Abstract
The ASCE superfamily of proteins consists of structurally similar ATPases associated with diverse cellular activities involving metabolism and transport of proteins and nucleic acids in all forms of life1. A subset of these enzymes are multimeric ringed pumps responsible for DNA transport in processes including genome packaging in adenoviruses, herpesviruses, poxviruses, and tailed bacteriophages2. While their mechanism of mechanochemical conversion is beginning to be understood3, little is known about how these motors engage their nucleic acid substrates. Do motors contact a single DNA element, such as a phosphate or a base, or are contacts distributed over multiple parts of the DNA? In addition, what role do these contacts play in the mechanochemical cycle? Here we use the genome packaging motor of the Bacillus subtilis bacteriophage φ294 to address these questions. The full mechanochemical cycle of the motor, whose ATPase is a pentameric-ring5 of gene product 16, involves two phases-- an ATP loading dwell followed by a translocation burst of four 2.5-bp steps6 triggered by hydrolysis product release7. By challenging the motor with a variety of modified DNA substrates, we find that during the dwell phase important contacts are made with adjacent phosphates every 10-bp on the 5’-3’ strand in the direction of packaging. In addition to providing stable, long-lived contacts, these phosphate interactions also regulate the chemical cycle. In contrast, during the burst phase, we find that DNA translocation is driven against large forces by extensive contacts, some of which are not specific to the chemical moieties of DNA. Such promiscuous, non-specific contacts may reflect common translocase-substrate interactions for both the nucleic acid and protein translocases of the ASCE superfamily1.
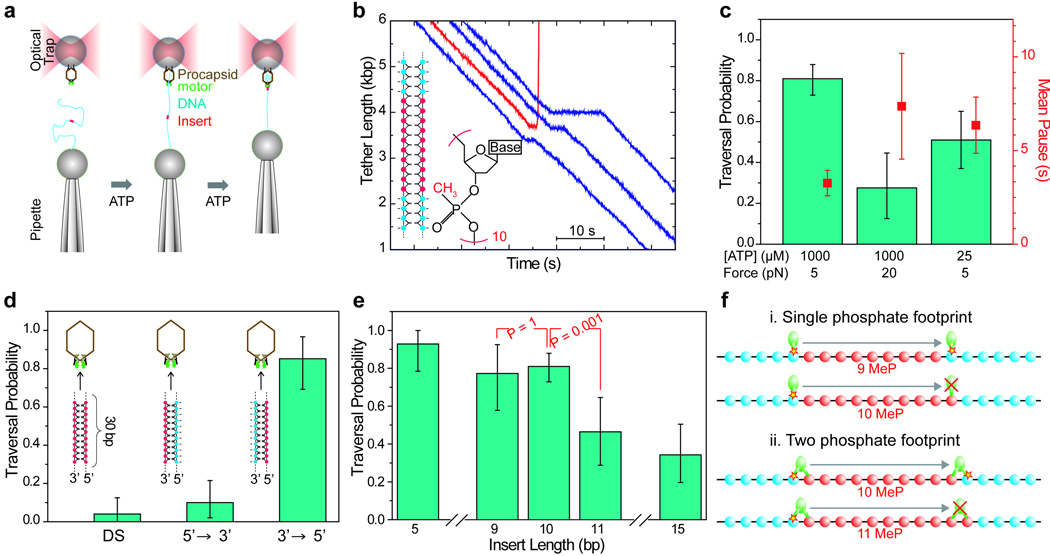
To test the role of the phosphate backbone charge in motor-DNA interactions we inserted a 10 bp region of double stranded methylphosphonate DNA (ds-MeP) into the middle of an ∼8Kbp native DNA molecule (Figure 1a) and followed the packaging of this molecule by a single φ29 prohead-motor complex using optical tweezers. In MeP the charged oxygen on DNA is replaced with an uncharged isosteric methyl group while conserving the B-form structure of DNA8,9 (Figure 1b, inset). Thus, it is possible to infer the role of this chemical modification in a native geometric context. Figure 1b shows sample packaging traces under 5 pN of constant tension and saturating [ATP] (1 mM). Packaging proceeds normally until the motor encounters the inserted modification where it pauses and then either successfully traverses the insert or completely dissociates. In stark contrast to related helicases, where disruption of a single charge-backbone interaction10–14 completely abolishes translocation, the packaging motor traverses 10 bp of neutral DNA with a probability of ∼80% under a tension of 5pN (Figure 1c).
