Abstract
Mechanical training of engineered tissue constructs is believed necessary to improve regeneration of cartilaginous grafts. Nevertheless, molecular mechanisms underlying mechanical activation are not clear. This is partly due to unavailability of appropriate scaffolds allowing exposure of cells to dynamic compressive strains (DCS) in vitro while permitting subsequent molecular analyses. We demonstrate that three-dimensional macroporous electrospun poly(ɛ-caprolactone) scaffolds can be fabricated that are suitable for the functional and molecular analysis of dynamically loaded chondrocytes. These scaffolds encourage chondrocytic proliferation promoting expression of collagen type II, aggrecan, and Sox9 while retaining mechanical strength after prolonged dynamic compression. Further, they exhibit superior infiltration of exogenous agents into the cells and permit easy retrieval of cellular components postcompression to allow exploration of molecular mechanisms of DCS. Using these scaffolds, we observed that chondrocytes responded to DCS in a magnitude-dependent manner exhibiting antiinflammatory and proanabolic responses at low physiological magnitudes. Proinflammatory responses and decreased cellular viability were observed at hyperphysiological magnitudes. These scaffolds provide a means of unraveling the mechanotransduction-induced transcriptional and posttranslational activities involved in cartilage regeneration and repair.
Introduction
Biomechanical forces are essential for the regulation of development, homeostasis, and regeneration of skeletal tissue. Therefore, mechanical activation of tissue-engineered constructs is currently being considered to augment their regenerative capacity. This is especially important in avascular tissues like cartilage, where such forces not only regulate metabolic activity but also facilitate the exchange of fluids, nutrients, and oxygen for resident chondrocytes. However, prior to their application in vitro and in vivo, it is critical to understand the mechanisms by which biomechanical forces may activate the regenerative potential of cells. In joints, cartilage is constantly exposed to an array of dynamic biomechanical stimuli exerting compressive, tensile, shear, hydrostatic, and osmotic forces during normal joint movement. Chondrocytes can perceive and respond to these biomechanical signals transmitted through extracellular matrix (ECM). Studies focusing on the mechanisms of actions of biomechanical forces have shown that dynamic tensile forces dramatically influence the biological responses of chondrocytes.1,2 However, the fundamental molecular mechanisms of dynamic compressive forces influencing on chondrocytes are yet to be unraveled due largely to a lack of appropriate in vitro models.
Since biomechanical forces are involved in cartilage homeostasis and repair, understanding their mechanisms has been a subject of intense inquiry. To study mechanotransduction in chondrocytes, cartilage explants have been particularly useful due to their physiologic simulation of in vivo biomechanics.3–9 Several studies have shown that the responses of the chondrocytes to mechanical forces are largely dependent on the magnitude, frequency, and duration of the applied forces.3,5–7,9,10 Generally, dynamic compression at low magnitude upregulates the anabolic metabolism like deposition of collagen type II and aggrecan4,8 while downregulating catabolic matrix degradation.4 On the other hand, compressive forces of high magnitudes are catabolic and upregulate matrix destruction.10,11
Although explants provide an outstanding model for the cartilage biomechanics, the complex environment of the tissue poses challenges when dissecting the fast evolving individual molecular events responding to the applied forces. To simplify this complexity, several in vitro models have been developed, including agarose and alginate gel systems12–18 and polymeric scaffolds.19–22 Hydrogels offer an especially amenable simulation of cartilage tissue as they surround embedded cells and provide an environment for sustaining chondrocytic phenotype in vitro.23 Using these hydrogels, dynamic compression of low magnitudes has been shown to suppress inflammation stimulated by exogenous interleukin-1β (IL-1β).13,14,24 However, the mechanical properties of hydrogels limit the extent of magnitude and frequency that can be applied due to the highly viscoelastic nature of the gels. Further, the rapid activation of embedded cells within the gels using exogenous inflammatory agents or growth factors to simulate in vivo–like environments cannot be carried out.
With these limitations in mind, we have developed a relatively open form of electrospun fiber matrix to function as a biological solid support amenable to the application of compressive forces to chondrocytes proliferating within the three-dimensional (3D) cell culture system. These scaffolds are uniquely suited to examine cellular responses under both high and low levels of dynamic compression. The scaffolds assist in maintaining the phenotypic stability of the chondrocytes in vitro and readily allow infiltration of soluble agents for the experimental manipulation of the embedded cells. Further, the mechanical properties of the fabricated scaffolds are stable and do not change over the period of sustained dynamic compression up to 40% strain for 4 h. In this report, we present data showing that these cell–scaffold constructs mimicking a full thickness of articular cartilage (∼3 mm) can be used to delineate the responses of chondrocytes to dynamic compressive forces. The effects of dynamic compressive strain (DCS) at various magnitudes in the absence or presence of an inflammatory agent were investigated to simulate conditions posed during the mechanical training of engineered tissue constructs or their implantation in diseased or inflamed joints.
Materials and Methods
Scaffold synthesis
Fifteen percent poly(ɛ-caprolactone) (PCL, Mw 65,000; Sigma-Aldrich, St. Louis, MO) dissolved in dichloromethane (Mallinckroff Baker, Phillipsburg, NJ) was electrospun onto an aluminum foil–wrapped 7.6 × 7.6 cm steel plate at −20 kV with a flow rate of 15 mL/h and a 30 cm tip-to-substrate distance. A voltage of 0 to +5 kV was gradually applied to the ground plate over the course of 1.5 h of electrospinning to compensate for the effect of insulation caused by the gradual thickening of the deposit. The as-spun fiber mesh was treated in a vacuum oven (<30 mmHg) at 45°C for 24 h to remove residual solvents.25 The approximately 3-mm-thick mesh was then cut into 6-mm-diameter cylinders using a biopsy punch (Miltex, York, PA).
Mechanical testing
Mechanical properties of scaffolds were measured under various magnitudes of DCS (15%, 30%, and 40%) with a 1 Hz saw-tooth profile on a load frame (Model 1322; Instron, Norwood, MA) using a 1 kg load cell (Model 31; Honeywell Sensotec, Chardon, OH). Prior to measurements, the samples were hydrated overnight in cell culture medium (CCM; Ham's F12 medium containing 10% fetal calf serum; 10 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine). All mechanical tests were performed under subaqueous condition in CCM.
Cell culture in 3D PCL scaffolds
Articular cartilage was harvested from the knees of 10–12-week-old female Sprague-Dawley rats (Harlan, Indianapolis, IN), and primary chondrocytes were isolated as previously described.26 All protocols were preapproved by the Institutional Animal Care and Use Committee at The Ohio State University. The cells were subcultured at a high seeding density of approximately 20,000 cells/cm2 in CCM at 37°C and 90% relative humidity. Passage 2 or 3 from primary chondrocytes was used in all experiments.
