rad18 in its function of DNA repair (47), and we find that Ho is stabilized in a rad18 mutant (Fig. 4 C and D). Mms2 and Ubc13 form an ubiquitin-conjugating complex that is a downstream component of the UBC2Rad6 error-free postreplication repair pathway (48, 49) but there was no effect on degradation of Ho in ubc13 mutant yeast (Fig. 4A).
Degradation of Ho via UBC3Cdc34.
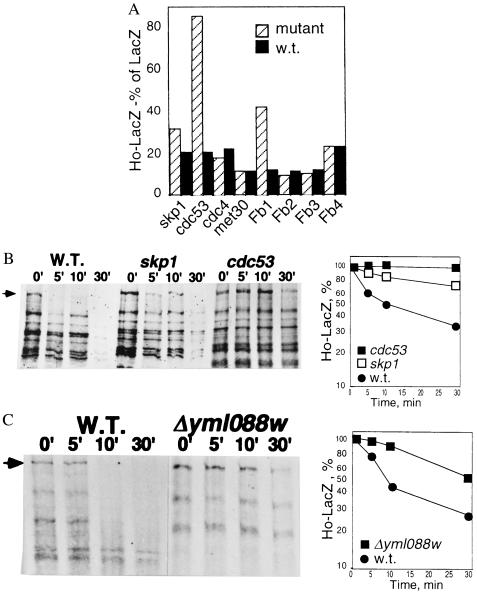
UBC3Cdc34 is activated by the SCF ubiquitin ligase complex that comprises Skp1, Cdc53, and Roc1 and a target-specific F-box protein (above). We therefore tested the half-life of Ho in yeast mutant for Cdc53 and Skp1. Ho is stabilized in cdc53 and skp1 mutant yeast, indicating a role for the SCF in its degradation (Fig. 5 A and B).
Figure 5.
(A) The steady-state level of Ho-LacZ is compared with that of LacZ by the ONPG assay in mutants of the SCF, skp1, cdc53, cdc4, met30, and their isogenic wild types. Fb1 is Δyml088w, Fb2 is Δynl311c, Fb3 is Δyjl149w, and Fb4 is Δyor080w. Ho is stabilized in skp1, cdc53, and the Δyml088w F-box mutant. (B) Gel with accompanying graph showing degradation of Ho in cdc53 and skp1 mutants compared with their congenic wild type. (C) Degradation of Ho is retarded in a strain deleted for the F-box ORF YML088w compared with its isogenic parent.
To identify the E3 component that recruits Ho to the SCF we measured the steady-state level of Ho in a number of known F-box mutants (cdc4 and met30) and in three F-box deletion strains listed in the blocks database, Δynl311c, Δyjl149w, and Δyml088w (50) and also in the F-box mutant, Δyor080w. Ho accumulates only in the Δyml088w mutant and not in any of the other F-box mutant strains (Fig. 5 A and C). These experiments identify the ORF YML088w as the F-box that recruits Ho for degradation.
Identification of a Protein Kinase Required for Ho Degradation.
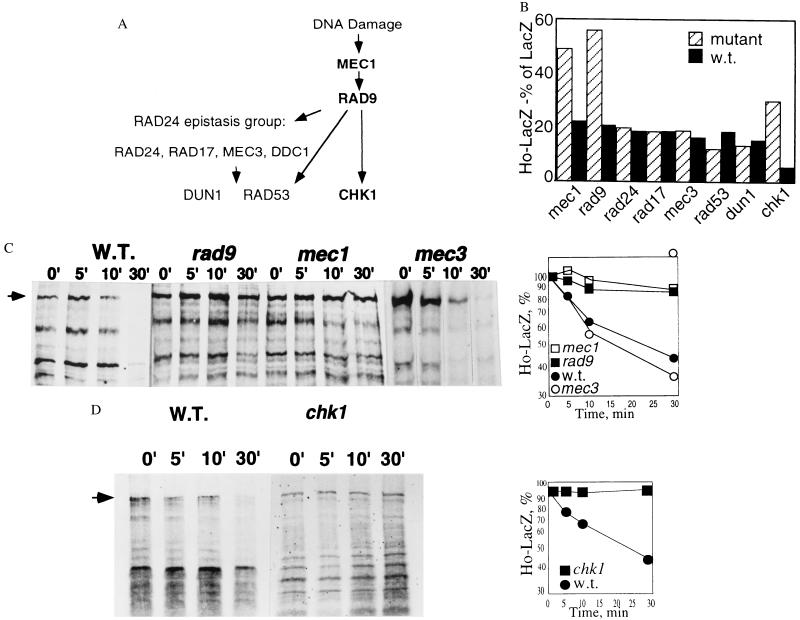
Having identified a role for the SCF in Ho degradation and a PEST sequence as a major degradation signal we then screened the stability of Ho in yeast mutant for various protein kinases that we thought might be relevant. In particular we checked nuclear protein kinases that are active at the G1/S stage, including those that have a role in cell cycle arrest in response to DNA damage (Fig. 6A). The initial screen was by the ONPG assay, subsequently the stability of Ho-LacZ was tested in promising candidates by pulse–chase and immunoprecipitation. We found that Ho is stabilized in mec1 and rad9 mutants, indicating a major role for the DDR pathway in degradation of Ho (Fig. 6 B and C). Mutants of the RAD24 epistasis group that we tested, rad24, rad17, and mec3, had no effect on the steady-state level or the half-life of Ho. The downstream kinases, rad53 and dun1, were likewise not necessary for degradation of Ho (Fig. 6 B and C). However, a significant stabilization of Ho was observed in a mutant of the Rad9-dependent chk1 protein kinase strain (Fig. 6 B and D). We tested a further group of mutants of nuclear protein kinases that are active at the G1/S stage, cdc7 (31), cak1 (43), pho85 (44), and hrr25 (45), but did not find any stabilization of Ho in these strains (not shown).