Figure 1. Packaging of neutral DNA analogs.
(a) A prohead-gp16-ATPase-motor complex bound to a microsphere is held in an optical trap while a micropipette holds a second microsphere bound to DNA containing a modified insert. A tether is formed and packaging is initiated when the beads are briefly brought into close proximity in the presence of ATP. (b) Representative packaging traces of DNA containing 10 bp ds-MeP at a constant load of 5pN. Blue traces show traversal following a pause, and the red trace shows a terminal dissociation event following a pause. Inset are a schematic of the insert, with MeP nucleotides in red and unmodified nucleotides in blue, and the chemical structure of a MeP nucleotide. (c) Force and ATP dependence of traversal probability and pause duration of 10 bp ds-MeP inserts. (d) Traversal probability of 30 bp ds-MeP and DNA-MeP hybrid inserts at 5 pN. (e) Traversal probability of ds-MeP inserts at 5 pN force as a function of insert length. P-values (Two-tailed Fisher exact test) between 9 and 10 bp, and 10 and 11 bp are indicated. (f) Translocation cycle length and footprint size limits from MeP length dependence. This scheme shows the position of a subunit that contacts the DNA before and after a full mechanochemical cycle, i.e. 10 bp. Contact with a single phosphate would produce a drop in traversal probability between 9 and 10 bp ds-MeP whereas contact with two phosphates would produce the observed drop between 10 and 11 bp ds-MeP. In c, d, and e the traversal probability is plotted using the Laplace best estimator26, with 95% confidence intervals from the adjusted Wald method27, and error bars of pause durations are the SEM.
To rule out the possibility that the motor crosses the neutral insert via diffusive fluctuations as opposed to making direct contact with uncharged moieties, we took advantage of the strong force dependence of diffusive traversal times15. Figure 1c shows that there is only a two-fold increase in pause duration with a 15 pN increase in force, much less than the 105-fold increase predicted for diffusion across a 10 bp distance (Supplementary Discussion). In addition, lowering [ATP] increases the pause duration and decreases the traversal probability, providing further support for an active, ATP-dependent crossing mechanism. Thus, the motor actively traverses the insert by making contacts with elements other than the phosphate charge, albeit with reduced efficiency, implying that native packaging involves both charge and non-charge contacts.
To determine if phosphate charges from both strands are equally important, we created hybrid inserts in which only one strand contains the MeP backbone. We used a 30-bp insert to accentuate the effect of the uncharged section since the traversal probability of a 30-bp dsMeP insert at 5 pN is ∼4%. At this force the traversal probability of the hybrid insert with MeP on the strand packaged from 3’–5’ is almost 90%, whereas that of the hybrid insert with MeP in the 5’-3’strand is reduced to 10% (Figure 1d). This result clearly indicates that the most important phosphate interactions are made with the 5’-3’ strand in the direction of packaging. Such preferential interaction with a single DNA strand has been shown for the monomeric dsDNA translocase EcoR124I16, though it is more surprising in this case of a ring-ATPase where multiple subunits of the ring are simultaneously in close proximity to both strands.
Next, we asked if a critical length is involved in the interaction of the motor and its DNA substrate. Figure 1e shows that as we increase the length of the double-stranded neutral insert from 5 to 10 bp there is no statistically significant change in traversal probability until the 1 bp increase from 10 to 11 bp that results in a two-fold reduction. However, further increasing the length to 15 bp does not produce a similar change. The location of this discrete change in traversal probability at first appears inconsistent with the 10 bp of DNA packaged by the motor each full mechanochemical cycle6, but these results are easily reconciled if the motor makes contact with two adjacent phosphates, with either contact being sufficient for packaging (Figure 1f). The co-crystal structure of the related BPV helicase E1 with its ssDNA substrate reveals simultaneous contact with adjacent phosphate charges17, lending support to this interpretation.