The scaffolds were sterilized overnight in 70% ethanol solution and washed with sterile water followed by overnight incubation in CCM to improve hydrophilicity. Prior to cell seeding, the medium present in the scaffolds was aspirated, and approximately 400,000 cells suspended in 40 μL of CCM were seeded into each scaffold placed in a 24-well plate. The cell–scaffold constructs were incubated for 4 h at 37°C, before supplementing an additional 2 mL of CCM into each well. The cell–scaffold constructs were cultured for 6 days prior to the application of compressive forces. Twelve hours prior to compression, the cell–scaffold constructs were serum-restricted by replacing CCM with CCM containing 1% FBS.
Application of dynamic compression
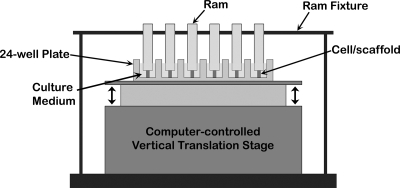
The cell–scaffold constructs were subjected to cyclic unconfined DCS at 1 Hz with a saw-tooth profile for 4 h, using a custom-designed, computer-controlled device consisting of a vertical translation stage and a servo controller (AVL125 and Soloist, respectively; Aerotech, Pittsburgh, PA) (Fig. 1). The samples were preloaded with a ram weighing approximately 2.5 g and fastened with a ram fixture. The loading with the ram resulted in only 0.8% strain on the scaffold; preliminary experiments showed no demonstrable differences in gene expression between samples with or without the rams. The entire setup was then placed in a cell culture incubator at 37°C during the application of compression. The cell–scaffold constructs were exposed to various DCS in the absence or presence of recombinant human interleukin-1β (rhIL-1β, 1 ng/mL; Calbiochem, San Diego, CA). The experimental protocols consisted of (i) control uncompressed cell–scaffold constructs; (ii, iii, and iv) cell–scaffold constructs exposed to 15%, 30%, or 40% DCS, respectively, without IL-1β supplementation; (v) uncompressed cell–scaffold constructs treated with IL-1β (1 ng/mL); (vi, vii, and viii) cell–scaffold constructs exposed to 15%, 30%, or 40% DCS, respectively, in the presence of IL-1β (1 ng/mL).
FIG. 1.
A schematic illustration of the device used for the dynamic compression of the cell–scaffold constructs.
Scanning electron microscopy
Scanning electron microscopy (SEM) was carried out as described previously.27 Briefly, cell–scaffold constructs cultured for 6 days were fixed with 10% formalin (Richard-Allen Scientific, Kalamazoo, MI) and dehydrated in a graded series of ethanol in deionized water (50%, 70%, 85%, 90%, and 100% ethanol) followed by removal of ethanol in a graded series of 25%, 50%, 75%, and 100% ethanol-hexamethyldisilazane (HMDS; Electron Microscopy Sciences, Hatfield, PA). The dehydrated samples were coated with an 8-nm-thick layer of osmium (OPC-80T; SPI Supplies, West Chester, PA). In addition, the cross section of acellular fibers was observed by fracturing in liquid nitrogen prior to osmium coating. An FEI XL-30 Sirion SEM (Hillsboro, OR) with a field emission gun source was used to examine the samples.
Cryosection and immunofluorescence microscopy
Following culture for 6 days, cell–scaffold constructs were fixed with 10% formalin followed by three 10 min rinses in phosphate-buffered saline (PBS; Gibco, Carlsbad, CA). The fixed samples were embedded in optimal cutting temperature (OCT) compound (Sakura Finetek, Torrence, CA) and then frozen at −80°C. The frozen samples were vertically cut to 12-μm sections using a cryostat (CM3050S; Leica Microsystems, Bannockburn, IL) and placed onto glass slides (ThermoFisher Scientific, Fairlawn, NJ).
The cryosectioned samples were rinsed four times with PBS to remove residual OCT and permeabilized with 0.2% TritonX-100 (Sigma-Aldrich) for 30 min. Subsequently, cells were stained with 16.6 pM of fluorescein isothiocyanate (FITC)–labeled phalloidin (Invitrogen, Carlsbad, CA) in PBS containing 0.5% bovine serum albumin and 0.02% sodium azide (Sigma-Aldrich) for 20 min. The stained samples were counterstained with 0.01% 4′,6-diamidino-2-phenyl-indole dihydrochloride (DAPI; Invitrogen) for 5 min and observed using an Axioplan2 epifluorescence microscope (Zeiss MicroImaging, Thornwood, NY).
For the observation of nuclear translocation of nuclear factor-kappa B (NF-κB), cell–scaffold constructs were fixed with 2% paraformaldehyde following exposure to IL-1β–containing CCM for 15 min. The fixed cell–scaffold constructs were horizontally cut in the middle followed by cell permeabilization. Cells were then stained with rabbit anti-NF-κB p65 primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and CY3 anti-rabbit secondary antibody (The Jackson Laboratory, Bar Harbor, ME). Subsequently, the samples were counterstained with DAPI, and observed using a confocal microscope (LSM510; Zeiss MicroImaging).
Real-time polymerase chain reaction
Two samples from each condition were pooled immediately after various treatments, and total RNA was extracted using the RNeasy Micro Kit (Qiagen, Valencia, CA). RNA (1 μg) was measured using the Nanodrop spectrophotometer (ND-1000; Nanodrop Tech, Wilmington, DE), and first-strand synthesis performed using the Superscript III Reverse Transcriptase Kit (Invitrogen). Custom-designed gene-specific primers were used to amplify the cDNA using the Bio-Rad SYBR Green Master Mix (Bio-Rad; Hercules, CA) in the iCycler iQ real-time polymerase chain reaction (RT-PCR) system (Bio-Rad). Primers used are shown in Table 1. The thermocycling protocol was 95°C for 3 min, 40 cycles of denaturation at 95°C for 30 s, annealing at 63.5°C for 30 s, and extension at 72°C for 30 s. Collected data were analyzed by the comparative threshold cycle (CT) method.28
Table 1.
Oligonucleotide Primers Used for Real-Time Polymerase Chain Reaction
| Gene | Sense (5′–3′) | Antisense (5′–3′) | Length (bp) | Accession no. |
|---|---|---|---|---|
| Rps18 | GCGGCGGAAAATAGCCTTCG | GGCCAGTGGTCTTGGTGTGCTG | 356 | NM213557 |
| Col2a1 | ATGAGGGCCGAGGGCAACAG | GATGTCCATGGGTGCAATGTCAA | 150 | NM012929 |
| Acan | GAGAGGGGTGAATGGAACGATGTC | ATCTTTCTTCTGCCCAAGGGTTCTG | 126 | NM022190 |
| Sox9 | ATCTGAAGAAGGAGAGCGAG | CAAGCTCTGGAGACTGCTGA | 264 | AB073720 |
| Nos2 | CACCTCACTGTGGCTGTGGTCAC | GCACCCAAACACCAAGGTCATG | 285 | NM012611 |
| Mmp13 | CATACGAGCATCCATCCCGAGAC | CTCAAAGTGAACCGCAGCACTGAG | 158 | XM343345 |
Western blot analysis
Protein synthesis in the cells under each condition was examined by Western blot analysis. Following the application of DCS for 4 h, cell–scaffold constructs were incubated for 20 h in CCM, and three samples exposed to the same conditions were pooled for protein extraction. Total cellular protein was extracted in radioimmunoprecipitation assay (RIPA; Santa Cruz Biotechnology) buffer supplemented with Protease Inhibitor Cocktail (Roche, Indianapolis, IN) and Phosphatase Inhibitor Cocktail 2 (Sigma-Aldrich). The extracted proteins were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and then electrophoretically transferred onto nitrocellulose membranes (Bio-Rad). The membranes were probed with rabbit anti-iNOS, rabbit anti-MMP13 or mouse anti-SOX9 antibodies (Santa Cruz Biotechnology). The binding of primary antibodies was detected with an IR-Dye 680–conjugated secondary antibody (LI-COR Biosciences, Lincoln, NE) and scanned using an infrared imaging system (Odyssey; LI-COR Biosciences). For the normalization of protein loading, the same membranes were subsequently probed with mouse anti-β-Actin (Sigma-Aldrich) and detected using an IR-Dye 800–conjugated secondary antibody.