Figure 6.
(A) Stability of Ho in mutants of the DDR pathway compared with their isogenic parents. Shown is a diagram of the DDR pathway based on integration of data from refs. 20 and 24; mutants in which Ho is stabilized are indicated in bold. (B) The steady-state level of Ho-LacZ is compared with that of LacZ in mutants of the DDR and their isogenic wild types. Ho is stabilized in mec1, rad9, and chk1 mutants but not in mutants of the RAD24 epistasis group, rad24, rad17, mec3 nor in rad53 or dun1. (C) Gel and graph of pulse–chase experiment of Ho in mec1, rad9, and mec3 mutants and their isogenic wild types. Ho is stabilized in mec1 and rad9 mutants but not in mec3, a member of the RAD24 epistasis group. Graphs show rate of degradation of Ho in mec1, rad9, and mec3 mutants compared with the congenic wild type. (D) Gel and graph showing rate of degradation of Ho in chk1 mutant compared with the isogenic wild type.
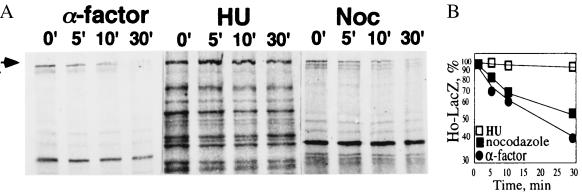
Chk1 is activated by DNA damage and is not activated in cells arrested in S phase with HU (22). We therefore measured the half-life of Ho by pulse–chase in cells arrested in S phase with HU. This was compared with the half-life of Ho in cells arrested in G1 with α factor and with that in cells arrested in G2 with nocodazole. Cells arrested with α factor and nocodazole degrade Ho almost as rapidly as asynchronous cultures whereas in cells arrested in S phase with HU Ho is stabilized (Fig. 7). This result supports a role for Chk1 in its degradation.
Figure 7.
Ho degradation in cells arrested in G1 with α factor, in S with HU, and in G2 with nocodazole. Ho is stabilized in cells arrested with HU.
Discussion
Our results suggest that rapid degradation of Ho contributes to the stringent temporal regulation of its activity, which is essential to protect the new MAT alleles from recleavage after repair of the Ho-induced DSB. We found that Ho is degraded via the ubiquitin-26S proteasome pathway and that functions of the DDR pathway target Ho for degradation.
Two ubiquitin-conjugating enzymes, UBC2Rad6 and UBC3Cdc34, are necessary for degradation of Ho. This finding is similar to reports indicating that both UBCs have a role in degradation of repressors of cAMP-induced transcription (51). Ho from which the putative PEST residues are deleted is stable for at least 60 min, suggesting that both UBC2Rad6 and UBC3Cdc34 may be recognizing a major degradation signal in the same region of the protein. The absence of either one of these UBCs results in stabilization of Ho. Ho degradation may be similar to that of the Matα2 repressor where two epistatic groups of UBCs are involved in its degradation (35).
Ho is stabilized in ubc9 mutants. Ho can induce a mating type switch in ubc9 mutants (not shown), indicating that it is being imported into the nucleus in these cells. The stabilization of Ho in the ubc9 mutant is not a result of the ubc9 G2 arrest phenotype as the half-life of Ho in nocodazole-arrested cells is similar to that seen in an asynchronous culture. Ubc9 conjugates Smt3 (36) and the stabilization of Ho may be an indirect effect caused by lack of activation of a component of the ubiquitin-26S proteasome system in these mutants.