We next investigated the specific role of these phosphates in the mechanochemical cycle by probing the base-pair-scale dynamics of the motor at an uncharged insert. Phosphate-motor interactions may serve two possible roles in the mechanochemical cycle: they may provide the long-lived contacts required to keep the enzyme attached to the DNA, or they may serve a sensory role, accelerating a chemical rate, such as ATP hydrolysis, upon detecting that the DNA is bound and properly oriented. These two roles of the phosphate charge, while not mutually exclusive, should be revealed by characteristic dynamics of the motor as it traverses the modified insert. If the phosphate provides load-bearing contacts, its absence will increase the dissociation rate of the motor, and the insert-induced pause will consist of a series of attempts to package followed by small slips. Alternatively, if the role of the phosphate is sensory, we expect the time between packaging steps to be lengthened, due to the decreased rate of catalytic turnover.
To reveal the dynamics of the motor as it crosses a neutral insert (10-bp ds-MeP), we followed packaging using a dual-trap optical tweezers with higher spatial and temporal resolution6. Figure 2a shows that the pauses observed at low resolution are actually remarkably dynamic events, containing two types of sub-pauses that occur at distinct locations along the modified DNA insert. The first type of sub-pause, which we term upstream pauses because they occur at longer DNA tether lengths, are followed by either brief disengagement of the motor (slips) or packaging attempts. These attempts are themselves followed by a second class of sub-pause, which we term downstream pauses. After slips from either the upstream or downstream pauses the motor typically recovers and repackages the DNA to the position of the upstream pause. Occasionally the motor does not recover from a slip, resulting in a terminal slip. Figure 2b contains the branching probabilities of these events.
Figure 2. High resolution dynamics at neutral-DNA insert.
(a) Base-pair-scale dynamics at 10 bp of ds-MeP consist of two classes of sub-pauses, upstream and downstream pauses, separated by attempts and punctuated by slips and repackaging events. (b) A cartoon model of the dynamics of these pauses with average lifetimes and inter-conversion probabilities. The full statistics of these states are in Supplementary Table 3. (c,d) Histograms of upstream and downstream pause durations. The distributions have nmin values—the ratio of the mean squared to the variance—of 1.1 ± 0.1 (s.d.) and 1.3 ± 0.4 (s.d.), respectively; and are thus well-described by single exponential decays. (e) Histogram of position changes in the upstream pause. (f) Histogram of distance between upstream and downstream pauses.
The upstream sub-pauses occur in a uniform position on a given tether, ± 1 bp (s.d.), and have longer average durations, 1.00 ± 0.08 s (s.e.m.; Figure 2c&e), while the downstream sub-pauses occur at the end of attempts of different sizes ± 3 bp (s.d.) and have shorter durations, 80 ± 10 ms (s.e.m.,Figure 2f&d). The upstream- and downstream-pause time distributions are both well-described by single exponential decays (Figure 2c&d), suggesting that each class of sub-pause corresponds to an individual kinetic process, as opposed to a weighted sum of different kinetic processes, and that each of them is governed by a single rate-limiting kinetic event. In combination with their distinct mean durations, these observations suggest that each class of sub-pause corresponds to one of the mechanochemical phases of the motor: ATP-loading dwell or stepping burst.
To identify the kinetic state of the motor associated with each class of sub-pause, we measured the dynamics of motors traversing 5 bp of neutral DNA, an insert shorter than the 10-bp span of the full mechanochemical cycle6. While contacts during the burst phase occur every 2.5-bp, contacts during the dwell phase should occur only every 10 bp. Thus all motors must make burst phase contacts with the neutral DNA while a fraction of the motors will never make dwell phase contacts with the neutral DNA. If the upstream pause occurs during the dwell phase, then only a fraction of the motors will display such a pause. Supporting Figure 1 shows that only ∼50% of the motors packaging 5 bp of ds-MeP display a long pause, strongly suggesting that the upstream pause corresponds to the dwell phase and hence the downstream pause to the burst phase. This conclusion is further supported by the average duration of the downstream pauses and the exponential nature of their distribution, which are both similar to those of the micro-dwells observed during the burst phase on charged DNA under similar conditions6. Finally, the highly uniform positions of the upstream pauses in a given tether indicate that this single-base-pair position is unique, suggesting that after slipping and repositioning, the motor resides at the boundary of the charged and neutral DNA during its dwell phase.