Cell viability
The cell viability of various samples was examined by the 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) cell proliferation assay (ATCC, Manassas, VA). The cell–scaffold constructs exposed to 15%, 30%, or 40% DCS for 4 h were incubated for 20 h in CCM prior to the viability test. Untreated samples were used as controls. Tetrazolium salt conversion to formazan was detected by absorbance at 450 nm using a Victor3 multilabel plate reader (PerkinElmer, Waltham, MA).
Statistical analysis
For the mechanical testing, seven acellular scaffolds per condition (15%, 30%, and 40% DCS) were used for data acquisition, and the data presented as mean ± SD (standard deviation). For all other experiments, at least three independent experiments were conducted using chondrocytes harvested from different animals. Data were analyzed using SPSS (v.15.0) by one-way ANOVA with Tukey's Honestly Significant Difference (HSD) post hoc test. p < 0.05 was regarded as statistically significant. The measurements for gene expression with RT-PCR and cell viability using the MTT assay were performed in duplicate in three separate experiments (n = 6), and presented as mean ± standard error of the mean (SEM).
Results
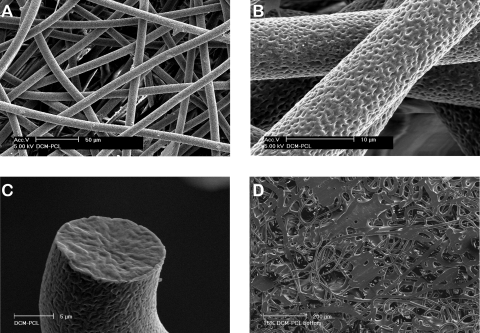
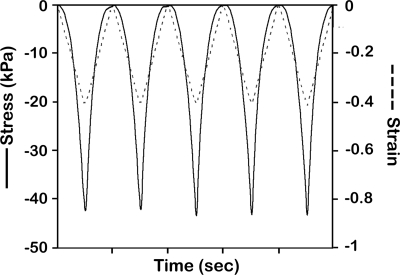
Unlike conventional nano-sized electrospun fibers, the modified electrospun PCL fibers exhibited an average fiber diameter of 10 μm with polygonal-shaped macropores ranging from 50 to 100 μm in size (Fig. 2A). This resulted in an approximately 90% relative porosity calculated from the measured dimension and weight of the scaffold. The process modification also generated a network of nanoscalar pores across the surface of the fibers that potentially enhanced cellular attachment (Fig. 2B). The observation of fracture surface revealed that the pores only present on the surface (Fig. 2C). The bottom side of the electrospun scaffolds was relatively dense, enabling the scaffold to retain both the cells and the CCM upon seeding (Fig. 2D). The baseline mechanical properties of approximately 3-mm-thick scaffolds were assessed (Fig. 3). Exposure of scaffolds to 15%, 30%, or 40% DCS for 4 h resulted in the peak stresses of 3.91 (±1.81), 18.40 (±3.81), and 45.01 (±7.75) kPa, respectively. These experiments revealed that a maximal dynamic strain of 40% could be reproducibly applied to the scaffolds without plastic deformation over the course of 4 h. Thus, the sample “lift-off ” problem often associated with hydrogel-based systems was not observed under any conditions employed in this study.16,17
FIG. 2.
SEM images of acellular electrospun scaffolds showing (A) the initial microstructure of the fibrous network, (B) nanoscalar pores on the surface of fibers that promote cell attachment, (C) a cross section of the fiber revealing the absence of pores in the core, and (D) the relatively dense flattened bottom surface of the scaffolds to retard cell migration to the culture plate.
FIG. 3.
Periodic stress variation experienced by the scaffold during 40% DCS at 1 Hz.
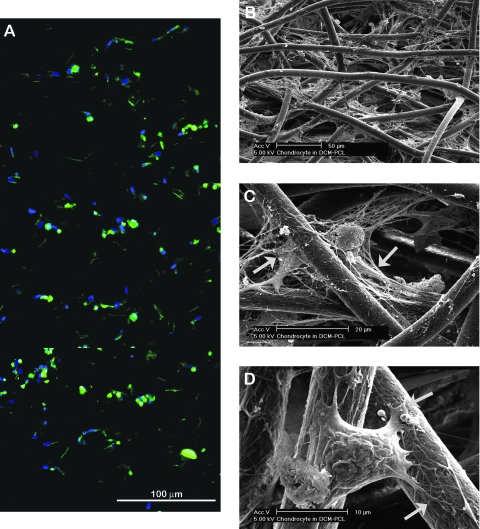
The distribution of cells in scaffolds was observed following staining of cellular actin with FITC phalloidin and nuclei with DAPI (Fig. 4A). Although the cell seeding was performed only under gravity, the capillary force generated by the network of randomly oriented fibers allowed efficient permeation of the cell/CCM mixture and resulted in a relatively uniform cellular distribution throughout the height of scaffolds. SEM revealed that an appreciable amount of ECM was deposited in the scaffolds (Fig. 4B, C). The cell-surrounding ECM interconnected adjacent PCL fibers suggests that matrix deposition may assist in the efficient transmission of the applied mechanical forces through the fibrous network. Figure 4D shows the nature of cellular adhesion on the fibers. The nanopores on the surface of fibers appear to be used as attachment/anchoring points as evident in the contact points of cellular processes.
FIG. 4.
Characteristics of chondrocyte distribution and matrix deposition in scaffolds. (A) A vertical cross-sectional image of a cell–scaffold construct at day 6 showing a relatively uniform cellular distribution throughout its height (actin, green; nuclei, blue). (B, C) SEM images of cell–scaffold construct at day 6 showing extensive ECM deposition connecting/surrounding cells and PCL fibers (indicated by asterisk and arrows, respectively). (D) A chondrocyte spanning over adjacent fibers and anchoring via nanopores to the fiber surface (indicated by arrows). Color images available online at www.liebertonline.com/ten.
