Abstract
NF-κB is a key activator of inflammatory and immune responses with important pathological roles in cancer, heart disease, and autoimmune diseases. Transcriptional activity of NF-κB is regulated by different posttranslational modifications. Here, we report a novel mechanism of NF-κB regulation through lysine monomethylation by SET9 methyltransferase. Set9 specifically methylates p65 at lysine 37. Both TNFα and IL-1β treatments induced methylation of p65. Methylated p65 is restricted to the nucleus and this modification regulates the promoter binding of p65. Moreover, Set9 mediated methylation of p65 is required for the expression of a subset of NF-κB target genes in response to TNFα stimulation.
Keywords: methylation, SET9
NF-κB is a transcription factor that plays a pivotal role in regulating multiple biological functions including inflammation, immunity, cell proliferation, and apoptosis. NF-κB represents a group of evolutionarily conserved and structurally related proteins. The 5 members of the mammalian NF-κB, p65 (RelA), RelB, cRel, p50/p105 (NF-κB1), and p52/p100 (NF-κB2), form homo- or heterodimers that bind to IκB family proteins in unstimulated cells (1). NF-κB is sequestered in the cytoplasm through its interaction with the IκB in resting cells. Stimulation of cells with a variety of ligands, such as tumor necrosis factor-α (TNFα), interleukin-1β (IL-1β), or pathogen-associated molecular patterns (PAMPs), leads to the rapid phosphorylation of IκB by the IκB kinase complex (IKK). The IKK kinase complex contains 2 catalytic subunits, IKKα and IKKβ, and the regulatory subunit, IKKγ. IKK catalyzes the phosphorylation of IκB at 2 serine residues in the N terminus. The phosphorylated IκB becomes ubiquitinated and subsequently degraded by 26S proteasome, thereby allowing NF-κB to enter the nucleus to turn on a large array of target genes (1, 2).
Although the activity of NF-κB is regulated by nuclear translocation, covalent modifications of the protein by various events including phosphorylation, ubiquitination, nitrosylation, and acetylation can affect its activity (3). These regulatory modifications have distinct functional consequences. For example, acetylation of p65 at K218 and K221 inhibits IκBα binding and enhances DNA-binding (4) whereas acetylation of p65 at K122 and K123 inhibits its transcriptional activating activity (5).
Proteins can be posttranslationally methylated at lysine, arginine, histidine, and dicarboxylic amino acids by highly specific methyltransferases (6). In the process of protein-lysine methylation, the addition of methyl groups to the ε-amine of a lysine residue results in the formation of mono-, di-, or trimethyllysine. This process can be reversed by demethylases. Histone is one of the best studied proteins that undergoes methylation (7). Specific sites of methylation on histones correlate with either activation or repression of transcription. Recently, several transcription factors, including p53 (8, 9), STAT1 (10), RARα (11), and ERα (12), have been shown to be methylated and their biological activity modified by the modification.
Set9 (also known as Set7) was initially purified as a H3K4 histone methyltransferase from HeLa nuclear extracts (13, 14). Biochemical and structural studies suggest that Set9 catalyzes monomethylation of H3K4 (15, 16). In addition, Set9 has been shown to methylate several nonhistone proteins such as p53 (8), TAF10 (17), and ER (12). Methylation of p53 and ER results in the stabilization of these proteins and the transcriptional activation. On the other hand, methylation of TAF10 increases the binding to RNA polymerase II. These results collectively suggest that Set9-mediated methylation of proteins other than histone may be more general.
In the present report, we demonstrate that NF-κB is also regulated by methylation. We show that p65 is monomethylated by Set9 at lysine 37. Both TNFα and IL-1β treatments induced methylation of p65. Methylated p65 is restricted to the nucleus. Moreover, Set9 catalyzed methylation of p65 is required for the induction of a subset NF-κB regulated genes. Methylation of p65 affects the stability of DNA-p65 complexes, which in turn regulates the recruitment of p65 to the promoter.
Results
NF-κB Is Regulated by Methylation.