The multiple slip/attempt phenotype observed in Figure 2a reveals that removing the phosphate charge reduces the processivity of the motor. Since the motor slips both from upstream and downstream pauses, load-bearing phosphate contacts are made during both the dwell and burst phase of the cycle. Interestingly, however, upstream pauses are on average 10-fold longer than dwells on normal DNA under the same experimental conditions ( ∼100 ms6); thus, the absence of the phosphate charge slows the dwell phase, revealing an additional sensory role for this contact. The fact that a single kinetic event dominates the upstream pause duration, in sharp contrast to the ∼4 kinetic events that are known to be rate-limiting for the dwell phase on charged DNA6, indicates that the mechanochemical cycle of the packaging motor contains a single kinetic checkpoint—a process that halts the chemical cycle until the DNA is correctly positioned. In contrast, the average duration of the contacts during the burst phase is not significantly modified on neutral DNA, hence these contacts likely do not involve a significant sensory role.
These dynamics suggest a mechanism for the motor to cross neutral inserts. Successful traversal is a kinetic competition between the increased off-rate of the motor on neutral DNA and the time it takes to successfully complete the mechanochemical events necessary to traverse this modified DNA. There is a small probability of loading the necessary ATPs and starting the burst, out-competing the increased slipping rate on the neutral DNA; however, as illustrated by the probabilities in Figure 2b, the large probability of recovering from a slip allows the motor to attempt to package the insert many times, amplifying this small probability to the large traversal probabilities observed in Figure 1. This kinetic competition also explains the dramatic decrease in the ability of the motor to package the 30 bp of neutral DNA since successful traversal would require multiple consecutive complete mechanochemical cycles to out-compete slipping. Finally, the branching probabilities observed in Figure 2b predict traversal probabilities consistent with the values in Figure 1, providing further support for this model (Supplementary Discussion).
The fact that the motor resides for a finite time in the neutral DNA (Figure 2) provides further evidence for additional, non-charge contacts. To test the role of sugars and bases in these interactions, we created an insert with these elements removed (Figure 3a). These chemical modifications also disrupt the helical geometry of the DNA; thus, to isolate the role of DNA geometry, we created inserts with no chemical modification yet with large disruptions to the helix; namely, single-stranded gaps (Figure 3b) and unpaired bulges (Figure 3c). Remarkably, the φ29 motor is capable of traversing all of these modifications (Figure 3b and 3c) and the force dependence of the mean pause durations again rules out a purely diffusive model (Supplementary Table 4). Notably, the motor shows lower traversal probability and higher pause durations for gaps on the 5’-3’ strand than those on the 3’-5’ strand, with a similar trend seen in pause durations of the bulges, consistent with the important 5’-3’ contacts discussed above. Our findings are consistent with the successful packaging of short unpaired bulges in phage lambda18, though they differ from reported results in φ2919 and T420 where single-stranded gaps were not packaged. However, disparate experimental methods likely account for these apparent differences (Supplementary Discussion).
Figure 3. Substrate promiscuity.
(a–d) The force dependence of traversal probabilities and pause durations of different modified DNA substrates. (a) 10 nt ds-abasic phosphate backbone. (b) 20 nt ssDNA (poly-AC) on 5’-3’ and 3’-5’ strands. (c) 10bp bulge (poly-AC) on 5’-3’ and 3’-5’ strands. (d) ds-linker. The traversal probabilities are plotted using the Laplace best estimator26, with 95% confidence intervals from the adjusted Wald method27.The error bars of the pause durations are the SEM. (The results for other sizes of bulges and gaps are listed in Supplementary Table 2.)
Many of the modifications we have probed (Figure 1 and Figure 3) change multiple features of the DNA simultaneously; thus, in order to extract the relative importance of the different chemical moieties while controlling for other changes, we performed a set of multivariate logistic regressions21 on different sub-sets of our data. This analysis provides a quantitative ranking of the importance of the different contacts and their force dependence (Figure 4 and Supplementary Figure 2). These regressions confirm the importance of the phosphates every 10 bp on the 5’-3’ strand and reveal that these contacts are only important under the application of force. Regression analysis also reveals less important contacts with phosphates on both strands on a smaller distance scale, which, remarkably, remain important when extrapolated to zero applied load. This residual importance may arise from the force the motor generates on the DNA, as opposed to the external load applied optically, supporting our conclusion that these contacts occur during the burst phase.
