Abstract
Mouse oocytes produce members of the transforming growth factor beta (TGFbeta) superfamily, including bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9), as well as fibroblast growth factors (FGFs). These growth factors cooperate to regulate cumulus cell function. To identify potential mechanisms involved in these interactions, the ability of fully grown oocytes to regulate expression of BMP or FGF antagonists in cumulus cells was examined. Oocytes promoted cumulus cell expression of transcripts encoding antagonists to TGFbeta superfamily members, including Grem2, Htra1, Htra3, and Nog mRNAs. In contrast, oocytes suppressed cumulus cell expression of Spry2 mRNA, which encodes a regulator of receptor tyrosine kinase signals, such as FGF and epidermal growth factor (EGF) receptor signals. The regulation of Spry2 mRNA levels in cumulus cells was studied further as a model for analysis of potential mechanisms for cooperativity of FGF/EGF signaling with oocyte-derived members of the TGFbeta superfamily. Oocytes suppressed basal and FGF-stimulated Spry2 mRNA levels in cumulus cells but promoted EGF-stimulated levels. Furthermore, recombinant TGFbeta superfamily proteins, including BMP15 and GDF9, mimicked these effects of oocytes. Elevated expression of Spry2 mRNA in cumulus and mural granulosa cells correlated with human chorionic gonadotropin-induced expression of mRNAs encoding EGF-like peptides. Therefore, oocyte-derived members of the TGFbeta superfamily suppress FGF-stimulated Spry2 mRNA levels before the luteinizing hormone surge but promote Spry2 mRNA levels stimulated by EGF receptor-mediated signals after the surge.
Keywords: BMP15, EGF, FGF8, follicular development, GDF9, gene regulation, granulosa cells, oocytes, sprouty 2
Oocyte-derived TGFβ superfamily members suppress FGF-stimulated Spry2 mRNA levels before the LH surge but promote Spry2 mRNA levels stimulated by EGF receptor-mediated signals after the surge.
INTRODUCTION
Mammalian oocytes play a key role in orchestrating the development and function of surrounding granulosa cells. For example, paracrine signals derived from oocytes affect proliferation, steroidogenesis, and several metabolic processes of granulosa cells and, at least in mouse, promote cumulus expansion [1–3]. Critical paracrine factors mediating the oocyte-to-granulosa cell communication include members of the transforming growth factor beta (TGFβ) superfamily, such as bone morphogenetic protein 15 (BMP15) and growth differentiation factor 9 (GDF9) [4–7]. Bmp15 and/or Gdf9 null mice exhibit reduced fertility, at least in part because of impaired functions of the granulosa cells and lower developmental competence of the oocytes [6–9]. Likewise, oocyte-secreted factors, BMP15 and GDF9, improved the developmental competence of bovine oocytes undergoing maturation in vitro [10]. Therefore, precise coordination of oocyte-derived paracrine signals in the follicles is essential for normal development of both granulosa cells and oocytes.
In addition to TGFβ superfamily members, oocytes as well as follicular somatic cells express various fibroblast growth factor (FGF) ligands and receptors in many mammalian species, including mouse [9, 11–13], rat [14–17], bovine [18–20], and human [21, 22]. In cultured granulosa cells, FGFs stimulate mitosis in rabbit, pig, guinea pig, and human [23]; alter steroidogenic activity in rat and bovine [24–28]; suppress luteinizing hormone (LH) receptor formation [24, 26]; and inhibit apoptosis in rat [15]. Furthermore, FGFs promote primordial follicle development in rat and human ovarian organ culture [17, 29, 30] and growth of bovine oocytes enclosed in granulosa cell complexes [31]. Therefore, it has become evident that FGF signals also play roles during mammalian follicular development. Interestingly, a recent study has shown that oocyte-derived FGFs, mainly FGF8, act in concert with BMP15 to induce glycolysis in cumulus cells, which are granulosa cells associated with oocytes [9]; however, the mechanisms by which these signals cooperate are unknown.
Cooperation of BMP and FGF signals is found in several tissues where both BMP and FGF signals are present. For example, during calvarial suture osteogenesis, increases in FGF signals augment BMP signals by suppressing expression of Nog transcript, encoding the BMP antagonist noggin [32]. Similarly, in chondrocytes, FGF signals suppress levels of Nog, Grem1, and Chrd mRNA encoding BMP antagonists, noggin, gremlin1, and chordin [33, 34]. Furthermore, during heart tract development, loss of FGF8 signal results in elevated expression of Grem2 mRNA, which is correlated with disrupted BMP and TGFB signals [35]. These reports suggest that suppression of transcripts encoding BMP/FGF antagonists might be involved in the cooperation of BMP and FGF signals in cumulus cells during ovarian follicular development.
To understand further how oocytes regulate development and function of surrounding granulosa cells, we sought to identify potential mechanisms involved in the signal cross talk between BMP and FGF signals in cumulus cells. We screened granulosa cell expression of antagonists of BMP or FGF action and found that Spry2 mRNA levels in cumulus cells were suppressed by oocyte-derived factors. Spry2 mRNA encodes sprouty 2 (SPRY2) protein, which regulates signals of receptor tyrosine kinases, including FGF and epidermal growth factor (EGF) receptors [36, 37]. In mammalian cells, SPRY protein inhibits FGF receptor signals but appears to potentiate EGF receptor signals [36, 37]. Because EGF receptor signals play a critical role after the LH surge [38–41], the effects of EGF, as well as FGFs, on Spry2 mRNA levels in cumulus cells were examined.
MATERIALS AND METHODS
Isolation and Culture of Cumulus Cell-Oocyte Complexes, Fully Grown Oocytes, Mural Granulosa Cells, and Oocytectomized Cumulus Cells
All experiments were conducted using B6SJLF1 mice maintained at The Jackson Laboratory according to the Guide for the Care and Use of Laboratory Animals (Institute for Learning and Animal Research).
Cumulus cell-oocyte complexes (COCs), fully grown oocytes, and mural granulosa cells were isolated from antral follicles of 22- to 24-day-old female mice that had been injected with 5 IU of equine chorionic gonadotropin (eCG) 44 to 48 h earlier as reported previously [42]. Oocytectomized (OOX) cumulus cells were produced by microsurgically removing oocytes, but not the zona pellucida, from the COCs [43]. In the experiments shown in Figure 4, COCs and mural granulosa cells were isolated from 22- to 24-day-old female mice injected with 5 IU of human chorionic gonadotropin (hCG) after the 48-h eCG injection as indicated in the figure legend.