Rad18 is a single-stranded DNA binding protein, and most cellular Rad18 is complexed with UBC2Rad6 (52). Our finding that Ho is stabilized in rad18 mutants may indicate that this ring finger protein is functioning as the E3 ubiquitin ligase in this pathway (53, 54). UBC3Cdc34 is essential for cell cycle progression at G1/S and is a component of the SCF (8). Our data indicate that Ho degradation involves the SCF because Ho is stabilized in skp1 and cdc53 mutants. In addition, we show that Ho is stabilized in mutants of the F-box protein YML088w. YML088w is an uncharacterized ORF transcription of which is elevated 4-fold in response to DNA damage by methyl methanesulfonate (55). Taken together our results indicate that Ho is recruited to the SCF by an F-box protein that may have a novel role as a component of the DDR.
The known substrates of the SCF ubiquitin ligase complex are phosphorylated and the PEST sequence we delineated between residues 216 and 236 is the major signal that determines the half-life of Ho. We found no clear effect on degradation of Ho in mutants of a number of nuclear protein kinases that are active at the G1/S phase of the cell cycle. These include cak1, cdc7, pho85, and hrr25.
Targeting of Ho for degradation depends on functions of the DDR pathway, specifically MEC1, RAD9, and CHK1. We find no role for functions of the RAD24 epistasis group, RAD24, RAD17, and MEC3 nor for RAD53 or DUN1. Chk1 is phosphorylated in response to DNA damage and this is MEC1- and RAD9-dependent but RAD53-independent, indicating the presence of a separate pathway of DDR (22). There is a very distinct effect of DNA structure on the activation of either Rad53 or Chk1 (56). Rad53 is activated by the stalled replication forks that accumulate in cells arrested by HU in S phase whereas Chk1 is specifically activated in response to DNA damage and is not activated in HU-arrested cells (22). Ho is stabilized in HU-arrested cells, providing further support of a role for Chk1 in its degradation. Our interpretation is that MEC1, RAD9, and CHK1 act upstream of UBC2Rad6 and UBC3Cdc34 and target Ho for ubiquitination.
Our data indicate that the rapid degradation of Ho by the ubiquitin-26S proteasome system involves several functions that mediate the DDR and cell cycle progression. The model emerging from our data is that Ho activity, by generating a DSB, induces the kinase Chk1. Chk1, either by directly phosphorylating Ho in the region 216–236 important for degradation, or indirectly, enables the recognition of Ho by the F-box protein Yml088w. This F-box protein recruits Ho for ubiquitination by the SCF complex. Ubiquitinated Ho is rapidly degraded by the proteasome so that after the DSB at MAT is repaired and the cell cycle progresses, no further DSBs are introduced into the switched MAT alleles, thereby ensuring that the mating type switch occurs only once in every generation. Thus temporal regulation of Ho activity involves both tight transcriptional regulation of the gene and DDR-mediated rapid degradation of the protein via the ubiquitin-26S proteasome system.
Acknowledgments
We gratefully acknowledge B. Garvick, G. Lucchini, R. Kulka, D. Finley, and Y. Sanchez for strains and advice. Special thanks go to Y. Sanchez for sharing unpublished data. D.R. thanks the Israel Cancer Research Fund for support. L.K. is a Kreitman Foundation Doctoral Fellow.