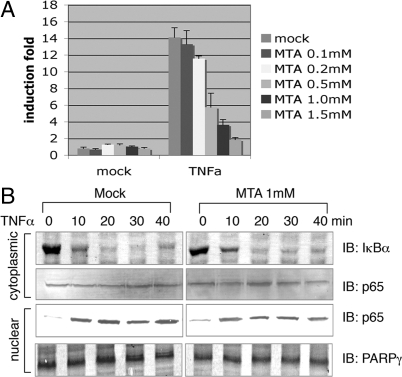
To test whether NF-κB is regulated by methylation, we blocked methylation reactions in cells with a broad methylation inhibitor, 5′-deoxy-5′-methylthioadenosine (MTA) (18) and observed that it inhibited the TNFα-induced response of an NF-κB-driven luciferase reporter in a dose-dependent manner (Fig. 1A). To further investigate the role of methylation in the NF-κB pathway, we examined IκBα degradation and p65 nuclear translocation, 2 key steps in NF-κB activation. HEK293 cells were treated with or without MTA (1 mM) for 1 h; cytoplasmic and nuclear extracts were prepared after treatment with TNFα for various time points. We found that MTA-treatment blocked neither the degradation of IκBα nor the nuclear translocation of p65, indicating MTA inhibits a step in NF-κB activation that is independent of the nuclear translocation process (Fig. 1B).
Fig. 1.
(A) MTA inhibits TNFα-induced NF-κB activation. HEK293 cells were transfected with a NF-κB-driven luciferase reporter. Twenty-four hours later, the cells were pretreated with or without various concentrations of MTA for 1 h. The cells were then treated with or without TNFα for 12 h. The luciferase activity was measured and normalized for transfection efficiency as determined by cotransfection of a constitutively expressed β-galactosidase reporter. (B) MTA does not affect IκBα degradation or p65 nuclear translocation in response to TNFα stimulation. HEK293 cells were treated with or without MTA (1 mM) for 1 h, cytoplasmic and nuclear extracts were prepared after treatment with TNFα for various time points as indicated, and were immunoblotted with indicated antibodies.
Set9 Methylates p65 in Vitro.
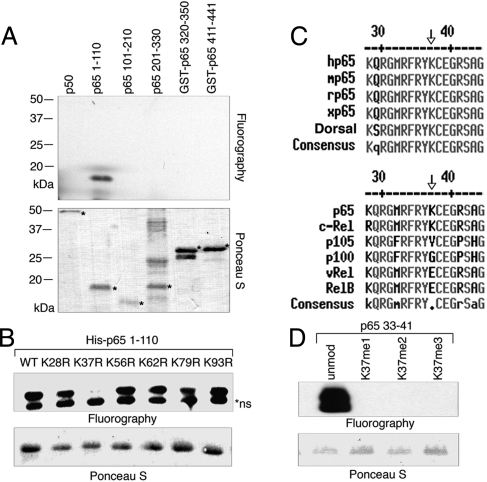
We then tested whether p65 can be methylated in vitro. It has been shown that Set9 methylates histone H3 (13, 14) and several nonhistone proteins such as p53 (8), TAF10 (17), and ER (12). We therefore purified recombinant Set9 from bacteria and tested whether it could methylate p65 in vitro. Using recombinant p65 fragments that cover all of the lysine residues of p65 as substrates, we performed in vitro methylation and found that the N terminus (1–110) of p65 was methylated by Set9 in vitro (Fig. 2A). Set9-mediated methylation was specific to p65, as p50 was not methylated under the same experimental condition (Fig. 2A). There are 6 lysine residues within this region, K28, K37, K56, K62, K79, and K93. To map the methylation site, we substituted each of these lysines with nonmethylatable arginines. A single point mutation at lysine 37 (K37) completely abolished the methylation by Set9 (Fig. 2B), suggesting that Set9 methylates p65 at K37. Lysine 37 is highly conserved from Drosophila to humans and is present only in p65 and c-Rel among NF-κB family proteins (Fig. 2C). Using p65 peptides (residues 33–41) with unmodified, mono-, di- or trimethylated K37, we further showed that Set9 only methylated unmodified p65 peptide suggesting that Set9 monomethylates p65 (Fig. 2D). This is consistent with studies showing that Set9 is exclusively a monomethyltransferase (15, 16).