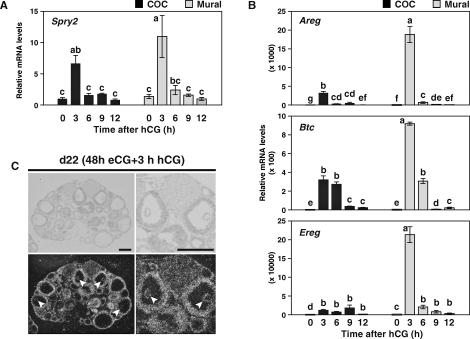
FIG. 4.
Correlation of Spry2 mRNA expression with transcripts encoding EGF-like growth factors. The eCG-primed mice were injected with hCG, and (A) kinetics of Spry2 expression levels as well as (B) Areg, Btc, and Ereg mRNA expression in COCs and mural granulosa cells were determined. Values indicated by different lowercase letters (a–g) are significantly different (P < 0.05). Also shown is (C) localization of Spry2 mRNA in ovaries 3 h after hCG treatment. Top) Brightfield images. Bottom) Darkfield images. Arrows indicate cumulus cells. d22 = Day 22. Bar = 500 μm.
The culture medium used was bicarbonate-buffered MEMα (Life Technologies, Inc.) with Earles salts, supplemented with 75 μg/ml of penicillin G, 50 μg/ml of streptomycin sulfate, 0.23 mM pyruvate, and 3 mg/ml of bovine serum albumin (Sigma-Aldrich Co.). Oocytes were maintained at the germinal vesicle stage by addition of the phosphodiesterase inhibitor milrinone (10 mM; Sigma-Aldrich) throughout the experiments. Milrinone was added to all culture medium even if no oocytes were present for experimental balance. Cultures were performed in drops of culture medium under washed mineral oil for the indicated period. All cultures were maintained at 37°C in a modular incubation chamber (Billups Rothenberg) infused with 5% O2, 5% CO2, and 90% N2.
In some experiments, culture medium was supplemented with recombinant human TGFB2 (10 ng/ml; Leinco Technologies), human BMP4 (100 ng/ml; R&D Systems), human BMP6 (100 ng/ml; R&D Systems), human BMP15 (500 ng/ml), mouse GDF9 (500 ng/ml), human FGF1 (Sigma), human FGF2 (BD Biosciences), human FGF4 (R&D Systems), mouse FGF8B (Sigma), or mouse EGF (BD Biosciences) as indicated in figure legends. Recombinant BMP15 and GDF9 were produced at Baylor College of Medicine by Q. Li and M.M. Matzuk or were obtained from R&D Systems. Control buffer from purification of recombinant proteins had no effect on transcript levels in cumulus cells. We obtained identical results using either preparation of recombinant GDF9 or BMP15.
Real-Time RT-PCR
Steady-state levels of transcripts were assessed using real-time RT-PCR as reported previously [9]. The results were first normalized to the expression levels of a housekeeping gene, ribosomal protein L19 (Rpl19), by the 2−ΔΔCt method [44] and then presented as the expression levels relative to the transcript amount of a control group. PCR primers used were shown in Table 1. Lhcgr, Pfkp, Rpl19, Areg, Btc, and Ereg primers were reported previously [9, 45]. To avoid false-positive signals, dissociation-curve analyses were performed at the end of analyses, and the PCR products were applied to agarose gel electrophoresis to confirm the sizes. Moreover, the PCR products were purified and sequenced to verify sequence identity. The reactions were conducted at least in duplicate.
TABLE 1.
Sequence of PCR primers used for real-time PCR.
In Situ Hybridization
A fragment of Spry2 cDNA (1152 bp, GenBank accession no. NM_011897) was amplified by PCR using the primers 5′-TTCCCCGTGGTTTTAATCCA-3′ and 5′-GGCCTGTTCACTCGGTCATAA-3′. The PCR product was cloned into pCRII–TOPO plasmid using the TOPO TA Cloning Kit (Invitrogen). The identity and orientation of the sequence were confirmed by sequencing. The plasmid DNA was digested with restriction enzyme XhoI (Roche Diagnostics) and used for in vitro transcription to make [33P]Cytidine-5′-triphosphate labeled riboprobes for in situ hybridization as reported previously [9]. No signal was detected with a Spry2 sense probe (see Supplemental Fig. S1 available online at www.biolreprod.org).
Statistical Analysis
All experiments were repeated at least three times. The Tukey-Kramer Honestly Significant Difference (HSD) test or a standard t-test were used for multiple or paired comparison, respectively, using JMP software (SAS Institute, Inc.). Relative fold-change values were log10 transformed to scale the data such that the normality and equal variance assumptions of the Tukey-Kramer HSD test were satisfied (see Fig. 4). A P value of less than 0.05 was considered to be statistically significant.
RESULTS
Expression of Transcripts Encoding BMP/FGF Antagonists in Granulosa Cells and Paracrine Regulation by Oocytes
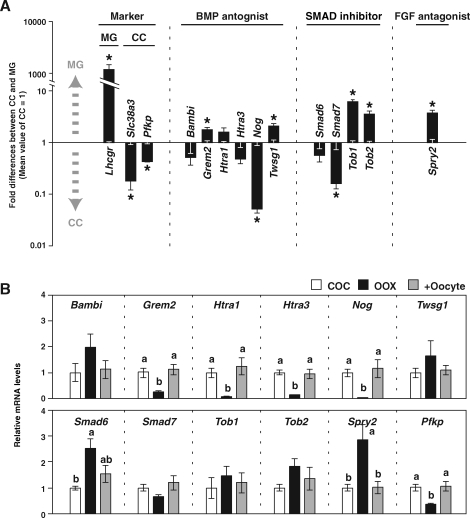
To identify transcripts encoding BMP/FGF antagonists for which the expression levels in granulosa cells are suppressed by oocytes, expression levels of mRNA encoding BMP antagonists (Bambi, Grem2, Htra1, Htra3, Nog, and Twsg1) and an FGF antagonist (Spry2) were compared between freshly isolated cumulus and mural granulosa cells. The rationale was as follows: Transcripts that are differentially expressed between cumulus cells, which are associated with oocytes, and mural granulosa cells, which are located far from oocytes, may be regulated in vivo by oocyte-derived factors. Because BMP signals in granulosa cells are mediated by SMAD proteins [46], levels of transcripts encoding SMAD inhibitors (Smad6, Smad7, Tob1, and Tob2) also were examined. These transcripts were selected as candidate transcripts based on their expression in ovary and cumulus cells, indicated by the publicly available GNF SymAtlas database (http://symatlas.gnf.org) and our microarray data [7], respectively. Efficient separation of cumulus and mural granulosa cells was confirmed by assessing mRNA levels of Slc38a3 and Pfkp, encoding an amino acid transporter and a glycolytic enzyme (markers of cumulus cells), respectively, and Lhcgr, encoding LH receptor (a marker of mural granulosa cells) [2] (Fig. 1A).