Figure 4. The Motor-DNA Contacts.
The relative importance of the motor-DNA contacts are marked using pymol with a quantitative color scale with magnitudes inferred from the traversal probabilities at 5 pN for the measured modifications (Figure 1–Figure 3; Supplementary Table 2, Supplementary Discussion; Supplementary Figure 2). The units of “contact importance” correspond to the inverse of the distance over which the removal of the specific moiety would reduce the traversal probability to 50% (Supplementary Discussion.) The mechanochemical phase of the motor as it moves along the DNA is indicated to the right.
The regression analysis also provides information on the nature of the non-charge contacts, revealing that important, but minor, contacts are made with bases or sugars (Supplementary Figure 2a). However, the analysis also predicts that removing all of the characteristic features of nucleic acid—phosphates, bases, sugars and native double-helix structure—will result in a reduced but finite traversal probability (Supplementary Discussion). Thus, a component of these additional, non-charge interactions is not specific to DNA. We tested this surprising result by challenging the motor with a polymer lacking any resemblance to nucleic acid. Remarkably, the motor packages this insert as well (Figure 3d), revealing a non-specific component to the motor-DNA interaction. Such interactions may correspond to chemical contacts, i.e. hydrogen bonds or hydrophobic interactions, which do not require a specific nucleic acid moiety. Alternatively, it is possible that these interactions are steric in nature. A prediction of such a steric drive mechanism is that amino acids essential for DNA translocation can be replaced by any residue with a bulky side chain, a behaviour suggested by mutational analysis of the DNA binding loop of the related hexameric helicase SV4022.
Taken together, our data indicate that the motor-DNA interaction involves a wide variety of contacts with the full complement of nucleic acid moieties, as well as contacts not specific to DNA. In addition, our data suggest that the type of motor-DNA interaction changes during the course of the mechanochemical cycle. During the dwell phase, when ATP is loaded, the motor maintains strong, load-bearing contact with the DNA via interactions with adjacent charges on the 5’-3’ strand every 10 bp. These contacts play a sensory role, coupling the mechanical and chemical cycles. In the burst phase, when the DNA is translocated, the motor makes contact with a variety of chemical moieties, including charges on both strands, bases or sugars, and additional non-DNA-specific contacts. Given the relatively long duration of the contacts during the dwell phase, specific, strong, ionic interactions may be preferable. In contrast, contacts during active translocation in the burst phase must be made and broken more quickly; thus, it would be advantageous to make weaker, more promiscuous contacts. Figure 4 summarizes the important motor-DNA interactions and the specific point in the mechanochemical cycle at which these contacts are made.
Our findings have broad implications for the mechanism of the packaging motor. In particular, non-specific contacts can serve an important role in generating a step size that is a non-integer repeat of the chemical periodicity of DNA6. Moreover, the observation that important contacts are made below distances of ∼10 bp provides evidence against an open ring model of translocation in which only two subunits contact the DNA, thereby limiting the mechanism by which a non-integer step size can be generated6. In parallel, our findings have broader implications for the family of ASCE ring-ATPases. The use of alternate contacts during different portions of the mechanochemical cycle as well as the use of sensory contacts could serve as a general mechanism by which multimeric motors synchronize and coordinate the hydrolysis cycles of their individual subunits with the position of their substrate. The use of non-specific, perhaps steric, contacts by nucleic acid and polypeptide translocases23 may reflect the existence of a conserved translocation mechanism shared by members of the ASCE superfamily; such non-specific contacts may have facilitated the evolution of peptide translocases from nucleic acid translocases24.
Methods Summary
Substrates were prepared by ligating custom oligonucleotide inserts with unmodified DNA. Single molecule packaging assays were performed as described6,7,25. Insert-induced pauses were identified according to their location in the packaging trace.