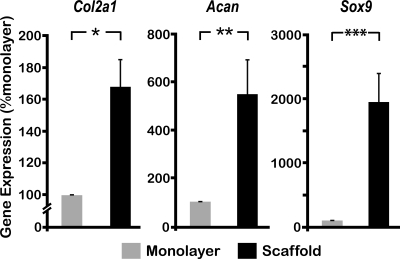
The phenotypic markers of chondrocytes in the cell–scaffold constructs were compared to those of cells cultured in a high-density monolayer (Fig. 5). The cells in scaffolds exhibited a significant upregulation of all the chondrocytic markers tested, including collagen type II (Col2a1), Aggrecan (Acan), and SRY (sex-determining region Y)-box 9 (Sox9). Of note is that the expression of Sox9, a transcription factor known to modulate chondrogenesis and directly regulate the transcription of Col2a1,29 was approximately 20-fold upregulated in the cell–scaffold construct.
FIG. 5.
Phenotypic characterization of chondrocytes. Chondrocytes were either grown in high-density monolayer or in scaffolds for 6 days. Subsequently, mRNA expression for Col2a1, Acan, and Sox9 was examined by RT-PCR, using the 18S ribosomal RNA gene (Rps18) as an internal control to normalize relative gene expression levels. Bars represent mean and SEM (n = 6), *p < 0.05, **p < 0.01, and ***p < 0.001.
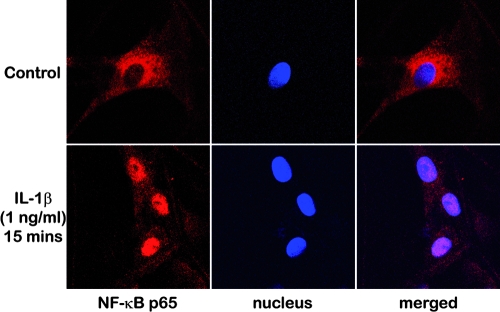
The permeability of exogenous agents was examined by exposing the constructs to IL-1β–containing CCM and observing the localization of NF-κB subunit p65 within the cells located in the center of the constructs (Fig. 6). Nuclear translocation of NF-κB was observed within 15 min of IL-1β stimulation, suggesting that the exogenous agent rapidly infiltrated into the constructs. On the other hand, the continuing presence of NF-κB in the cytoplasm was observed in the untreated control samples.
FIG. 6.
Confocal microscopy images (400×) of the cells located in the center of the cell–scaffold constructs revealing the intracellular distribution of NF-κB subunit p65 (p65, red; nuclei, blue); cytoplasmic NF-κB p65 observed in the untreated control samples translocated into nuclei following exposure to IL-1β containing CCM for 15 min. Color images available online at www.liebertonline.com/ten.
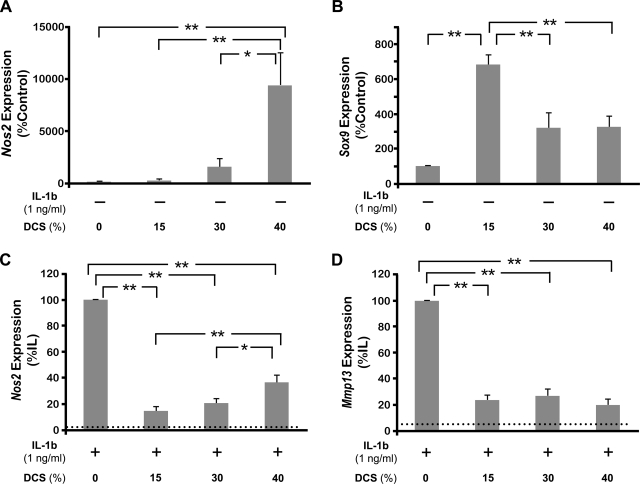
To investigate the feasibility of using these scaffolds to study the responses of the chondrocytes to biomechanical forces, DCS of various magnitudes was applied to the cell–scaffold constructs, and the expression of inducible nitric oxide synthase (iNOS/Nos2) and Sox9 assessed. Based on previous studies,30–32 15% was selected as a representative level of physiological strain, while 40%, the maximum magnitude applicable in this system, was chosen to represent a nonphysiological level of strain. In these experiments, 30% DCS represented an intermediate strain approximately twice physiological loading. As shown in Figure 7A, Nos2 mRNA induction increased with the magnitude of DCS. In fact, 40% DCS induced a significantly greater amount of this inflammatory molecule compared to the untreated chondrocytes–scaffold constructs and those exposed to 15% or 30% DCS. For the assessment of the anabolic effects of compressive force, the regulation of Sox9 by biomechanical forces was examined (Fig. 7B). Compressive forces significantly upregulated Sox9 mRNA expression following exposure to 15% DCS. Exposure to 30% or 40% DCS, however, did not result in comparable upregulation of Sox9.
FIG. 7.
Effect of DCS of various magnitudes on the gene expression of chondrocytes in the absence or presence of IL-1β. Cells were exposed to 15%, 30%, or 40% DCS at 1 Hz in the absence of IL-1β for 4 h and analyzed for the mRNA expression for (A) Nos2 and (B) Sox9. Alternatively, cells were exposed to compressive forces of various magnitudes in the presence of IL-1β for 4 h and the expression of mRNA for (C) Nos2 and (D) Mmp13 examined by RT-PCR. Dotted lines in (C) and (D) represent untreated control levels. Bars represent mean and SEM (n = 6), *p < 0.05 and **p < 0.01. DCS, dynamic compressive strain.
We next investigated the feasibility of using these scaffolds to study the responses of the cells to exogenously applied agents alone or in combination with compression in vitro. In these experiments, various magnitudes of DCS were applied to the cell–scaffold constructs following the addition of IL-1β in the medium to simulate inflammatory conditions. As shown in Figure 7C, Nos2 expression was significantly upregulated by IL-1β. The application of dynamic compression at all magnitudes suppressed IL-1β–induced Nos2 expression; however, a maximal suppression (88%) was observed at 15% DCS. Additionally, the amount of suppression inversely related to the magnitude of DCS applied; that is, 15% DCS suppressed significantly greater amount of Nos2 expression compared to 40% DCS. Matrix metalloproteinase 13 (Mmp13/MMP13), another inflammatory gene, was also suppressed by dynamic compression (Fig. 7D). However, there were no apparent differences in the amount of Mmp13 suppression in response to different magnitudes of applied DCS.