FIG. 1.
Expression of transcripts encoding BMP/FGF antagonists in granulosa cells (A) and paracrine regulation by oocytes (B). A) Fold-differences in transcript levels of BMP/FGF antagonist between mural granulosa cells (MG) and cumulus cells (CC) are presented using an exponential scale. Mean values of cumulus cells were normalized to one. Therefore, a value greater than one indicates that the transcript is enriched in mural granulosa cells, and a value smaller than one indicates that the transcript is enriched in cumulus cells. An asterisk indicates that the difference of expression levels between mural granulosa cells and cumulus cells is statistically significant (P < 0.05). B) Cumulus cells were cultured as intact COCs (COC) or OOX cumulus cells (OOX), or OOX cumulus cells were cocultured with oocytes (+Oocyte) and expression levels of transcripts encoding BMP/FGF antagonists in cumulus cells were examined after 20 h. Values indicated by different lowercase letters (a and b) are significantly different (P < 0.05). All values are presented as the mean ± SEM.
Expression levels of Nog and Smad7 mRNA were significantly higher in cumulus cells than in mural granulosa cells, suggesting a possibility that expression levels of these transcripts in cumulus cells are promoted by oocyte-derived factors (Fig. 1A). On the other hand, levels of Grem2, Twsg1, Tob1, Tob2, and Spry2 mRNA were significantly higher in mural granulosa cells than in cumulus cells; thus, oocyte-secreted factors may suppress expression levels of these transcripts in cumulus cells in vivo (Fig. 1A).
To address directly whether levels of these transcripts in cumulus cells are regulated by oocytes in vitro, the effect of oocyte removal (OOX) from COCs on the levels of these transcripts was determined. Cumulus cells were cultured as intact COCs, OOX cumulus cells, or OOX cumulus cells cocultured with denuded, fully grown, germinal vesicle-stage oocytes (2 oocytes/μl) for 20 h, and expression levels of these transcripts in cumulus cells were examined (Fig. 1B). Levels of Pfkp mRNA (a known transcript, the expression of which in cumulus cells is promoted by oocytes [42]) as well as levels of Grem2, Htra1, Htra3, and Nog mRNA were significantly down-regulated when oocytes were removed from COCs, and the levels found in cumulus cells cultured as intact COCs were restored when OOX cumulus cells were cocultured with oocytes. This indicates that steady-state levels of these transcripts in cumulus cells are promoted by oocytes in vitro. In contrast, levels of Smad6 and Spry2 transcripts were up-regulated in OOX cumulus cells compared with those in cumulus cells cultured as intact COCs, and these increases were prevented by coculturing OOX cumulus cells with oocytes. Therefore, oocytes suppress expression levels of Smad6 and Spry2 transcripts in cumulus cells in vitro.
Taken together, these results strongly suggest that Spry2 mRNA expression in cumulus cells is suppressed by oocyte-derived factors both in vivo and in vitro. Furthermore, expression of the other sprouty family members, Spry1, Spry3, and Spry4, was not detectable in granulosa cells (data not shown). Although Smad6 mRNA levels in cumulus cells were suppressed by oocytes in vitro, the expression levels in vivo were not significantly different between cumulus and mural granulosa cells. This suggests that the suppressive effect of oocytes on Smad6 mRNA expression may be masked by an unknown factor acting on both cumulus and mural granulosa cells. Therefore, we focused on the regulation of Spry2 expression in the present study.
Localization of Spry2 mRNA During Follicular Development
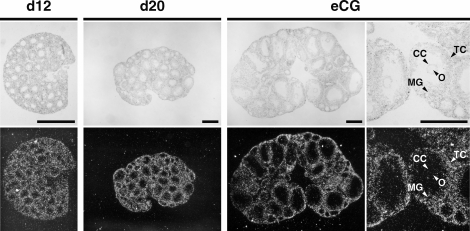
Localization of Spry2 transcripts during follicular development before the LH surge was determined by in situ hybridization (Fig. 2). Spry2 mRNA localization was detected in granulosa cells of preantral follicles (Fig. 2, d12) and early antral follicles (Fig. 2, d20). In well-developed antral follicles of ovaries from eCG-primed mice, signals were detected in mural granulosa cells at higher levels than in cumulus cells (Fig. 2, eCG), as was shown by real-time PCR as well (Fig. 1A). Spry2 mRNA localization also was detected in theca cells but was not detected in oocytes at any stage.
FIG. 2.
Localization of Spry2 mRNA during follicular development before the LH surge. Localization of Spry2 mRNA in ovaries was detected by in situ hybridization using 12-day-old (d12) and 20-day-old (d20) mice and 22-day-old eCG-primed (eCG) mice. Spry2 mRNA signals were not detectable in cumulus cells (CC). MG, mural granulosa cells; O, oocytes; TC, theca cells. Top) Brightfield images. Bottom) Darkfield images. Bar = 500 μm.
Effect of FGFs, Oocyte Coculture, and TGFβ Superfamily Proteins on Expression Levels of Spry2 mRNA in OOX Cumulus Cells
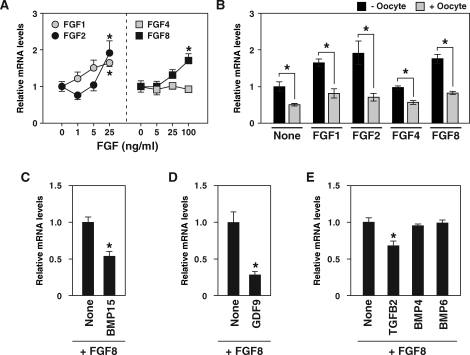
Because Spry2 expression in some nonovarian cell types is promoted by FGF ligands [36, 37], the effect of recombinant FGFs on levels of Spry2 mRNA in OOX cumulus cells was determined. OOX cumulus cells were cultured with recombinant FGF1, FGF2, FGF4, and FGF8, and levels of Spry2 mRNA were examined after 6 h of treatment (Fig. 3A). Treatment with FGF1, FGF2, and FGF8, but not with FGF4, increased Spry2 mRNA levels in OOX cumulus cells in a dose-dependent manner. These results agree with those of a previous report that FGF2, but not FGF4, promoted Spry2 expression in human granulosa-lutein cells [47].
FIG. 3.
