Abstract
Aims and Background
Model-based insulin sensitivity testing via the intravenous glucose tolerance test (IVGTT) or similar is clinically very intensive due to the need for frequent sampling to accurately capture the dynamics of insulin secretion and clearance. The goal of this study was to significantly reduce the number of samples required in intravenous glucose tolerance test protocols to accurately identify C-peptide and insulin secretion characteristics.
Methods
Frequently sampled IVGTT data from 12 subjects [5 normal glucose-tolerant (NGT) and 7 type 2 diabetes mellitus (T2DM)] were analyzed to calculate insulin and C-peptide secretion using a well-accepted C-peptide model. Samples were reduced in a series of steps based on the critical IVGTT profile points required for the accurate estimation of C-peptide secretion. The full data set of 23 measurements was reduced to sets with six or four measurements. The peak secretion rate and total secreted C-peptide during 10 and 20 minutes postglucose input and during the total test time were calculated. Results were compared to those from the full data set using the Wilcoxon rank sum to assess any differences.
Results
In each case, the calculated secretion metrics were largely unchanged, within expected assay variation, and not significantly different from results obtained using the full 23 measurement data set (P < 0.05).
Conclusions
Peak and total C-peptide and insulin secretory characteristics can be estimated accurately in an IVGTT from as few as four systematically chosen samples, providing an opportunity to minimize sampling, cost, and burden.
Keywords: C-peptide, insulin, insulin secretion, IVGTT, modeling
Introduction
Assessing pancreatic insulin secretion is important in the diagnosis and monitoring of type 2 diabetes mellitus (T2DM).1–4 Different tests and markers have been proposed to quantify prehepatic insulin secretion. These tests include intravenous tests, such as the hyperglycemic clamp5 and the intravenous glucose tolerance test (IVGTT),6 the oral glucose tolerance test,2,7 and fasting state assessments.8,9 They also vary in resolution and the range of information provided, with intravenous tests generally providing more details about biphasic secretory characteristics.2
A good estimation of prehepatic insulin secretion can be achieved by estimating C-peptide secretion through modeling of its kinetics.10–14 This approach is unbiased by first-pass hepatic extraction of insulin and is a valid marker due to the equimolar secretion of both peptides.11 A two compartment model initially proposed by Eaton and colleagues11 has been shown to represent C-peptide kinetics accurately. To avoid individual model parameter estimation, Van Cauter and associates12 proposed a regression model to calculate population parameters from known subject-specific characteristics, such as height, weight, age, gender, and diagnosis of diabetes. This population methodology has been validated in several studies with peak errors of 10–20%.12,15–17
Accurate estimation of the peak secretion rate and total first phase-secreted insulin (first 10 minutes) is currently only possible with very frequent sampling during this interval. However, precisely capturing the peak C-peptide concentration and timing is crucial for accurate assessment, especially given the relatively fast first phase secretion dynamics. Frequent sampling protocols during an IVGTT or similar test sample the C-peptide up to every minute, making these protocols burdensome to the patient and difficult and costly to perform, as well as requiring significant blood sampling.
For a method to be useful in a clinical diagnostic setting, simplicity, robustness, and cost of the protocol are important factors. In this study, a simple method to estimate C-peptide secretion is proposed, using integrals instead of a typical deconvolution approach. Furthermore, errors introduced by reduced sampling are assessed by comparing different reduced sampling approaches to the full, original frequently sampled data set estimations and values. Analysis is performed on frequently sampled C-peptide data during an IVGTT in five normal glucose-tolerant (NGT) and seven subjects with T2DM.
Subjects, Materials, and Methods
C-peptide data from IVGTT studies in this research were kindly provided by Dr. Andrea Mari (Institute of Biomedical Engineering, Padova, Italy) and Dr. Angelo Avogaro (Department of Clinical and Experimental Medicine, University of Padova, Italy). Data have been published previously,18 with a full description of subjects and experimental protocol. The critical aspects are briefly reproduced here for clarity.
Subjects
The study was performed on 12 subjects, 5 with normal glucose tolerance (age 24 ± 2, weight 73 ± 6 kg, fasting glucose 5.2 ± 0.1 mmol/liter, fasting insulin 50 ± 5 pmol/liter) and 7 with type 2 diabetes (age 49 ± 5, weight 81 ± 3 kg, fasting glucose 8.6 ± 0.8 mmol/liter, fasting insulin 125 ± 27 pmol/liter). Pharmacological treatment in T2DM was stopped 3 days before the study and all subjects received a 2000-kcal/day diet (50% carbohydrate, 35% fat, 15% protein) for at least 30 days prior to the study.
Experimental Protocol
An insulin-modified IVGTT was performed on all subjects in the morning after an overnight fast. After three fasting samples at –30, –15, and 0 minutes, a 0.3-g/kg glucose bolus was injected. At 20 minutes, insulin was infused for 5 minutes, totaling 0.03 U/kg (NGT) and 0.05 U/kg (T2DM). Blood samples were collected at 2, 3, 4, 5, 6, 8, 10, 15, 20, 25, 30, 40, 60, 80, 100, 120, 140, 160, 180, 210, and 240 minutes and were analyzed for C-peptide, glucose, and insulin concentrations. Only the C-peptide samples were of interest in this study.
