Abstract
How living cells deal with head-on collisions of the replication and transcription complexes has been debated for a long time. Even in the widely studied model bacteria Escherichia coli, the enzymes that take care of such collisions are still unknown. We report here that in vivo, the DinG, Rep and UvrD helicases are essential for efficient replication across highly transcribed regions. We show that when rRNA operons (rrn) are inverted to face replication, the viability of the dinG mutant is affected and over-expression of RNase H rescues the growth defect, showing that DinG acts in vivo to remove R-loops. In addition, DinG, Rep and UvrD exert a common function, which requires the presence of two of these three helicases. After replication blockage by an inverted rrn, Rep in conjunction with DinG or UvrD removes RNA polymerase, a task that is fulfilled in its absence by the SOS-induced DinG and UvrD helicases. Finally, Rep and UvrD also act at inverted sequences other than rrn, and promote replication through highly transcribed regions in wild-type E. coli.
Keywords: Escherichia coli, recombination, replication restart, ribosomal operon, SOS induction
Introduction
Replication fork arrest is a recognized source of genetic instability in all types of living cells. To limit the danger of replication arrest, eukaryotes induce checkpoint proteins that stabilize and protect blocked replication forks (reviewed in Branzei and Foiani, 2007; Tourriere and Pasero, 2007). Prokaryotes behave differently, they constitutively express replication restart proteins that are associated with the replication machinery (Sandler, 2000; Lecointe et al, 2007).
Replication arrest can occur for many reasons, including collisions with DNA-bound proteins and particularly transcription complexes. As bacterial chromosomes are simultaneously transcribed and replicated, and because the velocity of the replication machinery (800 NT/s) is more than 10 times higher than that of the transcription machinery (50 NT/s), the problem raised by collisions between replication and transcription has been studied for decades (French, 1992; reviewed in Mirkin and Mirkin, 2007; Wang et al, 2007). Several in vitro and in vivo studies, showing that co-directional collisions do not seriously impede replication progression, lead to the conclusion that the replication machinery is not inactivated when it encounters an RNA polymerase transcribing the leading strand template (Pomerantz and O'Donnell, 2008 and references therein). In contrast, it is well established that head-on collisions between replication and transcription, that is the presence of an active RNA polymerase on the lagging strand template, arrest replication forks in vitro and in vivo (Deshpande and Newlon, 1996; Takeuchi et al, 2003; Mirkin and Mirkin, 2005). Genetic instability following head-on collisions of replication and transcription has been documented in bacteria and yeast (Vilette et al, 1995; Torres et al, 2004; Prado and Aguilera, 2005).
To limit head-on collisions between replication forks and the highly expressed rRNA genes, yeasts and eukaryotic cells use replication fork barriers, which are DNA sites where binding of a specific protein prevents replication from entering the rDNA region in the direction opposed to transcription (Brewer et al, 1992). In bacteria, to avoid head-on collisions ribosomal operons (rrn) are transcribed in the direction of replication (Brewer, 1988; Rocha and Danchin, 2003). rrn operons are highly expressed and their promoter regions carry regulatory elements that adapt their level of expression to the growth rate, so that transcription is more efficient in rich than in minimal medium (MM) (Condon et al, 1992; reviewed in Paul et al, 2004). Ribosomal-RNA transcripts are not translated and premature transcription arrest is prevented by the association of the RNA polymerase with ‘an anti-termination' machinery, which increases the transcription speed to 90 NT/s (reviewed in Condon et al, 1995). The universality of the presence of rrn operons on the leading strand template in bacteria suggests that rrn inversion impairs bacterial growth. Surprisingly, Escherichia coli viability was not affected by inverting large chromosomal regions that carry several rrn operons, even when the main homologous recombination DNA repair protein, RecA, was inactivated (Esnault et al, 2007). This observation suggested that bacteria encode proteins other than RecA that facilitates the progression of replication forks through oppositely oriented highly transcribed genes. We describe here the identification of helicases that have such a role.
Helicases are enzymes that associate NTP hydrolysis with the capacity to translocate on DNA. Most helicases translocate on single-strand DNA (ssDNA) to unwind double-stranded DNA, several also unwind DNA–RNA hybrids, or can remove proteins from DNA during translocation. The first helicase described to remove an RNA polymerase from the path of replication forks was the T4 dda helicase (Bedinger et al, 1983). In yeast, this function is fulfilled by the superfamily 1 (SF1) helicase Rrm3, a 5′–3′ helicase required for efficient replication at numerous protein-bound sequences such as in rRNA and tRNA genes, centromeric and telomeric regions (Azvolinsky et al, 2006; reviewed in Boule and Zakian, 2006). In this study, we show that in bacteria the three helicases DinG, Rep and UvrD facilitate replication of the chromosome through oppositely oriented highly transcribed ribosomal operons.
DinG belongs to the SF2 family of helicases and translocates in the 5′–3′ direction on ssDNA (Voloshin et al, 2003). In vitro, it unwinds a wide variety of substrates with a preference for D-loops and R-loops (Voloshin and Camerini-Otero, 2007). DinG is present in most prokaryotes and is related to the DNA helicases Chl1 and Rad3 from Saccharomyces cerevisiae, Rad15 from Schizosaccharomyces pombe and the human helicases XPD and BACH1 (Koonin, 1993; Rudolf et al, 2006; Voloshin and Camerini-Otero, 2007; Liu et al, 2008). Rad3 and XPD are components of the transcription factor IIH, they function in transcription initiation and nucleotide excision repair, and XPD defects are responsible for several human diseases (Liu et al, 2008 and references therein). Although DinG is an SOS-inducible protein (Lewis et al, 1992; Courcelle et al, 2001), its absence does not render E. coli sensitive to DNA damaging agents and to date the function of DinG in vivo is totally unknown.