Fig. 2.
Set9 methylates p65 at K37 in vitro. (A) Autoradiogram (Top) and Ponceau S staining (Bottom) of in vitro methylation assay with recombinant Set9 and recombinant p65 fragments. (B) In vitro methylation assay with p65 (1–110) bearing K28R, K37R, K56R, K62R, K79R, and K93R substitutions. (C) Alignment of protein sequences adjacent to K37 among different species (Top) and NF-κB family members (Bottom). h, human; m, mouse; r, rat; x, Xenopus. (D) Autoradiograph (Top) of methylation assay with p65 peptides (33–41) containing unmodified (K37), monomethylated (K37Me1), dimethylated (K37Me2), and trimethylated (K37Me3) K37. Ponceau S staining of the methylation gel is shown above (Bottom).
Set9 Methylates p65 in Vivo.
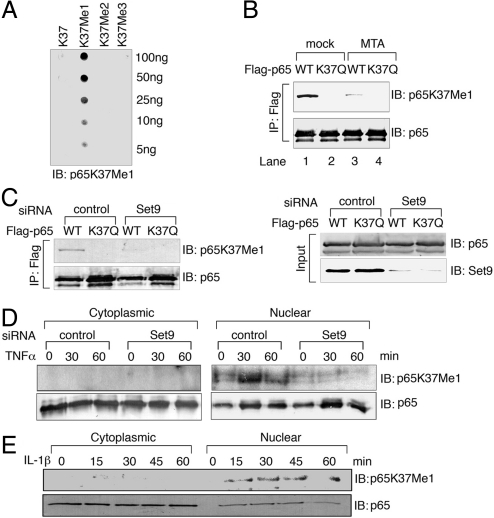
To facilitate studies on the role of Set9 methylation of p65 in vivo, we generated a polyclonal antibody that specifically recognized monomethylated p65 at K37 (p65K37me1). Rabbit antibodies were purified against unmodified-p65 peptide (negative selection) followed by monomethylated-p65 peptide (positive selection). The purified antibodies specifically recognized monomethylated p65-peptide, but failed to detect unmodified, di-, and trimethylated-p65 peptides (Fig. 3A).
Fig. 3.
Set9 methylates p65 in vivo. (A) Dotblot analysis with p65K37me1 antibody on p65 peptides. (B) Detection of p65K37me1 by Western blot. Expression vectors encoding Flag-p65 or Flag-p65 K37Q were transfected into HEK293 cells. Twenty-four hours later, cells were treated with or without MTA (1 mM) for 4 h. Eluates from Flag immunoprecipitation were subjected to Western blot analysis with p65 (Bottom) and p65K37me1 (Top) antibodies. (C) HEK293 cells were transfected with control or Set9-specific siRNAs. A day later the cells were retransfected with siRNAs along with Flag-p65 or Flag-p65K37Q. The cells were harvested the next day and eluates from Flag immunoprecipitation were analyzed with p65 and p65K37me1 antibodies. (D) HEK293 cells transfected with control or Set9-specific siRNAs were stimulated with TNFα and fractionated. P65 immunoprecipitates from cytoplasmic or nuclear extracts were immunoblotted with p65K37me1 (Top) or p65 (Bottom) antibodies. (E) HeLa cells were treated with or without IL-1β for the indicated time periods, and p65 immunoprecipitates from cytoplasmic or nuclear extracts were immunoblotted with p65K37me1 (Top) or p65 (Bottom) antibodies.