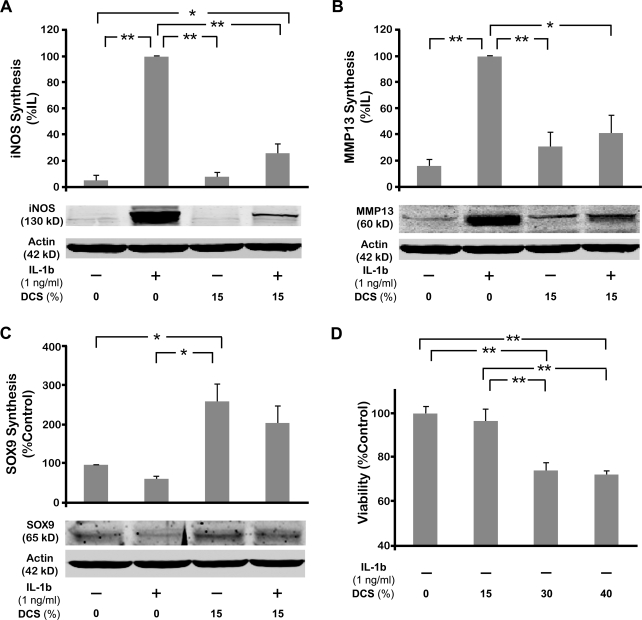
To confirm the changes in gene expression by dynamic compression, the protein syntheses of iNOS/NOS2, MMP13, and SOX9 were examined by Western blot analysis (Fig. 8A–C). As expected, iNOS synthesis was markedly upregulated by the presence of IL-1β (Fig. 8A). This upregulation of IL-1β–mediated iNOS synthesis was significantly inhibited by the application of dynamic compression as shown in 15% DSC. This trend also continued in case of MMP13 protein, demonstrating that DCS inhibited IL-1β–induced MMP13 synthesis (Fig. 8B). In contrast, SOX9 protein synthesis was appreciably downregulated by IL-1β (Fig. 8C). The suppression of SOX9 synthesis by IL-1β was recovered by the dynamic compression in appreciable amount. In addition, exposure of cells to 15% DCS alone significantly upregulated SOX9 synthesis consistent with the gene expression changes observed.
FIG. 8.
Effect of various magnitudes of DCS on the protein synthesis in chondrocytes in the absence or presence of IL-1β. Cells were exposed to 15% DCS in the absence or presence of IL-1β for 4 h, and allowed to rest in the IL-1β–containing medium for the next 20 h. Subsequently, (A) iNOS/NOS2, (B) MMP13, and (C) SOX9 protein synthesis was examined by Western blot analysis. (D) Viability of the cells following application of compressive forces. Cells were subjected to DCSs of various magnitudes for 4 h and allowed to rest for ensuing 20 h. The cell viability was analyzed by MTT assay. Bars represent mean and SEM in (A–C) (n = 3) and in (D) (n = 6). The Western blots in panels (A–C) are representative of one of three separate experiments. *p < 0.05, **p < 0.01. DCS, dynamic compressive strain.
Interestingly, no obvious regulation of protein synthesis was observed in chondrocytes exposed to 30% and 40% DCS. In fact, the concentration of proteins extracted from these samples was considerably lower (approximately 30%) than that of untreated control cells and cells exposed to 15% DCS. We speculated that the decrease in the concentration of proteins following treatment with 30% and 40% DCS may be due to decreased cellular viability during postcompression incubation. Hence, the viability of the cells under various magnitudes of DCS was examined at the same time point with the protein analysis (20 h after compression) (Fig. 8D). As expected, cellular viability significantly decreased up to 28% in the cells exposed to high levels of compression (30% and 40% DCS). Further, cells dynamically compressed at 15% DCS did not exhibit a significant decrease in the cellular viability, as compared to untreated controls.
Discussion
Electrospinning has drawn interest from scientists in the field of tissue engineering due to its ability to produce nano- to micron-sized fibrous structure that greatly resembles ECM. Especially for chondrocytes, electrospun matrices have been shown to promote proliferation while maintaining phenotype.33,34 However, the small fiber size that provides such advantages can itself hinder efficient cellular infiltration as the mean pore radius in these scaffolds varies with fiber diameter.35 For example, a 100 nm fiber diameter yields a mean pore radius <10 nm, which obstructs cellular infiltration, resulting in a nonuniform distribution of seeded cells. To circumvent this problem, several methods have been proposed, including the use of porogen to produce larger pores within the nano-fibrous structure27 and in situ cell seeding during electrospinning.36 In this study, we altered the “normal” electrospinning protocols to produce thicker fibers exhibiting considerably larger pore sizes by controlling solvent choice and polymer solution concentration. We have observed that the increased fiber diameter and pore size in the scaffolds rendered relatively uniform spatial distribution of chondrocytes throughout the full height of scaffolds. In addition, these scaffolds clearly promoted the phenotypic characteristics of chondrocytes as evident by the increased expression of the Sox9 gene, known to directly upregulate the transcriptional activity of collagen type II, the primary structural protein of cartilage.29 It is likely that the 3D microenvironment of these scaffolds may be important in promoting the chondrocytic phenotype. In fact, prechondrocytic cells grown in 3D micromass cultures have also been shown to differentiate along the chondrocytic lineage and express increased levels of Sox9, Col2a1, and Acan.37
We have observed that appreciable amount of ECM was deposited after 6 days of culture. A longer culture duration induced more extensive ECM deposition resulting in more rounded chondrocyte morphology surrounded by matrix usually found in a normal cartilage (data not shown). Further, it was observed that exogenous agents readily infiltrate into the constructs and activate residing chondrocytes based on the observation of NF-κB nuclear translocation, an early indication of cellular response to IL-1β stimulation.1,2 This allowed us to simulate an inflammatory condition posed during the implantation of engineered tissue constructs. In contrast, explants and hydrogel models often require prolonged incubation in an exogenous substance-containing medium due to their inherently poor permeability.38 A balance between permeability and mechanotransducibility is regulated by the density of either cell-secreted ECM or synthetic scaffold forms. From this perspective, the deposited ECM surrounding chondrocytes and connecting fibers in the present scaffolds appear to effectively transmit the applied forces and exogenous substances to the cells when applied alone or in combinations. An additional and significant advantage of this moderately open system is the simplicity of extracting cellular products for molecular analysis without interference or contamination of the components of the scaffolds. The average A260/280 absorption ratio of RNA extracts from 24 samples was 2.03 (±0.05), indicating that good-quality RNA was extracted from this system without contamination.
To examine how biomechanical forces affect the cells in inflammation, proinflammatory mediators such as Nos2 (iNOS) and Mmp13, and an anabolic gene like Sox9 were monitored in conjunction with DCS. IL-1 has been shown to mediate the cartilage degradation in inflammatory diseases such as osteoarthritis and rheumatoid arthritis,39 by upregulating several cartilage destructive genes, including Nos2 and Mmp13, and suppressing the expression of catabolic gene like Sox9. iNOS translated from Nos2 gene is crucial for the production of NO involved in cartilage catabolism.40 MMP13 is another proinflammatory protein that cleaves the ECM, especially collagen type II, to initiate cartilage pathologies.41 On the other hand, SOX9 is an essential transcription factor regulating chondrogenesis during development and is involved in the anabolic activity of chondrocytes by directly controlling collagen type II gene.29 In this study, chondrocytes cultured in these scaffolds perceived signals induced by IL-1β and responded by eliciting all of the above proinflammatory mediators, further demonstrating the utility of these scaffolds in tracking metabolic changes in chondrocytes.