C-Peptide Model
A well-accepted two compartment model of C-peptide kinetics was employed, as initially described by Eaton and colleagues.11 Equations describing the mass transport between compartments are defined:
| (1) |
| (2) |
where C(t) is the concentration in the central (or plasma) compartment (pmol/liter), ϒ(t) is the concentration in the peripheral (or interstitial) compartment (pmol/liter), k1 and k2 are transport rates between the compartments (1/minute), k3 is the renal loss from the central compartments (1/minute), S(t) is the pancreatic secretion rate (pmol/min), and Vc is the central distribution volume (liter). A priori identification of the kinetic parameters is done with known subject information, as described by Van Cauter and colleagues,12 which is a well-utilized, validated, and accurate methodology.15–17
Integral-Based Estimation of C-Peptide Secretion
Estimation of the C-peptide secretion rate S(t) is performed with an integral-based method, previously employed in real-time parameter identification in glycemic control trials in the critically ill19–21 and related biomedical applications. To best compute the integrals in all time steps, the profile of the C-peptide is approximated using linear interpolation between data points, which introduces no additional error over model error.21 Integral functions also have the advantage of being robust to noise in measured data, effectively providing a low-pass filter in the summations involved in numerical integrations.21
The C-peptide secretion rate, S(t), is estimated as a step function, with a step size of 1 minute. Thus, during any given 1-minute time interval, , S(t) is assumed constant. Integrating Equation (1) in the interval [t0, t1] yields:
| (3) |
Solving Equation (2) analytically for ϒ(t) yields:
| (4) |
where Cest represents interpolated C-peptide values estimated from discrete sampled measurements. Combining Equations (3) and (4) and solving for the assumed constant secretion rate S0,1 in this time interval yields:
| (5) |
Repeating this process for the intervals [t1, t2], [t2, t3], and so on results in a 1-minute stepwise constant secretion profile, S(t). This estimated S(t) profile is (physiologically) constrained to be nonnegative. Smoothing the estimated stepwise constant profile with a zero-phase, three-point moving average is done to avoid overfitting to noisy data and interpolated measurements.21 This particular filter was picked as a simple choice that does not require further assumptions and does not introduce a phase lag. This last step is not required in frequently sampled data, but results in a more physiological profile between more sparsely sampled data.
Points of Discontinuity
To minimize the number of samples required to describe secretion characteristics, it is crucial to identify key points of physiological discontinuity in the C-peptide concentration profile. These points of discontinuity are caused by sudden changes in the C-peptide concentration due to either endogenous or exogenous input. Common changes in C-peptide secretion that occur during an IVGTT are shown in Figure 1 and are defined as follow.
Injection of glucose (D1): A sudden increase in plasma glucose triggers a secretion burst of stored insulin (first phase) lasting 5–10 minutes, which is often reduced or blunted in type 2 diabetes.22,23 In the C-peptide concentration profile, this dynamic is seen as a very steep rise immediately after the administration of glucose. As glucose is administered between t = 0 and t = 1 minute, a lag of 1 minute is chosen here to account for glucose injection and pancreatic response time.
Peak first phase secretion rate (D2): The peak C-peptide secretion rate determines the peak C-peptide concentration during the first 10 minutes postglucose input. In the concentration profile, this point is the maximum value CPmax, located at , assumed between 0 and 10 minutes.
End of first phase/start of second phase secretion (D3): First phase secretion ends after approximately 10 minutes. If high glucose concentrations persist, pancreatic insulin secretion continues to rise or remains elevated over basal levels (second phase).24 In the concentration profile, this point can be identified as a local minimum around 10 minutes.
Injection of insulin (D4): A sudden increase in plasma insulin inhibits pancreatic insulin secretion.24 This response can be delayed significantly or not evident in type 2 diabetes.24 In the concentration profile, this point can be seen as a steepening of the negative slope soon after an exogenous insulin input.
Return to basal secretion rate (D5): This step varies widely in individuals, but is usually more gradual than the preceding factors. In the concentration profile, this change is evident when the slopes are tending toward zero and the C-peptide concentrations return to fasting values. To pick a clear point in the curve, this study uses the time of the first value to reach the fasting level.
Figure 1.
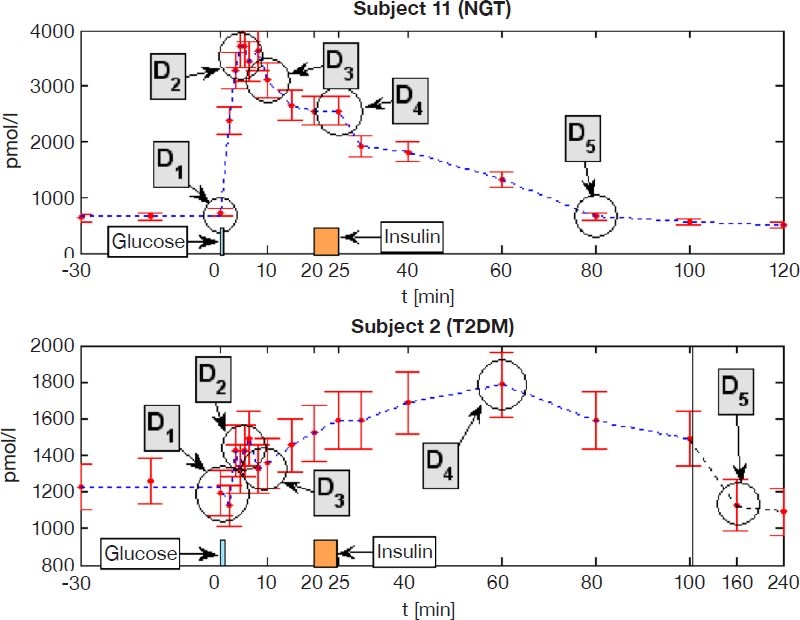
Example of points of discontinuity identified in the C-peptide profile during an IVGTT in NGT (top) and T2DM (bottom) subjects. The time axis in T2DM is not to scale between 100 and 240 minutes.
All five of these points are typically very pronounced and consistent in healthy individuals, but can be very gradual, blunted, or nonexistent in individuals with diabetes, who have an impaired first phase secretion and often have delays in the pancreatic response to glucose and insulin concentration changes. Figure 1 shows examples for NGT and T2DM subjects with the identified points of discontinuity. Note that points D2, D3, and D5 can be very variable in different individuals and may introduce errors when generic points are chosen.