Methods
DNA substrates
For each construct, two (or in the case of the gaps, three) modified or unmodified DNA oligonucleotides (listed in Supplementary Table 1) were purchased from Fidelity Systems or IDT. The oligos hybridize to form a DNA segment with three nucleotide overhangs. 4187 and 4008 bp DNA fragments with distinct non-palindromic 3-nucleotide overhangs were prepared by PCR, restriction digest, and agarose gel purification. The primer for the 4008 bp PCR fragment contains a terminal biotin. These fragments were ligated to the insert oligos and the 8.2 Kbp product was gel purified.
Single-molecule packaging
Single-molecule packaging assays were performed as previously described4,7,25,28 in packaging buffer containing 50 mM Tris-HCl pH 7.8, 50 mM NaCl, 5 mM MgCl2, and 1mM ATP unless mentioned otherwise. The biotinylated DNA is bound to a 2.1 µm streptavidin-coated polystyrene bead held by a micropipette and the prohead-gp16-ATPase complex is attached to a second bead held in an optical trap. Packaging is initiated in situ by bringing the beads together25,28. The instrument29 is run in force-feedback mode, where the separation between the beads is adjusted to maintain constant tension in the DNA molecule. High resolution packaging measurements were conducted on a dual trap instrument described previously30 with materials prepared in the same fashion as above, except 860 nm beads were used.
Analysis
The modified DNA insert is expected to reach the motor after packaging of 4.2 Kbp. However, variability in attachment geometry of the DNA to the beads and other systematic errors introduce uncertainty in the measurement of tether length, so the insert was considered to be between the 2.5 and 4.5 Kbp position. The longest pause in this interval was scored as the insert-induced pause. In traces in which a pause could not be identified a pause of 0.1 seconds, the temporal resolution, was assigned. The rate of pausing (0.017 kbp−1) and slipping (0.02 kbp−1) during normal packaging7 are small enough that these events do not significantly bias our measurements. All probabilities were calculated with the Laplace estimator26 which is given by pLaplace=(x+1)/(n+2) where x is the number of successes and n is the number of trials. This is a better estimator than the maximum-likelihood x/n, especially when n is small or the probabilities being estimated are near zero or one26. nmin values are a measure of the degree to which a distribution is exponential and provide a strict lower limit on the number of rate-limiting kinetic events6. These values were calculated by taking the ratio of the mean dwell time squared over the variance in the dwell times. nmin error bars are standard deviations and were calculated via a bootstrap method.
Supplementary Material
Acknowledgments
We thank C. L. Hetherington, M. Kopaczynska, A. Spakowitz, and J.M. Berger for critical discussions; D. Reid, M. T. Couvillon, and N. L. S. Chavez for preliminary work leading to this publication. KA acknowledges the PMMB fellowship through the Burroughs Wellcome Fund, ATP the NIH Molecular Biophysics Training Grant, AK the Human Frontier Science Program Cross-Disciplinary Fellowship, JRM the NSF Graduate Research Fellowship, and YRC the Burroughs Wellcome Fund Career Award at the Scientific Interface for funding. This research was supported in part by NIH grants GM-071552, DE-003606, and GM-059604. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
K.A., A.T.P, A.K, and J.R.M. conducted the experiments and K.A., A.T.P, A.K, J.R.M., and Y.R.C. performed the analysis, S. G., P. J. J., and D. A. prepared and provided experimental materials, and K.A., A.T.P, A.K, J.R.M., Y.R.C., S.G., P. J. J.,and C.B. wrote the paper. K.A., A.T.P., A.K and J.R.M contributed equally to this work.