Effect of FGFs (A), oocyte coculture (B), and TGFβ superfamily proteins (C–E) on expression levels of Spry2 mRNA in OOX cumulus cells. A) OOX cumulus cells were cultured with recombinant FGFs and Spry2 transcript levels were assessed after 6 h. A value with an asterisk is significantly different from the value of a group that was not treated with FGFs (P < 0.05). B) OOX cumulus cells were cultured with either FGF1 (25 ng/ml), FGF2 (25 ng/ml), FGF4 (100 ng/ml), or FGF8 (100 ng/ml), and the effect of oocytes (2 oocytes/μl) were examined. An asterisk indicates that the difference of expression levels in OOX cumulus cells cocultured with and without oocytes is statistically significant (P < 0.05). C–E) OOX cumulus cells were cultured with FGF8 (100 ng/ml) and with or without BMP15 (500 ng/ml), GDF9 (500 ng/ml), TGFB2 (10 ng/ml), BMP4 (100 ng/ml), and BMP6 (100 ng/ml), and Spry2 expression levels were assessed with real-time PCR. A value with an asterisk is significantly different from the value of a control group (None; P < 0.05). Values are presented as the mean ± SEM.
The effect of coculture of OOX cumulus cells with oocytes on levels of FGF-induced Spry2 expression was determined (Fig. 3B). Oocytes suppressed Spry2 mRNA levels that were elevated by FGF1, FGF2, and FGF8 as well as basal levels of Spry2 mRNA not stimulated by FGFs (Fig. 3B). Although mouse oocytes produce several FGFs, including FGF8 [9, 11], oocytes suppressed rather than promoted Spry2 mRNA expression in cumulus cells. Therefore, some other factors likely secreted from oocytes suppress Spry2 mRNA levels in cumulus cells.
To identify the oocyte factors that suppress FGF8-induced Spry2 expression in cumulus cells, the effects of the recombinant TGFβ superfamily proteins BMP15, GDF9, TGFB2, BMP4, and BMP6 on FGF8-induced Spry2 mRNA expression in OOX cumulus cells were determined (Fig. 3, C–E). In rodents, oocytes produce BMP6 [48–50], both oocytes and granulosa cells produce TGFB2 [51, 52], and theca cells produce BMP4 [53]. OOX cumulus cells were cultured with FGF8 alone or with both FGF8 and one of the TGFβ superfamily proteins, and Spry2 mRNA levels in cumulus cells were measured after 20 h of treatment. BMP15, TGFB2, and GDF9 suppressed Spry2 mRNA levels in FGF8-treated OOX cumulus cells, whereas BMP4 and BMP6 had little or no effect (Fig. 3, C–E). These results indicate that one or more oocyte-derived TGFβ superfamily ligands suppress Spry2 mRNA expression in cumulus cells before the LH surge.
Correlation of Spry2 mRNA Expression with Transcripts Encoding EGF-Like Growth Factors
During the ovulatory period, the LH surge induces robust expression of EGF-like growth factors, amphiregulin (AREG), betacellulin (BTC), and epiregulin (EREG), in granulosa cells. These EGF-like growth factors play critical roles in mediating LH action within the follicles [38, 39, 41]. In addition to FGFs, EGF receptor signals also induce Spry2 expression in some cell types, including human granulosa-lutein cells [47]. Therefore, it was of interest to determine the pattern of Spry2 expression after the LH surge, and the effect of hCG-treatment, which mimics the LH surge, on Spry2 mRNA levels was examined as well (Fig. 4). The eCG-primed mice were injected with hCG, and kinetics of Spry2, Areg, Btc, and Ereg mRNA expression in COCs and mural granulosa cells were determined (Fig. 4, A and B). Spry2 mRNA levels were low before hCG treatment (Fig. 4A, 0 h), but within 3 h of hCG treatment, dramatically increased Spry2 transcript levels were observed in both COCs and mural granulosa cells. Subsequently, Spry2 mRNA levels decreased by 12 h after hCG treatment to a level comparable with that found before hCG treatment. This pattern of Spry2 mRNA expression correlated well with those of Areg, Btc, and Ereg transcripts, suggesting that Spry2 mRNA expression after the LH surge may be regulated by these EGF-like growth factors (Fig. 4B).
Because Spry2 mRNA levels in COCs were greatest after 3 h of hCG treatment, localization of Spry2 transcripts in the ovary after 3 h of hCG treatment was determined by in situ hybridization (Fig. 4C). In contrast to eCG-primed ovary (Fig. 2, eCG), in the ovary 3 h after hCG treatment, comparable levels of Spry2 localization signals were found in both cumulus and mural granulosa cells in all antral follicles observed (Fig. 4C), and this was confirmed by real-time PCR (Fig. 4A, compare COC and mural granulosa cell at 3 h). These results suggest that oocytes may not suppress Spry2 mRNA expression in cumulus cells after the LH surge.
Effect of EGF, Oocyte Coculture, and TGFβ Superfamily Proteins on Expression Levels of Spry2 mRNA in OOX Cumulus Cells
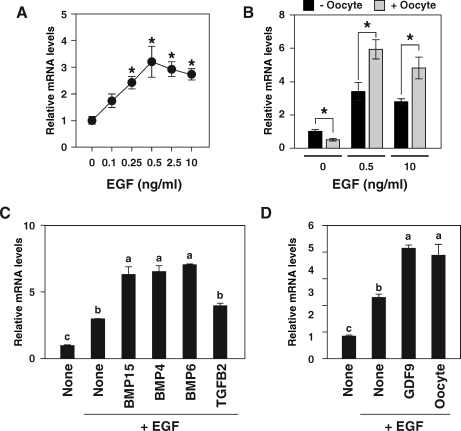
To test further the idea that EGF receptor signal regulates Spry2 mRNA levels in cumulus cells, the effect of EGF on Spry2 transcript levels in OOX cumulus cells was determined in vitro (Fig. 5A). OOX cumulus cells were cultured with recombinant EGF, and the steady-state levels of Spry2 mRNA were examined after 6 h of treatment (Fig. 5A). A dose-dependent increase of Spry2 mRNA levels in OOX cumulus cells was observed up to 0.5 ng/ml of EGF. Higher concentrations of EGF (up to 10 ng/ml) did not further stimulate Spry2 expression; therefore, the effect of EGF on Spry2 mRNA expression appeared to be saturated at concentrations of 0.5 ng/ml and higher.
FIG. 5.
Effect of EGF (A), oocyte coculture (B), and TGFβ superfamily proteins (C and D) on expression levels of Spry2 mRNA in OOX cumulus cells. A) OOX cumulus cells were cultured with recombinant EGF, and Spry2 transcript levels were assessed after 6 h. A value with an asterisk is significantly different from the value of the group which was not treated with EGF (P < 0.05). B) OOX cumulus cells were cultured with EGF, and the effect of oocytes (2 oocytes/μl) was examined. An asterisk indicates that the differences of expression levels in OOX cumulus cells cocultured with and without oocytes are statistically significant (P < 0.05). C and D) OOX cumulus cells were cultured with EGF (10 ng/ml) and with or without BMP15 (500 ng/ml), BMP4 (100 ng/ml), BMP6 (100 ng/ml), TGFB2 (10 ng/ml), and GDF9 (500 ng/ml), and Spry2 expression levels were assessed with real-time PCR. The values indicated by different lowercase letters (a–c) are significantly different (P < 0.05). Values are presented as the mean ± SEM.