Minimal Sampling Options
Minimal sample optimization analysis is performed in five steps. The original complete data set (step 1) is the reference to which all of the following steps or simplifications are compared to assess any loss in accuracy or utility. This full data set consists of 23 samples.
Steps 2 and 3 are sample reduced to keep only the optimal median points of discontinuity identified in NGT (step 2) and T2DM (step 3) subjects. These two reduced sets require only six samples. The points of discontinuity chosen are the median time point values observed over all subjects in the data set utilized. Using these median values over this diverse data set creates a generic approach that will generalize or extrapolate to any similar data set or study Step 4 analyzes a further reduction to four samples. This set thus keeps only the most critical points for identifying the dynamics. Specifically, the peak and the return to basal points.
Whereas steps 1–4 keep the maximal C-peptide sample (D2) during the first phase response, step 5 assesses a different approach. More specifically, it is a method that does not rely on capturing the peak concentration exactly. The first sample taken is the sample 2–3 minutes after the median peak time observed over all subjects. To correct for the missing peak sample and timing, an estimated “correction” sample is introduced at 3 minutes. This estimated point is given a value 10% larger than the actual sample taken 2–3 minutes later. Thus, this estimated value is used to increase the area under the concentration curve to a more physiological value without having to capture it explicitly. Note that while the timing of 3 minutes works well in data used in this study, this might not be the case for all people and could be a potential source of error. Its validity would have to be assessed in a larger validation study.
These five steps are clarified further in Figure 2 and are summarized as follow.
Step 1: Original data set without reduction of samples.
Step 2: Optimized for NGT subjects. Six samples at D1, , D3, D4, D5, tend.
Step 3: Optimized for T2DM subjects. Six samples at D1, , D3, D4, D5, tend.
Step 4: Further reduction of samples to only include most critical points. Four samples at D1, , D5, tend.
Step 5: Sampling missing peak by 2–3 minutes, with “correction” sample introduced at 2 minutes. Six samples at D1,( – 3), + 3, D3, D4, D5, tend.
Figure 2.
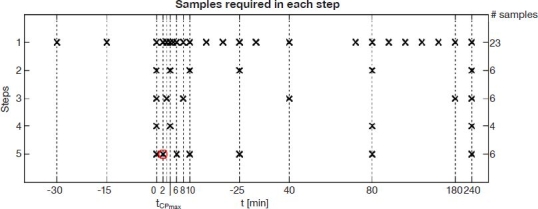
Sample optimization steps 1–5 and samples used for calculations in each step. Real samples are marked as ×, and the introduced “correction” sample in step 5 is marked with a circle.
Results from each are compared to step 1 to assess the performance of these reduced sampling schemes in comparison to the original data set.
Performance Metrics
The performance metrics used in this study try to capture all possible secretory characteristics of interest. The goal is to assess possible errors introduced by a reduced sampling protocol. These metrics are defined:
First phase peak secretion rate (Smax) and timing of peak (tpeak): Missing samples in the first minutes after glucose input can lead to large errors in the estimated peak secretion rate due to a slower observed increase in C-peptide concentration than actually occurs.
Total C-peptide secreted in first phase, 0–10 minutes (AUC10): The area under curve (AUC), also described as acute insulin response, is a common metric to describe total insulin secreted during the first phase response.2,9 It is calculated by integrating the estimated secretion rate between 0 and 10 minutes.
Total C-peptide secreted between glucose and insulin inputs, 0–20 min (AUC20): As the exogenous insulin inhibits pancreatic insulin secretion, it could be of interest to assess endogenously secreted insulin until it is inhibited by exogenous insulin.
Total C-peptide secreted during the IVGTT (AUCtotal): Calculated by integrating over the complete test, this metric assesses total pancreatic effort.
C-peptide assays also introduce errors for any data set that will affect the outcome values assessed. These expected error ranges are assessed by Monte Carlo analysis of the estimated secretion rate (104 runs), employing normally distributed, zero-mean noise with a coefficient of variation (CV) of 3%. This CV matches currently reported state-of-the-art assays25 and is thus a conservative choice, as older radioimmunoassays have CV values of up to twice this value,26 which would result in larger allowable errors from the reduced sampling protocol.
Therefore, Monte Carlo analysis provides an expected variation for the full set of step 1 results due to assay error. Reduced sampling schemes with results within this assay error range of step 1 results would be considered not different. Use of a small, state-of-the-art CV thus restricts this allowable variation to a minimum value.
Statistical Analysis
Nonparametric hypothesis testing with the Wilcoxon rank sum test is used to assess if steps 2–5 are significantly different to step 1. Normality of results is assessed by the single sample Kolmogorov–Smirnov test. Where results were log-normally distributed, the log-normal geometric mean and the multiplicative standard deviation27 are used and specifically noted in the respective results presented.
Results
The prehepatic insulin secretion rate was estimated well with the full data set using the integral-based method, resulting in the stepwise constant profiles in Figure 3. The qualitative shape of the secretory curves compares well to clinical data in the original publication.18 The mean peak secretion rate is slightly higher in this study for both subgroups, likely due to the smaller step size (1 minute vs 2 minutes) for the estimated secretion rate fitting in this study. Performance of the presented integral method is equivalent to the deconvolution method used by Mari,18 as seen by the matching metrics shown in Table 1.
Figure 3.