To test whether p65K37me1 antibody recognized methylated-p65 in vivo, we overexpressed Flag-tagged wild-type p65 or Flag-p65K37Q in HEK293 cells. Flag immunoprecipitates were subjected to Western blot analysis with p65K37me1 and anti-Flag antibodies. The p65K37me1 antibody was able to detect wild-type p65 but not the K37Q mutant (Fig. 3B, lanes 1 and 2). Moreover, detection of p65 by the antibody to p65K37me1 was greatly reduced when the cells were treated with MTA for 4 h (lanes 3 and 4). These results showed that p65K37me1 antibody detects the monomethylation of p65 at K37. More importantly these results indicate that p65 is methylated in vivo. To demonstrate that the methylation of p65 was mediated by Set9, we used RNA interference to silence the expression of Set9 in HEK293 cells and then transfected the cells with wild-type p65 or K37Q mutant expression vectors. A small interfering RNA (siRNA) targeted to Set9 decreased the expression of Set9 protein but not the expression of p65 (Fig. 3C, Lower panel). Overexpression of wild-type p65 led to methylation of p65 in cells transfected with control siRNA (Fig. 3C, Upper panel). In contrast, methylation of p65 was significantly reduced when Set9 was knocked down by siRNA, suggesting that Set9 is the major methyltransferase that monomethylates p65 at K37 in vivo.
We then examined whether Set9 could methylate endogenous p65 in response to TNFα stimulation. HEK293 cells were treated with control or Set9-specific siRNAs. Cytoplasmic and nuclear extracts were prepared from TNFα-treated and untreated cells followed by p65 immunoprecipitation. TNFα stimulation did induce methylation of p65 at K37 but the methylated p65 was only detected in the nucleus (Fig. 3D). Moreover, knockdown of expression of Set9 completely abolished the TNFα-induced methylation of p65. These in vivo data support our in vitro findings that Set9 monomethylates p65 at K37.
To test whether another NF-κB agonist induces methylation of p65 at K37, we treated HeLa cells with IL-1β for various time periods. We fractionated cell lysates into cytoplasmic and nuclear factions followed by p65 immunoprecipitation. Similar to TNFα stimulation, IL-1β treatment led to methylation of p65 at K37 (Fig. 3E). In addition, methylated p65 was again only present in the nuclear fraction. These findings collectively suggested that methylation of p65 in response to different forms of NF-κB stimulation is a general phenomenon, rather than a cell-type or agonist-specific event.
Set9 Is Required for the Expression of a Subset NF-κB-Regulated Genes in Response to TNFα Stimulation.
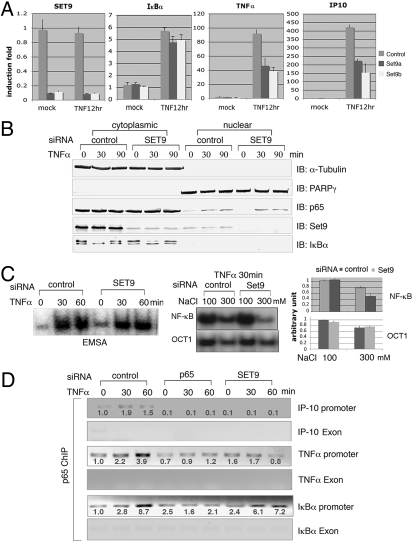
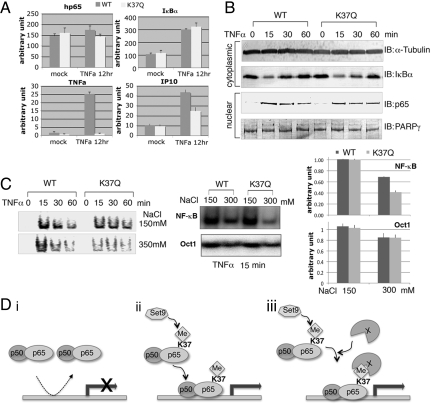
To characterize the physiological function of Set9-mediated p65-K37 methylation, we evaluated the effects of Set9 loss of function on TNFα-induced NF-κB-regulated gene expression. We silenced the expression of Set9 by RNAi, using 2 targeted siRNAs. Each of the siRNAs achieved 90% knockdown of Set9 mRNA as measured by RT-PCR (Fig. 4A, Left panel). TNFα-induced expression of both IP-10 and TNFα was reduced by 50−60% in cells transfected with Set9-specific siRNAs. In contrast, the induction of IκBα was not affected by knocking down Set9 expresssion. Similar results were obtained using HeLa cells (supporting information (SI) Fig. S1, Top Left panel). These results imply that Set9 is important for the expression of a subset NF-κB-dependent gene in response to TNFα stimulation. This finding is consistent with a recent microarray study showing that SET9 is required for the induction of 25% of genes activated by NF-κB in TNFα-treated human monocytes (19). To further investigate the role of Set9 in the NF-κB pathway, we examined IκBα degradation and p65 nuclear translocation, 2 key steps in NF-κB activation. We prepared cytoplasmic and nuclear extracts from Set9 knockdown and control HEK293 cells after treatment of TNFα for various time points. As expected, TNF induced degradation of IκBα and nuclear translocation of p65 in control cells. Neither of these steps was affected by silencing the expression of Set9 (Fig. 4B). These results indicate that Set9 might regulate the nuclear function of p65.