Abbreviations
- DSB
double-strand break
- DDR
DNA damage response
- HU
hydroxyurea
- ONPG
o-nitrophenyl β-d-galactoside
- PEST
proline/glutamate/serine/threonine/aspartic acid
- SCF
Skp1/Cdc53/F-box
References
- 1.Belfort M, Roberts R J. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breeden L, Nasmyth K. Cell. 1987;48:389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 3.Cosma M P, Tanaka T, Nasmyth K. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 4.King R, Deshaies R, Peters J, Kirschner M. Science. 1996;274:1652–1658. doi: 10.1126/science.274.5293.1652. [DOI] [PubMed] [Google Scholar]
- 5.Pagano M. FASEB J. 1997;11:1067–1075. doi: 10.1096/fasebj.11.13.9367342. [DOI] [PubMed] [Google Scholar]
- 6.Coux O, Tanaka K, Goldberg A. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 7.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 8.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 9.Goebl M G, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 10.Kishi T, Yamao F. J Cell Sci. 1998;111:3655–3661. doi: 10.1242/jcs.111.24.3655. [DOI] [PubMed] [Google Scholar]
- 11.Patton E E, Willems A R, Sa D, Kuras L, Thomas D, Craig K L, Tyers M. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seol J H, Feldman R M, Zachariae W, Shevchenko A, Correll C C, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, et al. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinney T, Chang F, Heintz N, Cross F. Genes Dev. 1993;7:833–843. doi: 10.1101/gad.7.5.833. [DOI] [PubMed] [Google Scholar]
- 14.Henchoz S, Chi Y, Catarin B, Herskowitz I, Deshaies R, Peter M. Genes Dev. 1997;11:3046–3060. doi: 10.1101/gad.11.22.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drury L S, Perkins G, Diffley J F. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cenciarelli C, Chiaur D S, Guardavaccaro D, Parks W, Vidal M, Pagano M. Curr Biol. 1999;9:1177–1179. doi: 10.1016/S0960-9822(00)80020-2. [DOI] [PubMed] [Google Scholar]
- 17.Hartwell L H, Weinert T A. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 18.Paulovich A G, Hartwell L H. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 19.Gardner R, Putnam C W, Weinert T. EMBO J. 1999;18:3173–3185. doi: 10.1093/emboj/18.11.3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 21.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge S J. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 23.Rotman G, Shiloh Y. Oncogene. 1999;18:6135–6144. doi: 10.1038/sj.onc.1203124. [DOI] [PubMed] [Google Scholar]
- 24.Longhese M P, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen-Fix O, Koshland D. Genes Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neecke H, Lucchini G, Longhese M P. EMBO J. 1999;18:4485–4497. doi: 10.1093/emboj/18.16.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinreich M, Stillman B. EMBO J. 1999;18:5334–5346. doi: 10.1093/emboj/18.19.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinert T A, Kiser G L, Hartwell L H. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 29.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 30.Strathern J N, Klar A J, Hicks J B, Abraham J A, Ivy J M, Nasmyth K A, McGill C. Cell. 1982;31:183–192. doi: 10.1016/0092-8674(82)90418-4. [DOI] [PubMed] [Google Scholar]
- 31.Strathern J, Hicks J, Herskowitz I. J Mol Biol. 1981;147:357–372. doi: 10.1016/0022-2836(81)90488-5. [DOI] [PubMed] [Google Scholar]
- 32.Gari E, Piedrafita L, Aldea M, Herrero E. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Adams A, Gottschling D, Kaiser C, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. [Google Scholar]
- 34.Kornitzer D, Raboy B, Kulka R G, Fink G R. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 36.Schwarz S E, Matuschewski K, Liakopoulos D, Scheffner M, Jentsch S. Proc Natl Acad Sci USA. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubin D M, Glickman M H, Larsen C N, Dhruvakumar S, Finley D. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman A M, Andrews B. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Espinoza F H, Farrell A, Nourse J L, Chamberlin H M, Gileadi O, Morgan D O. Mol Cell Biol. 1998;18:6365–6373. doi: 10.1128/mcb.18.11.6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larochelle S, Pandur J, Fisher R P, Salz H K, Suter B. Genes Dev. 1998;12:370–381. doi: 10.1101/gad.12.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meimoun A, Holtzman T, Weissman Z, McBride H J, Stillman D J, Fink G R, Kornitzer D. Mol Biol Cell. 2000;11:915–927. doi: 10.1091/mbc.11.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 44.Rechsteiner M, Rogers S W. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 45.Spence J, Sadis S, Haas A L, Finley D. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varshavsky A. Cell. 1992;69:725–735. doi: 10.1016/0092-8674(92)90285-k. [DOI] [PubMed] [Google Scholar]
- 47.Bailly V, Lauder S, Prakash S, Prakash L. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 48.Broomfield S, Chow B L, Xiao W. Proc Natl Acad Sci USA. 1998;95:5678–5683. doi: 10.1073/pnas.95.10.5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofmann R M, Pickart C M. Cell. 1999;96:645–653. doi: 10.1016/s0092-8674(00)80575-9. [DOI] [PubMed] [Google Scholar]
- 50.Pietrokovski S, Henikoff J G, Henikoff S. Nucleic Acids Res. 1996;24:197–200. doi: 10.1093/nar/24.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pati D, Meistrich M L, Plon S E. Mol Cell Biol. 1999;19:5001–5013. doi: 10.1128/mcb.19.7.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bailly V, Prakash S, Prakash L. Mol Cell Biol. 1997;17:4536–4543. doi: 10.1128/mcb.17.8.4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joazeiro C A, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 54.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jelinsky S A, Samson L D. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinho R G, Lindsay H D, Flaggs G, DeMaggio A J, Hoekstra M F, Carr A M, Bentley N J. EMBO J. 1998;17:7239–7249. doi: 10.1093/emboj/17.24.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]