Effect of coculture of OOX cumulus cells with oocytes on expression levels of EGF-stimulated Spry2 expression also was determined (Fig. 5B). Surprisingly, in contrast to FGF-induced Spry2 expression, EGF-simulated Spry2 expression in OOX cumulus cells was augmented by coculture with oocytes. This positive effect of oocytes on EGF-stimulated Spry2 mRNA expression was observed even when a concentration of EGF (10 ng/ml) higher than saturation was used (Fig. 5B).
To identify the oocyte factors that promote EGF-induced Spry2 expression in cumulus cells, effects of recombinant TGFβ superfamily proteins on EGF-induced Spry2 mRNA expression in OOX cumulus cells were determined (Fig. 5, C and D). Whereas TGFB2 had no effect, BMP15, BMP4, and BMP6 (Fig. 5C), as well as GDF9 (Fig. 5D), promoted Spry2 expression in OOX cumulus cells. Therefore, members of TGFβ superfamily derived from oocytes as well as from follicular somatic cells augment EGF-induced Spry2 expression in cumulus cells in vitro.
DISCUSSION
Precise coordination of multiple endocrine, autocrine, and paracrine growth factor signals is required for normal ovarian follicular development. However, the interactions of oocyte- and somatic cell-derived signaling pathways are far from well understood. The present results showed that several transcripts encoding antagonists of either BMP or FGF signaling are expressed in granulosa cells, and some of these are regulated by oocytes. Furthermore, before the LH surge, Spry2 mRNA was differentially expressed between cumulus and mural granulosa cells, probably because paracrine factors secreted from oocytes suppress Spry2 expression in cumulus cells. Although both FGFs and EGF promoted Spry2 mRNA expression in cultured cumulus cells, the effects of oocytes on FGF- and EGF-stimulated Spry2 mRNA expression were surprisingly different: Oocytes suppressed FGF-induced Spry2 mRNA expression in cumulus cells, but EGF-induced Spry2 mRNA expression was promoted by oocytes. Recombinant BMP15 and GDF9, as well as some of other TGFβ superfamily members, mimicked these effects of oocytes, suggesting that the oocyte factors affecting Spry2 expression in cumulus cells probably are members of the TGFβ superfamily, including BMP15 and GDF9. Therefore, we have concluded that oocyte-derived TGFβ superfamily members cooperate with FGFs and EGF receptor signals to regulate Spry2 mRNA levels in cumulus cells. Regulation of Spry2 expression by TGFβ superfamily signals may be one of the mechanisms by which oocyte-derived TGFβ superfamily signals cooperate with FGF/EGF signals in granulosa cells during ovarian follicular development.
The sprouty gene was first described in Drosophila sp. and the proteins characterized as antagonists of FGF receptor-mediated tracheal branching [54]. Subsequent studies have shown that sprouty proteins generally are inhibitors of receptor tyrosine kinase signals in both vertebrates and invertebrates [37]. In mammals, four sprouty genes, sprouty homolog 1 to sprouty homolog 4, have been identified. The present study focused on Spry2 mRNA regulation, because expression of the other members of sprouty family was not detectable in mouse granulosa cells (Sugiura and Eppig, unpublished results). Spry2 null mice exhibit a severe gastrointestinal phenotype, and approximately half of them die within 6 wk after birth [55, 56]. Remaining Spry2 null mice survived but were significantly smaller than wild-type littermates, and to our knowledge, their ovarian morphology and fertility have not been studied. In Drosophila ovaries, sprouty mRNA is expressed in follicle cells in a manner correlated with EGF receptor signals, and sprouty mRNA expression is reduced in mutants of gurken, one of the ligands of EGF receptor [57, 58]. Whereas overexpression of sprouty mRNA in follicle cells resulted in a defective ovary that resembled that of a reduced EGF receptor pathway, sprouty mutation resulted in a phenotype consistent with an increased activity of EGF receptor signals [58]. Therefore, Drosophila sprouty protein appears to be a key negative regulator of EGF receptor signals during ovarian development. Interestingly, in contrast to Drosophila sprouty protein, which inhibits EGF receptor signals [58–60], mammalian sprouty proteins appear to potentiate EGF receptor signals. This agonistic effect of mammalian SPRYs on EGF receptor signals is achieved by preventing ubiquitylation and degradation of activated EGF receptors [61–64]. Therefore, mammalian SPRY2 likely has a role distinct from that of Drosophila sprouty protein during ovarian follicular development.
How do members of TGFβ superfamily regulate Spry2 expression in cumulus cells? The present results showed that basal and FGF-induced Spry2 mRNA expression levels were down-regulated by members of TGFβ superfamily, including BMP15, GDF9, and TGFB2. Similar results have been reported using mesenchymal cells in which Spry2 mRNA expression was suppressed by TGFB1 [65]. That report also showed that inhibitors of histone deacetylases (HDACs) prevented TGFB1-induced Spry2 mRNA down-regulation, suggesting that HDAC activity was required for Spry2 down-regulation by TGFBs. Therefore, similar HDAC-dependent mechanisms may exist in cumulus cells. Furthermore, a recent study has shown that Spry2 is a direct target of forkhead transcription factors (FOXOs) in endothelial cells [66]. In addition, SMADs, mediators of TGFβ superfamily signals, are reported to associate with FOXOs and to promote gene expression in neuroepithelial and glioblastoma cells [67]. Therefore, up-regulation of EGF-stimulated Spry2 expression induced by members of TGFβ superfamily in cumulus cells may involve a similar FOXO-dependent mechanism. The reasons why the effects of TGFβ superfamily signals are different between FGF-induced and EGF-induced Spry2 expression remain to be resolved.