Fig. 4.
Set9 is required for TNFα-induced expression of a subset of NF-κB-depedent genes. (A) HEK293 cells transfected with control or Set9-specific siRNAs were stimulated with or without TNFα for 12 h. Expression of Set9, IκBα, TNFα, and IP-10 were measured by RT-PCR. (B) SET9 does not affect IκBα degradation or p65 nuclear translocation in response to TNFα stimulation. Cytoplasmic and nuclear extracts prepared from Set9 knockdown and control HEK293 cells after treatment of TNFα for various time points as indicated were immunoblotted with indicated antibodies. (C) Five micrograms of nuclear extracts from B were subjected to EMSA. Various salt concentrations were used to test the stability of p65-DNA complexes (Middle panel; 100 mM NaCl in the Left panel). Intensity of the bands was quantified using ImageQuant (GE); means of 3 independent experiments are shown in the Right panel (D) control, p65 and Set9 siRNAs transfected HeLa cells were treated with or without TNFα for the indicated time periods, and ChIP assays were performed with anti-p65 antibody. ChIP-enriched samples were analyzed by regular PCR using IP-10, TNFα, and IκBα promoter and exon primers. Intensity of the bands was quantified using ImageQuant and labeled as fold induction. Figures represent data from 2 independent experiments.
Set9 Regulates DNA Binding and Promoter Recruitment of p65.
To examine whether Set9 regulates the DNA binding of p65, we used the electrophoretic mobility shift assay (ESMA) with nuclear extracts prepared from cells transfected with control or Set9-specific siRNAs. As indicated in Fig. 4C (Left panel), TNFα induced strong p65 DNA binding in both control and Set9-deficient extracts. With 2 probes, 1 containing the NF-κB binding site found in Igκ (Fig. 4C), the other in the IL-8 promoter (Fig. S2), we observed no differences between control and Set9-depleted samples. However, when we increased the salt concentration in the EMSA reactions to 300 mM, there was a slightly reduced level of DNA-p65 complex formation in the control and a 35% further drop in the Set9-deficient extracts. Using an Oct1 probe, the increased salt had the same effect with or without Set9. (Fig. 4C Middle and Right panels). This result suggests that Set9 regulates the stability of DNA-p65 complexes, which might account for the reduction of expression of several genes in TNF-treated Set9-deficient cells. To explore this possibility, we analyzed the recruitment of p65 to promoters of IκBα, IP-10, and TNFα by chromatin immunoprecipitation (ChIP). As shown in Fig. 4D, p65 was recruited to promoters of IκBα, IP-10, and TNFα in response to TNFα stimulation. The recruitment of p65 to these promoters was abolished in the p65 siRNA transfected cells. On the other hand, in the absence of Set9, p65 was recruited to the promoter of IκBα but not to the promoters of IP-10 and TNFα. The amount of p65 recruited to the IκBα promoter was slightly less (15−20%) in Set9-deficient cells. However the slight reduction of p65 bound to IκBα did not affect the induction of IκBα by TNFα stimulation (Fig. 4A). These data suggest that methylation by Set9 regulates the affinity of p65 for DNA, which is a critical parameter for induction of a subset of NF-κB-dependent genes in response to TNFα stimulation.