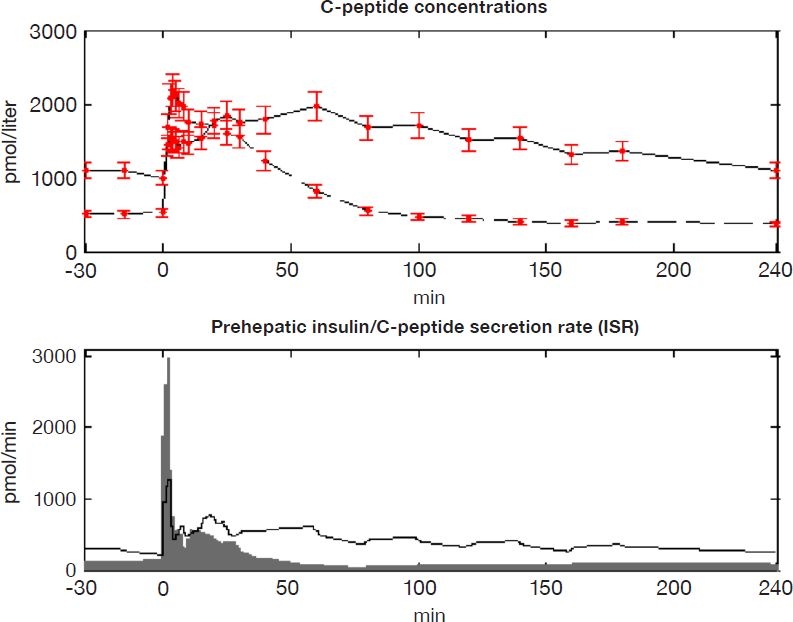
(Top) Mean C-peptide concentration in NGT (dashed) and T2DM (solid) subjects. Samples from Mari18 are shown with error bars of ± 2σ. (Bottom) Mean estimated C-peptide secretion rate (ISR) in NGT (gray area) and T2DM (solid line) subjects.
Table 1.
Integral Method Performance Compared to Metrics Obtained by Mari18 Using a Deconvolution Approach on Same Dataa
| ISRb | ISR1 | ISR2 | |
|---|---|---|---|
| Mean (SEMb) in pmol · min-1 · m-2 | |||
| NGT | |||
| Deconvolution | 71 (7) | 900 (233) | 127 (37) |
| Integral method | 71 (10) | 851 (216) | 132 (25) |
| Correlation (P < 0.001) | 0.93 | 1.00 | 0.95 |
| T2DM | |||
| Deconvolution | 141 (29) | 218 (120) | 121 (31) |
| Integral method | 136 (30) | 277 (136) | 130 (34) |
| Correlation (P < 0.001) | 0.98 | 1.00 | 0.99 |
ISRb is basal secretion rate, ISR1 is mean secretion rate over basal in the 6-minute postglucose injection, and ISR2 is mean secretion rate over basal from 7 minutes until glucose reaches basal levels. Values from this study are converted to match units used by Mari.18
Standard error of the mean.
Points of discontinuity are partly given by the protocol, as the timing of glucose and insulin inputs (D1 = 1 minute), and are otherwise identified from the sampled C-peptide profile in each subject (D2,D3,D4,D5). All identified points are given in Table 2. More variability in all points and especially a distinct lag in D4 (response to insulin input) and D5 (return to basal) are evident in T2DM subjects, as expected.
Table 2.
Points of Discontinuity Identified in All Subjectsa
| Subject | D1 | D2 | D3 | D4 | D5 |
|---|---|---|---|---|---|
| NGT | |||||
| 7 | 1 | 4 | 10 | 20 | 100 |
| 8 | 1 | 4 | 10 | 25 | 60 |
| 9 | 1 | 3 | 10 | 25 | 80 |
| 10 | 1 | 3 | 8 | 25 | 60 |
| 11 | 1 | 4 | 10 | 25 | 80 |
| Median | 1 | 4 | 10 | 25 | 80 |
| SD | 0.00 | 0.55 | 0.89 | 2.24 | 16.73 |
| T2DM | |||||
| 1 | 1 | 3 | 10 | 60 | 240 |
| 2 | 1 | 6 | 8 | 60 | 160 |
| 3 | 1 | 2 | 8 | 30 | 100 |
| 4 | 1 | 2 | 15 | 25 | 240 |
| 5 | 1 | 4 | 8 | 40 | 180 |
| 6 | 1 | 2 | 8 | 40 | 120 |
| 12 | 1 | 3 | 10 | 20 | 240 |
| Median | 1 | 3 | 8 | 40 | 180 |
| SD | 0.00 | 1.46 | 2.57 | 15.92 | 59.36 |
| Overall | |||||
| Median | 1 | 3 | 10 | 25 | 100 |
| SD | 0.00 | 1.15 | 1.98 | 14.22 | 71.07 |
Note that points D2–D5 have significant differences between subgroups. Median values are used in the generic selection of points for reduced sample analysis to enable a generalizable approach.
Resulting deviations in performance metrics for steps 2–5, compared to the original sample sets, are shown in Table 3. None of the metrics in steps 2–4 were significantly different to the corresponding reference metrics in step 1 in both subgroups (all P < 0.05). Distribution of the resulting performance metrics is log-normal and results are thus given using log-normal statistics. Relative differences are normally distributed and are described using normal statistics. Correlations of steps 2–5 compared to step 1 are also shown in Table 3.
Table 3.