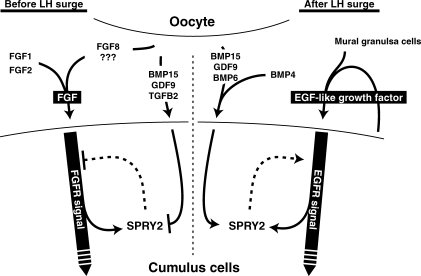
Based on the present results, together with well-established functions of mammalian sprouty proteins [37, 68], we suggest that SPRY2 may be a key regulator of ovarian follicular development. A working model explaining how oocyte-derived TGFβ superfamily signals might affect FGF and EGF receptor signals in cumulus cells is presented in Figure 6. Before the LH surge, oocyte-derived FGF8 and, perhaps, other FGFs produced by oocytes and somatic cells comprise a total FGF signal and activate FGF receptors on cumulus cells to affect development and function. However, at same time, these FGF signals induce elevated SPRY2 expression, which negatively affects the FGF signals. Consequently, oocyte-derived TGFβ superfamily proteins, including BMP15 and GDF9, suppress Spry2 expression; therefore, FGF signals can continue to affect the cumulus cell functions. On the other hand, after the LH surge, EGF-like growth factors produced by cumulus and mural granulosa cells act together to activate EGF receptor signals in cumulus cells. However, activated EGF receptor may be rapidly down-regulated by its degradation, which would negatively affect EGF-dependent cellular functions [69, 70]. Oocyte-derived TGFβ superfamily proteins, including BMP15 and GDF9, now promote Spry2 expression in cumulus cells and, therefore, prevent degradation of activated EGF receptor, which leads to a prolonged EGF signal in cumulus cells. This regulatory mechanism could explain why both FGFs and BMP15 are required for some cumulus cell function, such as elevated glycolysis, before the LH surge [9] and why both oocyte-derived factors and EGF receptor signals are required for post-LH-surge events, such as cumulus expansion. Genetic experiments are in progress to test this working model.
FIG. 6.
Schematic diagram of crosstalk between FGF/EGF signals and TGFβ superfamily signals in cumulus cells (see text for details).
Although SPRY2 may be one of the critical components involved in the cooperative action of TGFβ superfamily and FGF/EGFs during follicular development, other participants are likely to exist. In fact, the present results (Fig. 1) showed that several other BMP antagonists are expressed in granulosa cells, suggesting involvement of other BMP/FGF antagonists in this cooperation. In addition, agonists or receptors of TGFβ superfamily or FGFs may be involved. Furthermore, extracellular regulated kinases (MAPK3/1) downstream of receptor tyrosine kinases, such as the FGF and EGF receptors, can negatively affect transcriptional activity of SMADs by directly phosphorylating the linker region of SMADs in cultured epithelial cells [71], which may add further complexity in this model. Future studies of these signal interactions are required to resolve the complex cooperative pathways participating in oocyte control of follicular development.
Supplementary Material
Acknowledgments
Recombinant mouse GDF9 was a gift from R&D Systems. We thank Drs. Stephen A. Murray, Weidong Zhang, and Benjamin King for their helpful suggestions during preparation of the present paper.
Footnotes
Supported by grants HD23839 from the Eunice Kennedy Shriver NICHD and CA34196 from NCI.
REFERENCES
- Eppig JJ.Oocyte control of ovarian follicular development and function in mammals. Reproduction 2001; 122: 829–838.. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Eppig JJ.Society for Reproductive Biology Founders' Lecture 2005. Control of metabolic cooperativity between oocytes and their companion granulosa cells by mouse oocytes. Reprod Fertil Dev 2005; 17: 667–674.. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Eppig JJ.Mouse oocyte control of granulosa cell development and function: paracrine regulation of cumulus cell metabolism. Semin Reprod Med 2009; 27: 32–42.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Albertini DF, Nishimori K, Kumar TR, Lu N, Matzuk MM.Growth differentiation factor-9 is required during early ovarian folliculogenesis. Nature 1996; 383: 531–535.. [DOI] [PubMed] [Google Scholar]
- Elvin JA, Yan C, Wang P, Nishimori K, Matzuk MM.Molecular characterization of the follicle defects in the growth differentiation factor 9-deficient ovary. Mol Endocrinol 1999; 13: 1018–1034.. [DOI] [PubMed] [Google Scholar]
- Yan C, Wang P, DeMayo J, DeMayo FJ, Elvin JA, Carino C, Prasad SV, Skinner SS, Dunbar BS, Dube JL, Celeste AJ, Matzuk MM.Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol 2001; 15: 854–866.. [DOI] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Wigglesworth K, O'Brien MJ, Affourtit JP, Pangas SA, Matzuk MM, Eppig JJ.Oocyte regulation of metabolic cooperativity between mouse cumulus cells and oocytes: BMP15 and GDF9 control cholesterol biosynthesis in cumulus cells. Development 2008; 135: 111–121.. [DOI] [PubMed] [Google Scholar]
- Su YQ, Wu X, O'Brien MJ, Pendola FL, Denegre JN, Matzuk MM, Eppig JJ.Synergistic roles of BMP15 and GDF9 in the development and function of the oocyte-cumulus cell complex in mice: genetic evidence for an oocyte-granulosa cell regulatory loop. Dev Biol 2004; 276: 64–73.. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Su YQ, Diaz FJ, Pangas SA, Sharma S, Wigglesworth K, O'Brien MJ, Matzuk MM, Shimasaki S, Eppig JJ.Oocyte-derived BMP15 and FGFs cooperate to promote glycolysis in cumulus cells. Development 2007; 134: 2593–2603.. [DOI] [PubMed] [Google Scholar]
- Hussein TS, Thompson JG, Gilchrist RB.Oocyte-secreted factors enhance oocyte developmental competence. Dev Biol 2006; 296: 514–521.. [DOI] [PubMed] [Google Scholar]
- Valve E, Penttila TL, Paranko J, Harkonen P.FGF-8 is expressed during specific phases of rodent oocyte and spermatogonium development. Biochem Biophys Res Commun 1997; 232: 173–177.. [DOI] [PubMed] [Google Scholar]
- MacArthur CA, Shankar DB, Shackleford GM.Fgf-8, activated by proviral insertion, cooperates with the Wnt-1 transgene in murine mammary tumorigenesis. J Virol 1995; 69: 2501–2507.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puscheck EE, Patel Y, Rappolee DA.Fibroblast growth factor receptor (FGFR)-4, but not FGFR-3, is expressed in the pregnant ovary. Mol Cell Endocrinol 1997; 132: 169–176.. [DOI] [PubMed] [Google Scholar]