K37 Methylation of p65 Is Required for the Induction of a Subset NF-κB Regulated Genes.
To determine whether Set9 regulates the function of p65 specifically through K37 methylation, p65−/− MEF cells were infected with retroviruses encoding either wild-type p65 or p65K37Q. Stable reconstituted cell lines were established by puromycin selection. The expression level of wild type p65 and p65K37Q were comparable between 2 cell lines as determine by RT-PCR and Western blot (Fig. 5 A and B). Next we treated the cells with TNFα and measured expression of several NF-κB-dependent genes by RT-PCR. Consistent with the effects of Set9 loss of function, induction of IP-10 and TNFα was greatly reduced in p65K37Q-reconstituted cells, whereas the induction of IκBα was not affected (Fig. 5A). Furthermore, we prepared cytoplasmic and nuclear extracts from wild-type p65- and p65K37Q-reconstituted cells treated with or without TNFα and found that the IκBα degradation and the nuclear translocation of p65 were not affected in p65K37Q-reconstituted cells (Fig. 5B). We next analyzed whether methylation of p65 affects its DNA binding by EMSA. We performed EMSA using the nuclear extracts prepared as described in Fig. 4B. As expected, the nonmethylatable p65K37Q mutant formed less stable DNA-p65 complexes than the wild-type p65 (39% less, Fig. 5C) whereas no difference was observed when using Oct1 probe.
Fig. 5.
Methylation of p65 is critical for p65 function. (A) P65−/− MEF cells reconstituted with p65 or p65K37Q were treated with TNFα for 12 h. Expression of p65, IκBα, TNFα, and IP-10 was determined with RT-PCR. (B) Cytoplasmic and nuclear fractions from p65−/− MEF cells reconstituted with p65 or p65K37Q stimulated with TNFα for various time periods were subjected to immunoblotting with the indicated antibodies. (C) Five micrograms of nuclear extracts from B were subjected to EMSA. Intensity of the bands was quantified using ImageQuant; means of 3 independent experiments were shown in the Right panel. (D) Models of the mechanism of control by Set9-mediated methylation of p65: (i) P65 fails to form stable DNA-p65 complex on a subset of promoters (eg., TNFα, and IP-10) without methylation at K37. (ii) Methylation of p65 at K37 stabilizes the formation of DNA-p65 complex at the promoter. (iii) Methylation of p65 at K37 creates a docking site for recruiting coactivator X that leads to formation of a functional enhanceosome.
Although Set9 was first identified as a histone methyltransferase that monomethylates H3K4 (13, 14), it has been shown that Set9 is unable to methylate H3K4 assembled into nucleosomes (8). Furthermore, we noticed that neither silencing the expression of Set9 by RNAi nor TNFα stimulation affects the global level of H3K4me1 modification (Fig. S3A). More importantly the presence of H3K4me1 modification did not correlate with induction of TNFα and IP-10 in response to TNFα stimulation (Fig. S3B). As shown in Fig. S4B, H3K4me1 modification was detected at the promoters of TNFα and IP-10 in resting cells as determined by ChIP even though the mRNAs of these genes were barely detectable. Although TNFα stimulation induced the expression of IP-10 and TNFα, it failed to further increase H3K4m1 modification at their promoters. In contrast, p65 bound to these promoters only after TNFα stimulation. These findings support the idea that Set9-mediated K37 methylation of p65 but not H3K4 is required for maximal transcriptional activation of p65.