Further advantage of this scaffold is the feasibility of exposing cells to higher degree of strain, as compared to hydrogel systems. Although the hydrogel models provide an excellent tool for studying the responses of the cells under load,18,42 the maximum amount of strain that can be applied is limited by the high viscoelasticity of the gels.16,17 Mauck et al.16 have reported problems associated with lift-off of 2% agarose gel constructs, due to slow recovery of the sample height when exposed to 20% strain at 1 Hz. In this regard, we have observed that the electrospun PCL scaffold maintained its elasticity up to 40% DCS. These essential benefits appear to stem from both the physicochemical nature of the PCL and the highly pliable electrospun structure.43
High applicable strain in this system allowed us to observe the chondrocytes' responses to biomechanical forces in a magnitude-dependent manner. At low magnitudes (15% DCS), mechanical forces did not influence the Nos2 expression, while significant Nos2 mRNA (100-fold compared to control) was induced at higher magnitudes (40% DCS). This suggests that chondrocytes embedded in these scaffolds can recognize differences in the magnitudes of mechanical signals and respond to them by eliciting biological responses. Similarly, Sox9 was differentially regulated by the different magnitudes of DCS exhibiting the most significant induction at 15%. Interestingly, it was also upregulated at higher magnitudes compared to controls although they were not statistically significant (p-values of 0.106 and 0.098 for 30% and 40% DCS with respect to controls, respectively). This could be an injury response of chondrocytes for recovery attempt against the detrimental mechanical forces as it was shown that high magnitudes of DCS eventually led to decreased cellular viability. Salminen et al.44 observed a similar cellular response in a mouse model showing that some of the chondrocytes adjacent to cartilage lesions strongly express Sox9, while others are metabolically inactive.
While testing the simultaneous effects of IL-1β and DCS, we observed that 15% DCS was sufficient to inhibit approximately 88% of the IL-1β–induced Nos2 expression. These findings are similar to those observed previously.14 Interestingly, we have observed that cells subjected to DCS of 30% or 40% also exhibited a decrease in NOS2 and MMP13 expression. Since approximately 30% of the cells were dead by following the application of these forces, it is likely that the remaining cells were also damaged and thus did not respond to IL-1β. Although it was shown that low physiological forces do not affect IL-1 receptor2 the effects of high magnitudes are yet to be seen. Alternatively, there may be a force gradient that cells perceive in a scaffold during higher dynamic compression; that is, the cells near the bottom may not experience levels of compression similar to those at the top. This observation coincides with the depth-dependent anisotropic response of chondrocytes in cartilage explants under compression. Longitudinal anisotropy of cartilages has been experimentally observed in cartilage explants with depth-dependent differential strain45 and modulus,46 and has been computationally simulated.47,48 Further, Wong et al.49 have shown a correlation between the zonal strain and the differential biosynthesis of chondrocytes in cartilage explants under static loading. Torzilli et al.50 also demonstrated that cellular viability was affected only near the articular surface when various static loads were applied to bovine articular cartilage. Further, it was shown that the depth of cell death remains stagnant until the strain was applied over 50%,50 similar to our observation of comparable cellular viability at 30% and 40%. We believe that the zonal analysis of the cell–scaffold construct in our study may resemble the depth-dependent differential responses of chondrocytes.
Armstrong et al.30 and Macirowski et al.31 reported up to 20% strain in vitro and an order of 10% strain in vivo in the articular cartilage of the human hip joint under physiological loads, respectively. Similarly, Park et al.32 showed that peak compressive strain amplitudes of 15.8% at 0.1 Hz and 12.7% at 1 Hz using bovine articular cartilage explants. In this context, our observations showing magnitude-dependent induction/suppression of the inflammatory mediators suggest that the chondrocytes in the scaffolds mimic responses to loading conditions similar to that observed in vivo and in vitro in above studies. For example, 15% DCS is close to physiological levels and exhibits antiinflammatory actions by suppressing NOS2 mRNA upregulation, whereas 30% or 40% DCS appears to be hyperphysiologic and is detrimental to the cells.
Our data show that these scaffolds also allow examination of the relative extent of cellular viability using established procedures. At the functional level, our results were able to show that DCS at high magnitudes is detrimental, and the observed decrease in the protein synthesis in response to 30% or 40% DCS is likely due to cell death. On the other hand, a physiological level of DCS promotes anabolic activities of chondrocytes and suppresses inflammation. In tissue engineering perspective, these findings suggest that the application of a proper magnitude of mechanical signals would enhance the regenerative capacity of the cell–scaffold constructs. Appropriate biomechanical signals would upregulate synthesis of anabolic proteins associated with tissue regeneration, while they inhibit inflammation at the surgical site. In the future, these scaffolds could allow further examination of chondrocyte signaling pathways under dynamic compression, thus providing a molecular basis for biomechanical activation of cell–scaffold constructs as a means of augmenting the regenerative potential of tissue-engineered scaffolds.
Conclusions
Overall, present study demonstrated synthesis and characterization of an in vitro culture system using 3D electrospun PCL scaffolds, which can be used to examine the functional and molecular consequences of dynamic compressive loading. This specific form of electrospun macroporous PCL scaffolds are capable of addressing the issues associated with efficient cell seeding, dispersion and attachment, exogenous agent permeability, chondrocytic phenotypic maintenance, and biomechanical dynamic stress responsiveness. Based on our findings, we believe that 3D scaffolds of electrospun fibers can be utilized in bioreactors to enhance biomechanical functionalization of engineered tissues. These cell–scaffold constructs exhibit excellent characteristics for the mechanical activation of cells as well as the examination of functional and molecular changes induced by dynamic compressive forces. Further, these scaffolds are permeable to soluble activating agents that allow the easy examination of the effects of exogenous stimuli and compressive forces simultaneously. More importantly, by utilization of these scaffolds, the magnitude-dependent responses of cells can be examined at the biochemical and molecular levels due to the relative ease of extracting high-quality mRNA, proteins, and cellular metabolites.
Acknowledgments
This study was supported by National Institutes of Health Grants NCCAM AT000646, NIDCR DE015399, and NIAMS AR048781.