Outcomes of Sample Reduction Stepsa
| Steps (# samples) | 1 (23) | 2 (6) | 3 (6) | 4 (4) | 5 (6) | ||
|---|---|---|---|---|---|---|---|
| Reference | Percentile change [%] | ||||||
| NGT | |||||||
| Smax | Mean (geom) | 2578.8 | Mean | –5.52 | –7.58 | –5.52 | –1.80 |
| (pmol/min) | SD (multipl) | 1.8 | SD | 1.61 | 5.68 | 1.59 | 5.27 |
| correlation | 1.00 | 0.99 | 1.00 | 1.00 | |||
| tpeak | Median | 3 | Median | 0 | 0 | 0 | 0 |
| (min) | SD | 0.0 | SD | 0.0 | 0.0 | 0.0 | 0 |
| AUC10 | Mean (geom) | 10301.8 | Mean | –5.21 | 2.23 | 10.10 | –0.71 |
| (pmol) | SD (multipl) | 1.8 | SD | 1.56 | 3.10 | 9.28 | 2.243 |
| correlation | 1.00 | 1.00 | 0.99 | 1.00 | |||
| AUC20 | Mean (geom) | 15110.8 | Mean | –3.24 | –0.54 | 8.40 | –0.62 |
| (pmol) | SD (multipl) | 1.7 | SD | 9.97 | 9.84 | 9.34 | 9.56 |
| correlation | 0.97 | 0.97 | 0.97 | 0.97 | |||
| AUCtotal | Mean (geom) | 42648.9 | Mean | 4.95 | 19.44 | 9.30 | 5.70 |
| (pmol) | SD (multipl) | 1.4 | SD | 4.42 | 6.53 | 3.98 | 3.76 |
| correlation | 0.99 | 0.99 | 1.00 | 1.00 | |||
| T2DM | |||||||
| Smax | Mean (geom) | 826.3 | Mean | –15.64 | –2.36 | –15.30 | –0.61 |
| (pmol/min) | SD (multipl) | 2.5 | SD | 14.32 | 12.07 | 13.91 | 21.47 |
| correlation | 1.00 | 1.00 | 1.00 | 0.96 | |||
| tpeak | Median | 3 | Median | 0 | 0 | 0 | 0 |
| (min) | SD | 0.8 | SD | 0.8 | 0.8 | 3.0 | 1.11 |
| AUC10 | Mean (geom) | 4600.7 | Mean | –6.40 | –0.73 | 5.42 | –2.39 |
| (pmol) | SD (multipl) | 2.5 | SD | 5.83 | 9.27 | 27.14 | 10.88 |
| correlation | 1.00 | 1.00 | 0.99 | 1.00 | |||
| AUC20 | Mean (geom) | 9439.8 | Mean | –4.23 | –4.47 | –6.76 | –2.23 |
| (pmol) | SD (multipl) | 2.4 | SD | 9.38 | 12.74 | 20.27 | 8.66 |
| correlation | 0.99 | 0.98 | 0.95 | 0.99 | |||
| AUCtotal | Mean (geom) | 82523.7 | Mean | 0.43 | –0.52 | –1.32 | 0.63 |
| (pmol) | SD (multipl) | 2.2 | SD | 5.18 | 4.36 | 6.21 | 5.39 |
| correlation | 1.00 | 1.00 | 1.00 | 1.00 | |||
Relative percentile changes of steps 2–5 are shown compared to reference step 1 (tpeak is given as absolute difference in minutes). Results from step 1 have a log-normal distribution and are described by the log-normal geometric mean (geom) and the multiplicative standard deviation (multipl). Relative changes in steps 2–5 are distributed normally and are described by the mean and standard deviation (SD). Correlations shown are steps 2–5 compared to step 1 (P < 0.001).
Errors in performance metrics due to assay errors were assessed by Monte Carlo analysis (104 runs) and are given as a CV for each metric, with the median and 100% range over all 12 subjects:
Smax: CV = 5.47%, range 2.97–11.01%
AUC10: CV = 4.10%, range 1.92–9.39%
AUC20: CV = 3.13%, range 1.90–4.26%
AUCtotal: CV = 1.11%, range 0.97–1.25%
Reconstruction of C-peptide concentrations from the identified secretion profiles during analyzed steps resulted in the residuals shown in Figure 4. Residuals are given as relative values (decimal percentages), compared to the complete sampling protocol of step 1. Deviations from the original sample set are caused by smoothing of the estimated secretion profile, by errors introduced through linear interpolation, and, obviously, by the reduced number of sampling steps being examined. The ideal goal is to have all variation within the dashed lines due to assay error.
Figure 4.
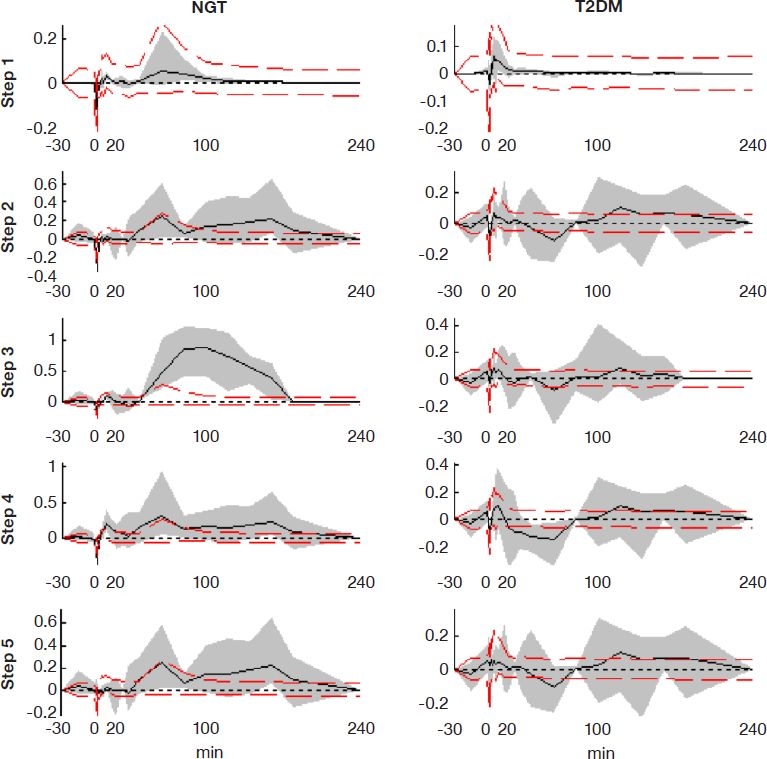
Residuals introduced when reproducing the full C-peptide sample profile with sample reduction steps. The solid line shows mean residuals, and the gray area shows the full 100% range of residuals in that step. Dashed lines show the 95% range of residuals for step 1 (full sample set) introduced by assay error, as estimated by Monte Carlo analysis.