Discussion
In this study, we report a signal-dependent methylation of p65. We showed that Set9 mediates methylation of p65 in response to TNFα and IL-1β stimulation. Furthermore, Set9-catalyzed methylation of p65 is required for the induction of a subset of NF-κB-regulated genes. Methylation of p65 affects the stability of DNA-p65 complexes, which in turn regulates the recruitment of p65 to the promoter. What is the mechanism for methylation-induced stabilization of DNA-p65 complexes? One of the possibilities is that K37 is within the loop structure of p65 that is critical for its DNA binding. Several amino acids adjacent to K37 have been shown to interact directly with DNA (20). Although K37 does not interact directly with DNA, methylation of K37 may change the conformation of the loop and increase the stability of the DNA-p65 complexes (Fig. 5D).
Another possibility is that the methylation of K37 creates a docking site to recruit a coactivator (Fig. 5D). Several protein domains, such as Tudor, Chromo, MBT, and PHD domain, have been identified as methylated-lysine binding domains (21). Most proteins containing these domains are involved in epigenetic modification and regulation of chromosomal structure. In addition, different coactivators have been shown to regulate induction of different subsets of NF-κB regulated genes. For example, we have previously shown that IRF3 and Bcl3 function as coactivator for induction of IP-10 but not MCP-1 in response to LPS and TNFα stimulation, respectively (22), while others have reported that RPS-3 is critical for induction of a subset of NF-κB regulated genes upon TCR stimulation (23). Methylation of p65 may serve as direct or indirect interacting interface for recruiting these coactivators to form an enhanceosome to activate gene transcription. Either model could lead to less stable DNA-p65 interactions because a coactivator could stabilize the complex. Further experimentation will be needed to differentiate among these possibilities.
Two recent reports have also shown that NF-κB is regulated by Set9 (19, 24). In the first article, the authors show that Set9 regulates the expression of a subset of NF-κB regulated genes in response to TNFα stimulation (19), which is consistent with our findings. However, the authors failed to detect any methylation of p65 using a commercially available methylated-lysine-specific antibody and they concluded that the histone methyltransferase activity of Set9 modulates NF-κB downstream genes. We acquired several commercially available methylated-lysine-specific antibodies and showed that none of them were able to detect p65 peptides with monomethylated K37 (Fig. S4). On the other hand, our highly specific p65K37me1 antibody enables us to directly detect methylation of p65 in vivo. Although methylation of H3K4 has been linked to transcriptional activation in a variety of eukaryotic species, it is the di- and trimethylation of H3K4 that are enriched at the promoter of active genes (21). Furthermore, it has been shown that Set9 catalyzed exclusively monomethylation of H3K4. More importantly, we show that the presence of monomethylation of H3K4 at the promoters of TNFα and IP-10 does not correlate with the expression of TNFα and IP-10 (Fig. S3B).
In the second study, the authors show that Set9 methylates p65 at K315 and K316 and negatively regulates the transcriptional activity of p65 (24). The differences in the in vitro data might result from different epitope tags used in generating the recombinant p65 fragments. For example, the N-terminal GST tag used in the study might block the access of Set9 to K37 while GST-induced dimerization might facilitate the methylation of K315 and K316. The in vivo methylation results are more controversial as the antibody used in the study recognized dimethylated lysine according to the company Website (http://www.abcam.com/pan-methyl-Lysine-methyl-K-pan-antibody-ChIP-Grade-ab7315.html) whereas both K315 and K316 are monomethylated by Set9 (24). Furthermore, we tested the same antibody against p65 peptides (residues 33–41) with unmodified, mono-, di- or trimethylated K37 and found that the antibody recognized exclusively the dimethylated K37 p65 peptide (Fig. S4). Finally, different cellular contexts may determine the role of Set9 in regulating p65. In THP-1 (19), HEK293, and HeLa cells, Set9 functions as a positive regulator whereas in U2OS and A549 cells it functions as a negative regulator. One possible explanation is that in THP-1, HEK293, and HeLa cells K37 methylation of p65 is exclusively catalyzed by Set9, whereas in the other 2 types of cells it can also be catalyzed by other methyltransferases. In fact, we found that knocking down the expression of Set9 in U2OS cells has little or no effect on TNF-induced methylation of p65 at K37 (Fig. S5). Moreover, the methylation of K37 has faster kinetics (peaks at 30 min, Fig. 3 D and E) than the methylation of K315 and K316 (peaks at 60 min) (24). Thus it is likely that K37 modification occurs before K315 and K316 modification. Although K37 methylation is required for the gene activation, K315 and K316 methylation is required for the termination of NF-κB activity. Further experimentation will be needed to fully understand these relationships.