- Asakai R, Song SY, Itoh N, Yamakuni T, Tamura K, Okamoto R.Differential gene expression of fibroblast growth factor receptor isoforms in rat ovary. Mol Cell Endocrinol 1994; 104: 75–80.. [DOI] [PubMed] [Google Scholar]
- Peluso JJ, Pappalardo A.Progesterone maintains large rat granulosa cell viability indirectly by stimulating small granulosa cells to synthesize basic fibroblast growth factor. Biol Reprod 1999; 60: 290–296.. [DOI] [PubMed] [Google Scholar]
- Shikone T, Yamoto M, Nakano R.Follicle-stimulating hormone induces functional receptors for basic fibroblast growth factor in rat granulosa cells. Endocrinology 1992; 131: 1063–1068.. [DOI] [PubMed] [Google Scholar]
- Nilsson E, Parrott JA, Skinner MK.Basic fibroblast growth factor induces primordial follicle development and initiates folliculogenesis. Mol Cell Endocrinol 2001; 175: 123–130.. [DOI] [PubMed] [Google Scholar]
- Buratini J, Jr, Glapinski VF, Giometti IC, Teixeira AB, Costa IB, Avellar MC, Barros CM, Price CA.Expression of fibroblast growth factor-8 and its cognate receptors, fibroblast growth factor receptor (FGFR)-3c and −4, in fetal bovine preantral follicles. Mol Reprod Dev 2005; 70: 255–261.. [DOI] [PubMed] [Google Scholar]
- Berisha B, Sinowatz F, Schams D.Expression and localization of fibroblast growth factor (FGF) family members during the final growth of bovine ovarian follicles. Mol Reprod Dev 2004; 67: 162–171.. [DOI] [PubMed] [Google Scholar]
- Buratini J, Jr, Pinto MG, Castilho AC, Amorim RL, Giometti IC, Portela VM, Nicola ES, Price CA.Expression and function of fibroblast growth factor 10 and its receptor, fibroblast growth factor receptor 2B, in bovine follicles. Biol Reprod 2007; 77: 743–750.. [DOI] [PubMed] [Google Scholar]
- Ben-Haroush A, Abir R, Ao A, Jin S, Kessler-Icekson G, Feldberg D, Fisch B.Expression of basic fibroblast growth factor and its receptors in human ovarian follicles from adults and fetuses. Fertil Steril 2005; 84(suppl 2):1257–1268.. [DOI] [PubMed] [Google Scholar]
- Knee RS, Pitcher SE, Murphy PR.Basic fibroblast growth factor sense (FGF) and antisense (gfg) RNA transcripts are expressed in unfertilized human oocytes and in differentiated adult tissues. Biochem Biophys Res Commun 1994; 205: 577–583.. [DOI] [PubMed] [Google Scholar]
- Gospodarowicz D, Bialecki H.Fibroblast and epidermal growth factors are mitogenic agents for cultured granulosa cells of rodent, porcine, and human origin. Endocrinology 1979; 104: 757–764.. [DOI] [PubMed] [Google Scholar]
- Baird A, Hsueh AJ.Fibroblast growth factor as an intraovarian hormone: differential regulation of steroidogenesis by an angiogenic factor. Regul Pept 1986; 16: 243–250.. [DOI] [PubMed] [Google Scholar]
- Adashi EY, Resnick CE, Croft CS, May JV, Gospodarowicz D.Basic fibroblast growth factor as a regulator of ovarian granulosa cell differentiation: a novel nonmitogenic role. Mol Cell Endocrinol 1988; 55: 7–14.. [DOI] [PubMed] [Google Scholar]
- Oury F, Darbon JM.Fibroblast growth factor regulates the expression of luteinizing hormone receptors in cultured rat granulosa cells. Biochem Biophys Res Commun 1988; 156: 634–643.. [DOI] [PubMed] [Google Scholar]
- McAllister JM, Byrd W, Simpson ER.The effects of growth factors and phorbol esters on steroid biosynthesis in isolated human theca interna and granulosa-lutein cells in long term culture. J Clin Endocrinol Metab 1994; 79: 106–112.. [DOI] [PubMed] [Google Scholar]
- Portela VM, Goncalves PB, Veiga AM, Nicola E, Buratini J, Jr, Price CA.Regulation of angiotensin type 2 receptor in bovine granulosa cells. Endocrinology 2008; 149: 5004–5011.. [DOI] [PubMed] [Google Scholar]
- Kezele P, Nilsson EE, Skinner MK.Keratinocyte growth factor acts as a mesenchymal factor that promotes ovarian primordial to primary follicle transition. Biol Reprod 2005; 73: 967–973.. [DOI] [PubMed] [Google Scholar]
- Garor R, Abir R, Erman A, Felz C, Nitke S, Fisch B.Effects of basic fibroblast growth factor on in vitro development of human ovarian primordial follicles. Fertil Steril 2009; 91: 1967–1975.. [DOI] [PubMed] [Google Scholar]
- Cho JH, Itoh T, Sendai Y, Hoshi H.Fibroblast growth factor 7 stimulates in vitro growth of oocytes originating from bovine early antral follicles. Mol Reprod Dev 2008; 75: 1736–1743.. [DOI] [PubMed] [Google Scholar]
- Warren SM, Brunet LJ, Harland RM, Economides AN, Longaker MT.The BMP antagonist noggin regulates cranial suture fusion. Nature 2003; 422: 625–629.. [DOI] [PubMed] [Google Scholar]
- Reinhold MI, Abe M, Kapadia RM, Liao Z, Naski MC.FGF18 represses noggin expression and is induced by calcineurin. J Biol Chem 2004; 279: 38209–38219.. [DOI] [PubMed] [Google Scholar]
- Matsushita T, Wilcox WR, Chan YY, Kawanami A, Bukulmez H, Balmes G, Krejci P, Mekikian PB, Otani K, Yamaura I, Warman ML, Givol D, Murakami S.FGFR3 promotes synchondrosis closure and fusion of ossification centers through the MAPK pathway. Hum Mol Genet 2009; 18: 227–240.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM.An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development 2008; 135: 3599–3610.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Bar-Sagi D.Modulation of signalling by Sprouty: a developing story. Nat Rev Mol Cell Biol 2004; 5: 441–450.. [DOI] [PubMed] [Google Scholar]
- Mason JM, Morrison DJ, Basson MA, Licht JD.Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol 2006; 16: 45–54.. [DOI] [PubMed] [Google Scholar]
- Park JY, Su YQ, Ariga M, Law E, Jin SL, Conti M.EGF-like growth factors as mediators of LH action in the ovulatory follicle. Science 2004; 303: 682–684.. [DOI] [PubMed] [Google Scholar]
- Hsieh M, Lee D, Panigone S, Horner K, Chen R, Theologis A, Lee DC, Threadgill DW, Conti M.Luteinizing hormone-dependent activation of the epidermal growth factor network is essential for ovulation. Mol Cell Biol 2007; 27: 1914–1924.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Hernandez-Gonzalez I, Gonzalez-Robayna I, Richards JS.Paracrine and autocrine regulation of epidermal growth factor-like factors in cumulus-oocyte complexes and granulosa cells: key roles for prostaglandin synthase 2 and progesterone receptor. Mol Endocrinol 2006; 20: 1352–1365.. [DOI] [PubMed] [Google Scholar]