Discussion
Estimating prehepatic insulin secretion through modeling of C-peptide kinetics has been a common methodology and is relatively less invasive to perform in research settings.12–14,28 In particular, the population method proposed by Van Cauter and colleagues12 enables estimation of the secretion rate with a single experiment. By employing this method, model parameters are consistent across studies, enabling a better comparison, as trade-offs between estimated parameters and secretion rates are reduced. Nonetheless, estimation of the peak secretion rate and insulin secreted during the first phase is still highly dependent on assay errors and sampling frequency during the initial minutes. Ideally, sampling should be performed every minute to assess an accurate profile, which introduces significant labor, cost, and burden, as well as reducing the robustness of the method.
It is important to keep in mind that significant errors are also introduced because of assay inaccuracy. Thus, for example, the peak estimated secretion rate, Smax, has a median CV = 5.47% and can therefore vary between ±11% (± 2σ), even with a 1-minute step sampling protocol. Most of the performance metrics are within or slightly outside of ± 2σ of assay error. This result implies that they are, in fact, just within the natural variability that can be identified.26
Using the same model and the parameter estimation method of Van Cauter and associates,12 estimation of the secretion rate has been proposed previously by deconvolution11 and a more elaborate constrained regularization method.14 The main drawbacks of these methods are the individualized method adjustments required for each subject, including knot placements for cubic spline interpolations,11 or a separate step to find the optimal proportionality constant in each subject.14 All of these extra steps introduce time, computation, and human variability into the results. While criteria are available to determine optimal regularization parameters, such as maximum likelihood, these methods require underlying a priori assumptions about the parameter solutions and thus add complexity to the approach.
In contrast, the integral-based method described in this study is a single step, computationally convex and fast method that only requires linearly interpolated data. By constraining the resulting linear least-squares estimation to nonzero values and smoothing the estimated secretion rate, the resulting profile is physiologically accurate and the effects caused by noisy data are reduced.21 First and second phase secretion characteristics were clearly identified, with slight quantitative, but not qualitative, deviations from the profiles originally reported with these data by Mari.18 In addition, these smaller deviations can be explained readily by the longer step size used in that study. In a quantitative comparison, the secretion metrics obtained with the integral-based method compare very well to the secretion metrics calculated by Mari18 using a deconvolution method, as seen in Table 1. Correlations of the subgroup results are all very high, showing equality in performance of both methods when the full sample data set is used.
The reduction of samples was approached by identifying key points of discontinuity and reducing the sample set to those points. Points of discontinuity varied only slightly during first phase secretion in both subgroups, but were delayed significantly in T2DM during the second secretion phase. This delay can be attributed to a delay in the pancreatic response to insulin input in the case of D424 and an increased total demand and production rate during this stage in D5.1
Comparing steps 2 and 3, which are optimized for the NGT and T2DM subgroups, respectively, it can be seen that the maximum secretion rate, Smax, is more accurate in the subgroup for which it was optimized. This result is especially valid for the T2DM subjects in step 3. This result also holds for total secreted C-peptide AUCtotal in NGT and T2DM, but is not the case in the other metrics.
The standard deviations of the metrics are mostly very broad in T2DM, especially in the metrics during the first minutes. This result indicates a very broad variability in the estimated metric. This variability may be due in part to the strongly blunted first phase response in T2DM, resulting in a weak signal-to-noise ratio and thus exaggerating the effects of assay errors. Nonetheless, none of the sample reduced steps were statistically significantly different than reference step 1 (P < 0.05).
As seen in the residuals reported in Figure 4, step 2 has a clear advantage over step 3 in NGT subjects. In T2DM subjects, a slight advantage for step 3 is evident between 0 and 20 minutes, but the remaining time is equivalent to step 2. This behavior could be due to the fact that the points of discontinuity in the time after insulin input are not as distinct in T2DM and thus not as critical if chosen inaccurately. Larger residuals appear after t = 20 minutes in all cases, where sampling is less frequent. During the first section up to t = 20 minutes, residuals are mostly within the assay variation bounds shown for the full sample set (step 1), giving accurate estimations of the most dynamic secretory characteristics. Overall, step 2 seems to be the better choice if one generic setting were chosen for both types of subjects examined.
In step 4, where samples are further reduced to a total of four, residuals are more variable, but still within similarly tight ranges, as in the previous steps. In particular, the first phase section is well represented and captured. In NGT subjects, residuals are even tighter than in step 3, which has two additional samples that are not optimally placed for this group.
Finally, step 5 analyzes a different approach by introducing a calculated “correction” sample to make up for the missing concentration peak sample. This step appears to give the tightest residuals during the first phase, even tighter than the full sampling set. This unexpected result is due to a more accurate fast rise in concentration, as the sample is introduced at t = 2 minutes, resulting in a higher secretory peak. Without this correction sample, linear interpolation from 1 to 6 minutes would result in a far slower secretion rate increase and a more constant and nonphysiological estimated secretion rate during these initial 5 minutes. Hence, the resulting area under the concentration curve is more physiological, which results in a more accurate integrated secretion curve and thus better residuals. During the later phase of these tests, residuals are identical to step 2 because the same sample timings are used.