Materials and Methods
Antibodies.
Antibodies against p65 (F6, C20), IκBα (C21), PARPγ (5A5), and α-tubulin (TU-02) were purchased from Santa Cruz Biotechnology. Antibodies against FLAG (M2, Sigma), HA (Covance), H3K4me1 (Abcam), and Set9 (Upstate Laboratories) were purchased from the respective commercial sources.
Peptides.
The following peptides were chemically synthesized for antibody production and dot blot analysis: P65 unmodified, NH2-RFRYKCEGR-COOH; p65 monomethyl-K37, NH2-RFRYK-(Me)CEGR-COOH; p65 dimethyl-K37, NH2-RFRYK-(Me2)CEGR-COOH; and p65 trimethyl-K37, NH2-RFRYK-(Me3)CEGR-COOH.
Methylation Assay.
The samples were incubated at 30 °C for 60 min in a reaction buffer containing 50 mM Tris-HCl (pH 8.5), 5 mM MgCl2, 4 mM DTT, and 1 μM 3H-labeled SAM (Amersham Pharmacia Biotech). One microgram of His6-p65 or GST–p65 fusion proteins were used as substrates. The total volume of a reaction mixture was adjusted to 10 μL. The reaction was stopped by the addition of SDS sample buffer and then fractionated on a 15% SDS/PAGE. Separated His6 or GST fusion proteins were then transferred onto an Immobilon-P membrane (Millipore) and visualized by the Ponceau S staining. The membrane was sprayed with the EN3HANCE (NEN) and exposed to Kodak XAR film overnight.
Supplementary Material
Acknowledgments.
We thank Dr. Danny Reinberg (NYU) for Set9 plasmids and Dr. Shengli Hao and Dr. Parameswaran Ramakrishnan for critically reading the manuscript.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910439106/DCSupplemental.
References
- 1.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 2.Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 3.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 4.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kiernan R, et al. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem. 2003;278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 6.Paik WK, Paik DC, Kim S. Historical review: The field of protein methylation. Trends Biochem Sci. 2007;32:146–152. doi: 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Margueron R, Trojer P, Reinberg D. The key to development: Interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Chuikov S, et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- 9.Huang J, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 10.Mowen KA, et al. Arginine methylation of STAT1 modulates IFNalpha/beta-induced transcription. Cell. 2001;104:731–741. doi: 10.1016/s0092-8674(01)00269-0. [DOI] [PubMed] [Google Scholar]
- 11.Huq MD, Tsai NP, Khan SA, Wei LN. Lysine trimethylation of retinoic acid receptor-alpha: A novel means to regulate receptor function. Mol Cell Proteomics. 2007;6:677–688. doi: 10.1074/mcp.M600223-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Subramanian K, et al. Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell. 2008;30:336–347. doi: 10.1016/j.molcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishioka K, et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479–489. doi: 10.1101/gad.967202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- 15.Xiao B, et al. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, et al. Structural basis for the product specificity of histone lysine methyltransferases. Mol Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouskouti A, Scheer E, Staub A, Tora L, Talianidis I. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol Cell. 2004;14:175–182. doi: 10.1016/s1097-2765(04)00182-0. [DOI] [PubMed] [Google Scholar]
- 18.Williams-Ashman HG, Seidenfeld J, Galletti P. Trends in the biochemical pharmacology of 5′-deoxy-5′-methylthioadenosine. Biochem Pharmacol. 1982;31:277–288. doi: 10.1016/0006-2952(82)90171-x. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, et al. Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem. 2008;283:26771–26781. doi: 10.1074/jbc.M802800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 21.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Wan F, et al. Ribosomal protein S3: A KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131:927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Yang XD, et al. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: Temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.