References
- 1.Dossumbekova A. Anghelina M. Madhavan S. He L.L. Quan N. Knobloch T. Agarwal S. Biomechanical signals inhibit IKK activity to attenuate NF-kappa B transcription activity in inflamed chondrocytes. Arthritis Rheum. 2007;56:3284. doi: 10.1002/art.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Madhavan S. Anghelina M. Sjostrom D. Dossumbekova A. Guttridge D.C. Agarwal S. Biomechanical signals suppress TAK1 activation to inhibit NF-kappa B transcriptional activation in fibrochondrocytes. J Immunol. 2007;179:6246. doi: 10.4049/jimmunol.179.9.6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehrenbacher A. Steck E. Rickert M. Roth W. Richter W. Rapid regulation of collagen but not metalloproteinase 1, 3, 13, 14 and tissue inhibitor of metalloproteinase 1, 2, 3 expression in response to mechanical loading of cartilage explants in vitro. Arch Biochem Biophys. 2003;410:39. doi: 10.1016/s0003-9861(02)00658-6. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald J.B. Jin M. Grodzinsky A.J. Shear and compression differentially regulate clusters of functionally related temporal transcription patterns in cartilage tissue. J Biol Chem. 2006;281:24095. doi: 10.1074/jbc.M510858200. [DOI] [PubMed] [Google Scholar]
- 5.Giannoni P. Siegrist M. Hunziker E.B. Wong M. The mechanosensitivity of cartilage oligomeric matrix protein (COMP) Biorheology. 2003;40:101. [PubMed] [Google Scholar]
- 6.Murata M. Bonassar L.J. Wright M. Mankin H.J. Towle C.A. A role for the interleukin-1 receptor in the pathway linking static mechanical compression to decreased proteoglycan synthesis in surface articular cartilage. Arch Biochem Biophys. 2003;413:229. doi: 10.1016/s0003-9861(03)00129-2. [DOI] [PubMed] [Google Scholar]
- 7.Valhmu W.B. Stazzone E.J. Bachrach N.M. Saed-Nejad F. Fischer S.G. Mow V.C. Ratcliffe A. Load-controlled compression of articular cartilage induces a transient stimulation of aggrecan gene expression. Arch Biochem Biophys. 1998;353:29. doi: 10.1006/abbi.1998.0633. [DOI] [PubMed] [Google Scholar]
- 8.Sauerland K. Raiss R.X. Steinmeyer J. Proteoglycan metabolism and viability of articular cartilage explants as modulated by the frequency of intermittent loading. Osteoarthritis Cartilage. 2003;11:343. doi: 10.1016/s1063-4584(03)00007-4. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald J.B. Jin M. Dean D. Wood D.J. Zheng M.H. Grodzinsky A.J. Mechanical compression of cartilage explants induces multiple time-dependent gene expression patterns and involves intracellular calcium and cyclic AMP. J Biol Chem. 2004;279:19502. doi: 10.1074/jbc.M400437200. [DOI] [PubMed] [Google Scholar]
- 10.Thibault M. Poole A.R. Buschmann M.D. Cyclic compression of cartilage/bone explants in vitro leads to physical weakening, mechanical breakdown of collagen and release of matrix fragments. J Orthop Res. 2002;20:1265. doi: 10.1016/S0736-0266(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 11.Piscoya J.L. Fermor B. Kraus V.B. Stabler T.V. Guilak F. The influence of mechanical compression on the induction of osteoarthritis-related biomarkers in articular cartilage explants. Osteoarthritis Cartilage. 2005;13:1092. doi: 10.1016/j.joca.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 12.Chowdhury T.T. Appleby R.N. Salter D.M. Bader D.A. Lee D.A. Integrin-mediated mechanotransduction in IL-1 beta stimulated chondrocytes. Biomech Model Mechanobiol. 2006;5:192. doi: 10.1007/s10237-006-0032-3. [DOI] [PubMed] [Google Scholar]
- 13.Chowdhury T.T. Bader D.L. Lee D.A. Anti-inflammatory effects of IL-4 and dynamic compression in IL-1 beta stimulated chondrocytes. Biochem Biophys Res Commun. 2006;339:241. doi: 10.1016/j.bbrc.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Chowdhury T.T. Bader D.L. Lee D.A. Dynamic compression inhibits the synthesis of nitric oxide and PGE(2) by IL-1 beta-stimulated chondrocytes cultured in agarose constructs. Biochem Biophys Res Commun. 2001;285:1168. doi: 10.1006/bbrc.2001.5311. [DOI] [PubMed] [Google Scholar]
- 15.Mauck R.L. Byers B.A. Yuan X. Tuan R.S. Regulation of cartilaginous ECM gene transcription by chondrocytes and MSCs in 3D culture in response to dynamic loading. Biomech Model Mechanobiol. 2007;6:113. doi: 10.1007/s10237-006-0042-1. [DOI] [PubMed] [Google Scholar]
- 16.Mauck R.L. Soltz M.A. Wang C.C.B. Wong D.D. Chao P.H.G. Valhmu W.B. Hung C.T. Ateshian G.A. Functional tissue engineering of articular cartilage through dynamic loading of chondrocyte-seeded agarose gels. J Biomech Eng Trans ASME. 2000;122:252. doi: 10.1115/1.429656. [DOI] [PubMed] [Google Scholar]
- 17.Mouw J.K. Connelly J.T. Wilson C.G. Michael K.E. Levenston M.E. Dynamic compression regulates the expression and synthesis of chondrocyte-specific matrix molecules in bone marrow stromal cells. Stem Cells. 2007;25:655. doi: 10.1634/stemcells.2006-0435. [DOI] [PubMed] [Google Scholar]
- 18.Sharma G. Saxena R.K. Mishra P. Differential effects of cyclic and static pressure on biochemical and morphological properties of chondrocytes from articular cartilage. Clin Biomech. 2007;22:248. doi: 10.1016/j.clinbiomech.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Grad S. Gogolewski S. Alini M. Wimmer M.A. Effects of simple and complex motion patterns on gene expression of chondrocytes seeded in 3D scaffolds. Tissue Eng. 2006;12:3171. doi: 10.1089/ten.2006.12.3171. [DOI] [PubMed] [Google Scholar]
- 20.Hunter C.J. Imler S.M. Malaviya P. Nerem R.M. Levenston M.E. Mechanical compression alters gene expression and extracellular matrix synthesis by chondrocytes cultured in collagen I gels. Biomaterials. 2002;23:1249. doi: 10.1016/s0142-9612(01)00245-9. [DOI] [PubMed] [Google Scholar]
- 21.Lee C.R. Grodzinsky A.J. Spector A. Biosynthetic response of passaged chondrocytes in a type II collagen scaffold to mechanical compression. J Biomed Mater Res A. 2003;64A:560. doi: 10.1002/jbm.a.10443. [DOI] [PubMed] [Google Scholar]
- 22.Xie J. Han Z.Y. Kim S.H. Kim Y.H. Matsuda T. Mechanical loading-dependence of mRNA expressions of extracellular matrices of chondrocytes inoculated into elastomeric microporous poly (L-lactide-co-epsilon-caprolactone) scaffold. Tissue Eng. 2007;13:29. doi: 10.1089/ten.2006.0060. [DOI] [PubMed] [Google Scholar]
- 23.Aydelotte M.B. Kuettner K.E. Differences between sub-populations of cultured bovine articular chondrocytes.1. Morphology and cartilage matrix production. Connective Tissue Res. 1988;18:205. doi: 10.3109/03008208809016808. [DOI] [PubMed] [Google Scholar]