- Panigone S, Hsieh M, Fu M, Persani L, Conti M.Luteinizing hormone signaling in preovulatory follicles involves early activation of the epidermal growth factor receptor pathway. Mol Endocrinol 2008; 22: 924–936.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura K, Pendola FL, Eppig JJ.Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: energy metabolism. Dev Biol 2005; 279: 20–30.. [DOI] [PubMed] [Google Scholar]
- Buccione R, Schroeder AC, Eppig JJ.Interactions between somatic cells and germ cells throughout mammalian oogenesis. Biol Reprod 1990; 43: 543–547.. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001; 25: 402–408.. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Su YQ, Eppig JJ.Targeted suppression of Has2 mRNA in mouse cumulus cell-oocyte complexes by adenovirus-mediated short-hairpin RNA expression. Mol Reprod Dev 2009; 76: 537–547.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimasaki S, Moore RK, Otsuka F, Erickson GF.The bone morphogenetic protein system in mammalian reproduction. Endocr Rev 2004; 25: 72–101.. [DOI] [PubMed] [Google Scholar]
- Haimov-Kochman R, Ravhon A, Prus D, Greenfield C, Finci-Yeheskel Z, S Goldman-Wohl D, Natanson-Yaron S, Reich R, Yagel S, Hurwitz A.Expression and regulation of Sprouty-2 in the granulosa-lutein cells of the corpus luteum. Mol Hum Reprod 2005; 11: 537–542.. [DOI] [PubMed] [Google Scholar]
- Elvin JA, Yan C, Matzuk MM.Oocyte-expressed TGF-beta superfamily members in female fertility. Mol Cell Endocrinol 2000; 159: 1–5.. [DOI] [PubMed] [Google Scholar]
- Lyons K, Graycar JL, Lee A, Hashmi S, Lindquist PB, Chen EY, Hogan BL, Derynck R.Vgr-1, a mammalian gene related to Xenopus Vg-1, is a member of the transforming growth factor beta gene superfamily. Proc Natl Acad Sci U S A 1989; 86: 4554–4558.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons KM, Pelton RW, Hogan BL.Patterns of expression of murine Vgr-1 and BMP-2a RNA suggest that transforming growth factor-beta-like genes coordinately regulate aspects of embryonic development. Genes Dev 1989; 3: 1657–1668.. [DOI] [PubMed] [Google Scholar]
- Schmid P, Cox D, van der Putten H, McMaster GK, Bilbe G.Expression of TGF-beta s and TGF-beta type II receptor mRNAs in mouse folliculogenesis: stored maternal TGF-beta 2 message in oocytes. Biochem Biophys Res Commun 1994; 201: 649–656.. [DOI] [PubMed] [Google Scholar]
- Ghiglieri C, Khatchadourian C, Tabone E, Hendrick JC, Benahmed M, Menezo Y.Immunolocalization of transforming growth factor-beta 1 and transforming growth factor-beta 2 in the mouse ovary during gonadotrophin-induced follicular maturation. Hum Reprod 1995; 10: 2115–2119.. [DOI] [PubMed] [Google Scholar]
- Shimasaki S, Zachow RJ, Li D, Kim H, Iemura S, Ueno N, Sampath K, Chang RJ, Erickson GF.A functional bone morphogenetic protein system in the ovary. Proc Natl Acad Sci U S A 1999; 96: 7282–7287.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA.Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 1998; 92: 253–263.. [DOI] [PubMed] [Google Scholar]
- Shim K, Minowada G, Coling DE, Martin GR.Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell 2005; 8: 553–564.. [DOI] [PubMed] [Google Scholar]
- Taketomi T, Yoshiga D, Taniguchi K, Kobayashi T, Nonami A, Kato R, Sasaki M, Sasaki A, Ishibashi H, Moriyama M, Nakamura K, Nishimura J, Yoshimura A.Loss of mammalian Sprouty2 leads to enteric neuronal hyperplasia and esophageal achalasia. Nat Neurosci 2005; 8: 855–857.. [DOI] [PubMed] [Google Scholar]
- Peri F, Bokel C, Roth S.Local Gurken signaling and dynamic MAPK activation during Drosophila oogenesis. Mech Dev 1999; 81: 75–88.. [DOI] [PubMed] [Google Scholar]
- Reich A, Sapir A, Shilo B.Sprouty is a general inhibitor of receptor tyrosine kinase signaling. Development 1999; 126: 4139–4147.. [DOI] [PubMed] [Google Scholar]
- Kramer S, Okabe M, Hacohen N, Krasnow MA, Hiromi Y.Sprouty: a common antagonist of FGF and EGF signaling pathways in Drosophila. Development 1999; 126: 2515–2525.. [DOI] [PubMed] [Google Scholar]
- Casci T, Vinos J, Freeman M.Sprouty, an intracellular inhibitor of Ras signaling. Cell 1999; 96: 655–665.. [DOI] [PubMed] [Google Scholar]
- Hall AB, Jura N, DaSilva J, Jang YJ, Gong D, Bar-Sagi D.hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Curr Biol 2003; 13: 308–314.. [DOI] [PubMed] [Google Scholar]
- Rubin C, Litvak V, Medvedovsky H, Zwang Y, Lev S, Yarden Y.Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Curr Biol 2003; 13: 297–307.. [DOI] [PubMed] [Google Scholar]
- Egan JE, Hall AB, Yatsula BA, Bar-Sagi D.The bimodal regulation of epidermal growth factor signaling by human Sprouty proteins. Proc Natl Acad Sci U S A 2002; 99: 6041–6046.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, Fong CW, Lim J, Yusoff P, Low BC, Langdon WY, Guy GR.Sprouty2 attenuates epidermal growth factor receptor ubiquitylation and endocytosis, and consequently enhances Ras/ERK signalling. EMBO J 2002; 21: 4796–4808.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Shi W, Bellusci S, Groffen J, Heisterkamp N, Minoo P, Warburton D.Sprouty2 down-regulation plays a pivotal role in mediating crosstalk between TGF-beta1 signaling and EGF as well as FGF receptor tyrosine kinase-ERK pathways in mesenchymal cells. J Cell Physiol 2007; 212: 796–806.. [DOI] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell 2007; 128: 309–323.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le HV, Shen L, Anderson SA, Massague J.Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 2004; 117: 211–223.. [DOI] [PubMed] [Google Scholar]
- Cabrita MA, Christofori G.Sprouty proteins, masterminds of receptor tyrosine kinase signaling. Angiogenesis 2008; 11: 53–62.. [DOI] [PubMed] [Google Scholar]
- Sorkin A, Waters CM.Endocytosis of growth factor receptors. Bioessays 1993; 15: 375–382.. [DOI] [PubMed] [Google Scholar]
- Schmid SL.Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem 1997; 66: 511–548.. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Massague J.Opposing BMP and EGF signaling pathways converge on the TGF-beta family mediator Smad1. Nature 1997; 389: 618–622.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.