Footnotes
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
References
- 1.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burroughs AM, Iyer LM, Aravind L. Basel: Karger; 2007. Comparative Genomics and Evolutionary Trajectories of Viral ATP Dependent DNA-Packaging Systems; pp. 48–65. [DOI] [PubMed] [Google Scholar]
- 3.Hopfner KP, Michaelis J. Mechanisms of nucleic acid translocases: lessons from structural biology and single-molecule biophysics. Curr Opin Struct Biol. 2007;17:87–95. doi: 10.1016/j.sbi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Smith DE, et al. The bacteriophage phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 5.Morais MC, et al. Defining molecular and domain boundaries in the bacteriophage phi29 DNA packaging motor. Structure. 2008;16:1267–1274. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffitt JR, et al. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chemla YR, et al. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 8.Thiviyanathan V, et al. Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry. Biochemistry. 2002;41:827–838. doi: 10.1021/bi011551k. [DOI] [PubMed] [Google Scholar]
- 9.Strauss JK, Maher LJ., 3rd DNA bending by asymmetric phosphate neutralization. Science. 1994;266:1829–1834. doi: 10.1126/science.7997878. [DOI] [PubMed] [Google Scholar]
- 10.Eoff RL, Spurling TL, Raney KD. Chemically modified DNA substrates implicate the importance of electrostatic interactions for DNA unwinding by Dda helicase. Biochemistry. 2005;44:666–674. doi: 10.1021/bi0484926. [DOI] [PubMed] [Google Scholar]
- 11.Kawaoka J, Jankowsky E, Pyle AM. Backbone tracking by the SF2 helicase NPH-II. Nat Struct Mol Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- 12.SenGupta DJ, Borowiec JA. Strand-specific recognition of a synthetic DNA replication fork by the SV40 large tumor antigen. Science. 1992;256:1656–1661. doi: 10.1126/science.256.5064.1656. [DOI] [PubMed] [Google Scholar]
- 13.Mancini EJ, et al. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Dillingham MS, Soultanas P, Wigley DB. Site-directed mutagenesis of motif III in PcrA helicase reveals a role in coupling ATP hydrolysis to strand separation. Nucleic Acids Res. 1999;27:3310–3317. doi: 10.1093/nar/27.16.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard J. xvi. Sunderland, Mass: Sinauer Associates, Pub; 2001. Mechanics of motor proteins and the cytoskeleton; p. 367. [Google Scholar]
- 16.Stanley LK, et al. When a helicase is not a helicase: dsDNA tracking by the motor protein EcoR124I. Embo J. 2006;25:2230–2239. doi: 10.1038/sj.emboj.7601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 18.Pearson RK, Fox MS. Effects of DNA heterologies on bacteriophage lambda packaging. Genetics. 1988;118:5–12. doi: 10.1093/genetics/118.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moll WD, Guo P. Translocation of nicked but not gapped DNA by the packaging motor of bacteriophage phi29. J Mol Biol. 2005;351:100–107. doi: 10.1016/j.jmb.2005.05.038. [DOI] [PubMed] [Google Scholar]
- 20.Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agresti A. Categorical data analysis. Wiley-Interscience; 2003. [Google Scholar]
- 22.Shen J, Gai D, Patrick A, Greenleaf WB, Chen XS. The roles of the residues on the channel beta-hairpin and loop structures of simian virus 40 hexameric helicase. Proc Natl Acad Sci U S A. 2005;102:11248–11253. doi: 10.1073/pnas.0409646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barkow SR, Levchenko I, Baker TA, Sauer RT. Polypeptide Translocation by the AAA+ ClpXP Protease Machine. Chemistry & Biology. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulkidjanian AY, Makarova KS, Galperin MY, Koonin EV. Inventing the dynamo machine: the evolution of the F-type and V-type ATPases. Nature Reviews Microbiology. 2007;5:892–899. doi: 10.1038/nrmicro1767. [DOI] [PubMed] [Google Scholar]
- 25.Fuller DN, et al. Ionic effects on viral DNA packaging and portal motor function in bacteriophage phi 29. Proc Natl Acad Sci U S A. 2007;104:11245–11250. doi: 10.1073/pnas.0701323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis J, Sauro J. When 100% Really Isn’t 100%: Improving the Accuracy of Small-Sample Estimates of Completion Rates. Journal of Usability studies. 2006;1:136–150. [Google Scholar]
- 27.Agresti A, Coull B. Approximate Is Better Than” Exact” for Interval Estimation of Binomial Proportions. The American Statistician. 1998;52 [Google Scholar]
- 28.Rickgauer JP, et al. Portal Motor Velocity and Internal Force Resisting Viral DNA Packaging in Bacteriophage φ29. Biophys J. 2008;94:159–167. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith SB, Cui Y, Bustamante C. Optical-Trap Force Transducer that Operates by Direct Measurement of Light Momentum. Methods in Enzymology. 2003;361:134–162. doi: 10.1016/s0076-6879(03)61009-8. [DOI] [PubMed] [Google Scholar]
- 30.Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci U S A. 2006;103:9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.