- 24.Chowdhury T.T. Bader D.L. Lee D.A. Dynamic compression counteracts IL-1 beta induced iNOS and COX-2 activity by human chondrocytes cultured in agarose constructs. Biorheology. 2006;43:413. [PubMed] [Google Scholar]
- 25.Nam J. Huang Y. Agarwal S. Lannutti J.J. Materials selection and residual solvent retention in biodegradable electrospun fibers. J Appl Polym Sci. 2008;107:1547. [Google Scholar]
- 26.Madhavan S. Anghelina M. Rath-Deschner B. Wypasek E. John A. Deschner J. Piesco N. Agarwal S. Biomechanical signals exert sustained attenuation of proinflammatory gene induction in articular chondrocytes. Osteoarthritis Cartilage. 2006;14:1023. doi: 10.1016/j.joca.2006.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nam J. Huang Y. Agarwal S. Lannutti J.J. Improved cellular infiltration in electrospun fiber via engineered porosity. Tissue Eng. 2007;13:2249. doi: 10.1089/ten.2006.0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak K.J. Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Bell D.M. Leung K.K.H. Wheatley S.C. Ng L.J. Zhou S. Ling K.W. Sham M.H. Koopman P. Tam P.P.L. Cheah K.S.E. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 30.Armstrong C.G. Bahrani A.S. Gardner D.L. In vitro measurement of articular cartilage deformations in the intact human hip joint under load. J Bone Joint Surg Am. 1979;61:744. [PubMed] [Google Scholar]
- 31.Macirowski T. Tepic S. Mann R.W. Cartilage stresses in the human hip joint. J Biomech Eng. 1994;116:10. doi: 10.1115/1.2895693. [DOI] [PubMed] [Google Scholar]
- 32.Park S. Hung C.T. Ateshian G.A. Mechanical response of bovine articular cartilage under dynamic unconfined compression loading at physiological stress levels. Osteoarthritis Cartilage. 2004;12:65. doi: 10.1016/j.joca.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Li W.-J. Danielson K.G. Alexander P.G. Tuan R.S. Biological response of chondrocytes cultured in three-dimensional nanofibrous poly(e-caprolactone) scaffolds. J Biomed Mater Res A. 2003;67A:1105. doi: 10.1002/jbm.a.10101. [DOI] [PubMed] [Google Scholar]
- 34.Li W.-J. Tuli R. Huang X. Laquerriere P. Tuan R.S. Multilineage differentiation of human mesenchymal stem cells in a three-dimensional nanofibrous scaffold. Biomaterials. 2005;26:5158. doi: 10.1016/j.biomaterials.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Eichhorn S.J. Sampson W.W. Statistical geometry of pores and statistics of porous nanofibrous assemblies. J R Soc Interface. 2005;2:309. doi: 10.1098/rsif.2005.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stankus J.J. Guan J.J. Fujimoto K. Wagner W.R. Microintegrating smooth muscle cells into a biodegradable, elastomeric fiber matrix. Biomaterials. 2006;27:735. doi: 10.1016/j.biomaterials.2005.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denker A.E. Nicoll S.B. Tuan R.S. Formation of cartilage-like spheroids by micromass cultures of murine C3H10T1/2 cells upon treatment with transforming growth-factor-beta-1. Differentiation. 1995;59:25. doi: 10.1046/j.1432-0436.1995.5910025.x. [DOI] [PubMed] [Google Scholar]
- 38.Pluen A. Netti P.A. Jain R.K. Berk D.A. Diffusion of macromolecules in agarose gels: comparison of linear and globular configurations. Biophys J. 1999;77:542. doi: 10.1016/S0006-3495(99)76911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dingle J.T. Page Thomas D.P. King B. Bard D.R. In vivo studies of articular tissue damage mediated by catabolin/interleukin 1. Ann Rheum Dis. 1987;46:527. doi: 10.1136/ard.46.7.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martel-Pelletier J. Mineau F. Jovanovic D. Di Battista J.A. Pelletier J.P. Mitogen-activated protein kinase and nuclear factor kappaB together regulate interleukin-17-induced nitric oxide production in human osteoarthritic chondrocytes: possible role of transactivating factor mitogen-activated protein kinase-activated protein kinase (MAPKAPK) Arthritis Rheum. 1999;42:2399. doi: 10.1002/1529-0131(199911)42:11<2399::AID-ANR19>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 41.Ahmad R. Sylvester J. Zafarullah M. MyD88, IRAK1 and TRAF6 knockdown in human chondrocytes inhibits interleukin-1-induced matrix metalloproteinase-13 gene expression and promoter activity by impairing MAP kinase activation. Cell Signal. 2007;19:2549. doi: 10.1016/j.cellsig.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Knight M.M. Toyoda T. Lee D.A. Bader D.L. Mechanical compression and hydrostatic pressure induce reversible changes in actin cytoskeletal organisation in chondrocytes in agarose. J Biomech. 2006;39:1547. doi: 10.1016/j.jbiomech.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Johnson J. Ghosh A. Lannutti J. Microstructure-property relationships in a tissue-engineering scaffold. J Appl Polym Sci. 2007;104:2919. [Google Scholar]
- 44.Salminen H. Vuorio E. Saamanen A.M. Expression of Sox9 and type IIA procollagen during attempted repair of articular cartilage damage in a transgenic mouse model of osteoarthritis. Arthritis Rheum. 2001;44:947. doi: 10.1002/1529-0131(200104)44:4<947::AID-ANR152>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 45.Clark A.L. Barclay L.D. Matyas J.R. Herzog W. In situ chondrocyte deformation with physiological compression of the feline patellofemoral joint. J Biomech. 2003;36:553. doi: 10.1016/s0021-9290(02)00424-4. [DOI] [PubMed] [Google Scholar]
- 46.Schinagl R.M. Gurskis D. Chen A.C. Sah R.L. Depth-dependent confined compression modulus of full-thickness bovine articular cartilage. J Orthop Res. 1997;15:499. doi: 10.1002/jor.1100150404. [DOI] [PubMed] [Google Scholar]
- 47.Han S.K. Federico S. Grillo A. Giaquinta G. Herzog W. The mechanical behaviour of chondrocytes predicted with a micro-structural model of articular cartilage. Biomech Model Mechanobiol. 2007;6:139. doi: 10.1007/s10237-006-0016-3. [DOI] [PubMed] [Google Scholar]
- 48.Wu J.Z. Herzog W. Analysis of the mechanical behavior of chondrocytes in unconfined compression tests for cyclic loading. J Biomech. 2006;39:603. doi: 10.1016/j.jbiomech.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Wong M. Wuethrich P. Buschmann M.D. Eggli P. Hunziker E. Chondrocyte biosynthesis correlates with local tissue strain in statically compressed adult articular cartilage. J Orthop Res. 1997;15:189. doi: 10.1002/jor.1100150206. [DOI] [PubMed] [Google Scholar]
- 50.Torzilli P.A. Deng X.H. Ramcharan M. Effect of compressive strain on cell viability in statically loaded articular cartilage. Biomech Model Mechanobiol. 2006;5:123. doi: 10.1007/s10237-006-0030-5. [DOI] [PubMed] [Google Scholar]