Overall, it can be seen that reduced sampling does not necessarily compromise the information that can be gathered from such a test. This is clearly visible by the very high correlations shown in Table 3 between the full and the sample reduced steps. However, smart sample placement is critical and needs to be chosen correctly according to the secretory information of interest to the researcher. Steps 2 and 3 propose optimized sampling protocols for NGT and T2DM subgroups, respectively, enabling the investigator to decide on an optimal sampling schedule when designing a test protocol. Even a heavily reduced and generic protocol using only 4 (17%) of the original 23 samples (step 4) results in acceptable accuracy in the stated performance metrics, most of which are still within reported assay errors.
While the methods developed in this study performed well on the presented data set, it could be argued that the number of subjects is insufficient to validate the approach. We acknowledge that the number of subjects used to derive the presented method is limited. The goal of the study was not to validate the method clinically, but to derive and present a new method to estimate insulin secretion that is more robust and automated compared to previously presented methods. In that sense, it should be regarded as a pilot study to derive a new method. This new method would have to be validated in a separate study on a different data set to prove its validity. In addition, the use of physiologically relevant points of discontinuity that are readily recognizable and well accepted adds weight to the underlying assumption that the results of this limited pilot analysis would carry through in a larger study.
In a similar approach, the analysis presented in this study could also be applied to a C-peptide data set without insulin administration. New points of discontinuity would have to be defined as they could differ slightly, particularly around the time when insulin is administered. We believe that the approach would work just as well on such a data set, but it is out of the scope of this study to analyze different trial protocols. This could be analyzed in a separate study with a corresponding data set.
We believe that our approach is novel compared to other methods used to estimate insulin secretion presented in the past. Strong emphasis was placed on developing a robust and convex method that would allow automated analysis of C-peptide data without requiring manual intervention or a priori assumptions about the solutions. While methods presented in the past have focused primarily on the accuracy of full data sets, our approach has been primarily on a method that could be applied to reduced data sets and thus be more useful in routine clinical testing environments, where time and cost contribute greatly to the success of a test.
Conclusions
Estimation of prehepatic insulin or C-peptide secretion can be achieved using an easy-to-apply population model in combination with a simple and consistent integral-based deconvolution method. A reduction of samples to reduce test complexity, clinical burden, and cost can be done without significantly reducing the accuracy of the test. If smart sample placements are chosen by identifying key points of discontinuity, these reductions are readily enabled, saving significant cost and burden.
The approaches presented in this study include sampling optimized for NGT or T2DM subjects (six samples), a further reduction to four samples, and a final option that does not require samples during the first 5 minutes after glucose administration by introducing an additional calculated “correction” sample. Each step further reduces the sampling stress, cost, and blood taken. Overall, results show that reduced sampling has no clinical or research “cost” in the outcome metrics derived, as shown by Monte Carlo and statistical results, but can enable significantly simpler test protocols.
Acknowledgements
The authors thank Professor Andrea Mari and Professor Angelo Avogaro for kindly providing IVGTT data used in this study. Uli Göltenbott was supported with a Baden-Württemberg-Scholarship from the Landesstiftung Baden-Württemberg, Germany.
Abbreviations
- AUC
area under curve
- CV
coefficient of variation
- IVGTT
intravenous glucose tolerance test
- NGT
normal glucose-tolerant
- T2DM
type 2 diabetes mellitus
References
- 1.Ferrannini E, Gastaldelli A, Miyazaki Y, Matsuda M, Mari A, DeFronzo RA. beta-Cell function in subjects spanning the range from normal glucose tolerance to overt diabetes: a new analysis. J Clin Endocrinol Metab. 2005;90(1):493–500. doi: 10.1210/jc.2004-1133. [DOI] [PubMed] [Google Scholar]
- 2.Ferrannini E, Mari A. Beta cell function and its relation to insulin action in humans: a critical appraisal. Diabetologia. 2004;47(5):943–956. doi: 10.1007/s00125-004-1381-z. [DOI] [PubMed] [Google Scholar]
- 3.Stumvoll M, Fritsche A, Haring HU. Clinical characterization of insulin secretion as the basis for genetic analyses. Diabetes. 2002;51(Suppl 1):S122–S129. doi: 10.2337/diabetes.51.2007.s122. [DOI] [PubMed] [Google Scholar]
- 4.Bergman RN, Ader M, Huecking K, Van Citters G. Accurate assessment of beta-cell function: the hyperbolic correction. Diabetes. 2002;51(Suppl 1):S212–S220. doi: 10.2337/diabetes.51.2007.s212. [DOI] [PubMed] [Google Scholar]
- 5.DeFronzo RA, Tobin JD, Andres R. Glucose clamp technique: a method for quantifying insulin secretion and resistance. Am J Physiol. 1979;237(3):E214–E223. doi: 10.1152/ajpendo.1979.237.3.E214. [DOI] [PubMed] [Google Scholar]
- 6.Bergman RN, Phillips LS, Cobelli C. Physiologic evaluation of factors controlling glucose tolerance in man: measurement of insulin sensitivity and beta-cell glucose sensitivity from the response to intravenous glucose. J Clin Invest. 1981;68(6):1456–1467. doi: 10.1172/JCI110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stumvoll M, Mitrakou A, Pimenta W, Jenssen T, Yki-Jarvinen H, Van Haeften T, Renn W, Gerich J. Use of the oral glucose tolerance test to assess insulin release and insulin sensitivity. Diabetes Care. 2000;23(3):295–301. doi: 10.2337/diacare.23.3.295. [DOI] [PubMed] [Google Scholar]
- 8.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 9.Pacini G, Mari A. Methods for clinical assessment of insulin sensitivity and beta-cell function. Best Pract Res Clin Endocrinol Metab. 2003;17(3):305–322. doi: 10.1016/s1521-690x(03)00042-3. [DOI] [PubMed] [Google Scholar]
- 10.Faber OK, Hagen C, Binder C, Markussen J, Naithani VK, Blix PM, Kuzuya H, Horwitz DL, Rubenstein AH, Rossing N. Kinetics of human connecting peptide in normal and diabetic subjects. J Clin Invest. 1978;62(1):197–203. doi: 10.1172/JCI109106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton RP, Allen RC, Schade DS, Erickson KM, Standefer J. Prehepatic insulin production in man: kinetic analysis using peripheral connecting peptide behavior. J Clin Endocrinol Metab. 1980;51(3):520–528. doi: 10.1210/jcem-51-3-520. [DOI] [PubMed] [Google Scholar]
- 12.Van Cauter E, Mestrez F, Sturis J, Polonsky KS. Estimation of insulin secretion rates from C-peptide levels. Diabetes. 1992;41(3):368–377. doi: 10.2337/diab.41.3.368. Comparison of individual and standard kinetic parameters for C-peptide clearance. [DOI] [PubMed] [Google Scholar]
- 13.Polonsky KS, Licinio-Paixao J, Given BD, Pugh W, Rue P, Galloway J, Karrison T, Frank B. Use of biosynthetic human C-peptide in the measurement of insulin secretion rates in normal volunteers and type I diabetic patients. J Clin Invest. 1986;77(1):98–105. doi: 10.1172/JCI112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovorka R, Soons PA, Young MA. ISEC: a program to calculate insulin secretion. Comput Methods Programs Biomed. 1996;50(3):253–264. doi: 10.1016/0169-2607(96)01755-5. [DOI] [PubMed] [Google Scholar]
- 15.Hovorka R, Koukkou E, Southerden D, Powrie JK, Young MA. Measuring pre-hepatic insulin secretion using a population model of C-peptide kinetics: accuracy and required sampling schedule. Diabetologia. 1998;41(5):548–554. doi: 10.1007/s001250050945. [DOI] [PubMed] [Google Scholar]
- 16.Jones CN, Pei D, Staris P, Polonsky KS, Chen YD, Reaven GM. Alterations in the glucose-stimulated insulin secretory dose-response curve and in insulin clearance in nondiabetic insulin-resistant individuals. J Clin Endocrinol Metab. 1997;82(6):1834–1838. doi: 10.1210/jcem.82.6.3979. [DOI] [PubMed] [Google Scholar]
- 17.Toffolo G, De Grandi F, Cobelli C. Estimation of beta-cell sensitivity from intravenous glucose tolerance test C-peptide data. Diabetes. 1995;44(7):845–854. doi: 10.2337/diab.44.7.845. Knowledge of the kinetics avoids errors in modeling the secretion. [DOI] [PubMed] [Google Scholar]
- 18.Mari A. Assessment of insulin sensitivity and secretion with the labelled intravenous glucose tolerance test: improved modelling analysis. Diabetologia. 1998;41(9):1029–1039. doi: 10.1007/s001250051027. [DOI] [PubMed] [Google Scholar]
- 19.Wong XW, Chase JG, Shaw GM, Hann CE, Lotz T, Lin J, Singh-Levett I, Hollingsworth LJ, Wong OS, Andreassen S. Model predictive glycaemic regulation in critical illness using insulin and nutrition input: a pilot study. Med Eng Phys. 2006;28(7):665–681. doi: 10.1016/j.medengphy.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Chase JG, Shaw GM, Lin J, Doran CV, Hann C, Lotz T, Wake GC, Broughton B. Targeted glycemic reduction in critical care using closed-loop control. Diabetes Technol Ther. 2005;7(2):274–282. doi: 10.1089/dia.2005.7.274. [DOI] [PubMed] [Google Scholar]
- 21.Hann CE, Chase JG, Lin J, Lotz T, Doran CV, Shaw GM. Integral-based parameter identification for long-term dynamic verification of a glucose-insulin system model. Comput Methods Programs Biomed. 2005;77(3):259–270. doi: 10.1016/j.cmpb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Davies MJ, Rayman G, Grenfell A, Gray IP, Day JL, Hales CN. Loss of the first phase insulin response to intravenous glucose in subjects with persistent impaired glucose tolerance. Diabet Med. 1994;11(5):432–436. doi: 10.1111/j.1464-5491.1994.tb00302.x. [DOI] [PubMed] [Google Scholar]
- 23.Del Prato S, Marchetti P, Bonadonna RC. Phasic insulin release and metabolic regulation in type 2 diabetes. Diabetes. 2002;51(Suppl 1):S109–S116. doi: 10.2337/diabetes.51.2007.s109. [DOI] [PubMed] [Google Scholar]
- 24.Jefferson LS, Cherrington A. Oxford: Oxford University Press; 2001. The endocrine pancreas and regulation of metabolism. Handbook of physiology: the endocrine system. [Google Scholar]
- 25.Roche. C-peptide immunoassay, Elecsys 1010/2010/Modular Analytics E170. Mannheim, Germany: Roche Diagnostics; 2005. Data sheet. Technical report, 03184897 190. [Google Scholar]
- 26.Clark PM. Assays for insulin, proinsulin(s) and C-peptide. Ann Clin Biochem. 1999;36(Pt 5):541–564. doi: 10.1177/000456329903600501. [DOI] [PubMed] [Google Scholar]
- 27.Limpert E, Stahel WA, Abbt M. Log-normal distributions across the sciences: keys and clues. Bioscience. 2001;51(5):341–352. [Google Scholar]
- 28.Watanabe RM, Volund A, Roy S, Bergman RN. Prehepatic beta-cell secretion during the intravenous glucose tolerance test in humans: application of a combined model of insulin and C-peptide kinetics. J Clin Endocrinol Metab. 1989;69(4):790–797. doi: 10.1210/jcem-69-4-790. [DOI] [PubMed] [Google Scholar]