Rep and UvrD are also the founders of a large family of helicases, homologous to Srs2 in yeast. They belong to the SF1 superfamily, share 40% identity and translocate in the 3′–5′ direction on ssDNA. The uvrD gene was originally identified for its crucial role in nucleotide excision repair and mismatch repair. In addition, UvrD (but not Rep) can remove the replication terminator Tus protein from its cognate site, Ter, and the recombination protein RecA from ssDNA (Flores et al, 2005; Veaute et al, 2005; Bidnenko et al, 2006). Rep assists replication because in its absence chromosome replication takes twice as long when compared with wild-type cells, and arrested replication forks undergo a remodelling reaction called replication fork reversal (Lane and Denhardt, 1975; Seigneur et al, 1998). Rep was hypothesized to facilitate replication across DNA-bound proteins because it can dislodge a DNA-bound repressor during translocation in vitro (Yancey-Wrona and Matson, 1992). The rep uvrD double mutant is lethal and rescued by the inactivation of the pre-synaptic recombination proteins RecQ, RecJ and RecFOR (Petit and Ehrlich, 2002; Lestini and Michel, 2008); one of the physiological roles of UvrD is thus to remove RecQJFOR-dependent RecA filaments from stalled replication forks, or to prevent their formation. Finally, replication forks that have been inactivated restart with the use of the major restart protein PriA; in a priA mutant, replication restart is catalysed by an Rep-PriC-dependent pathway (Sandler, 2000; Heller and Marians, 2005).
In this study, we show that chromosomal inversion, including E. coli rRNA operon(s) renders DinG essential for growth in rich medium. Moreover, the inactivation of the helicases DinG, Rep and UvrD has synergistic effects on replication blockage at an inverted rRNA locus. In the natural chromosome configuration, E. coli cells lacking these three helicases are viable only if the stability of the RNA polymerase is compromised and RecA binding is prevented by an RecF mutation. These results suggest that these helicases exert a fork-clearing function at inverted rrn loci and also at other transcription units.
Results
dinG inactivation confers rich medium sensitivity to strains that carry inverted rrn operons
The lambda attR and attL attachment sites were used to construct strains carrying a chromosome inversion (Valens et al, 2004; Esnault et al, 2007). The InvA mutant carries a 18 kb inversion encompassing the rrnA operon (Figure 1). It carries only 11 genes in addition to rrnA, among which 4 are naturally oriented in opposition to replication. rrnA is the only transcription unit that is highly expressed and whose expression is increased in rich medium in InvA strains (Corbin et al, 2003; Lopez-Campistrous et al, 2005). The InvBE mutant carries a 138.3 kb inversion containing rrnB and rrnE; about 100 genes are present in the inverted region, among which 67% are originally co-directional with replication and may be sites of transcription–replication collisions after inversion. As rrn expression is growth-rate regulated, these two Inv mutants allowed the analysis of three kinds of head-on replication–transcription collisions: (i) in highly expressed rrn, (ii) in moderately expressed rrn, (iii) in genes other than rrn. As previously observed for similar inversions (Esnault et al, 2007), InvA and InvBE were fully viable on MM and on rich medium (Luria broth, LB) (Figure 2A and B; Supplementary Table S2). DinG, Rep and UvrD were inactivated in InvA and InvBE mutants to test whether these helicases are required for replication across oppositely oriented genes.
Figure 1.
Schematic representation of the inverted region in the mutants InvA (top) and InvBE (bottom). Numbers indicate the sequence coordinates in the wild-type E. coli MG1655 chromosome. The large black arrows indicate the inversion end points (lambda att sites). The grey arrows indicate the position of rrn operons (the coordinates of rrnA, and of rrnE and rrnB 3′ ends are indicated). The vertical arrows show the position of NotI sites (used for PFGE).
Figure 2.
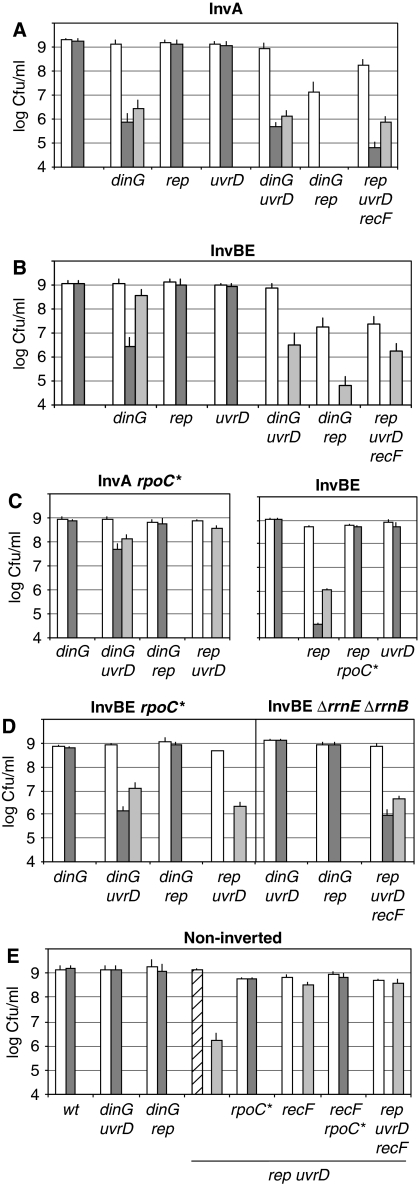
The helicases Rep, UvrD and DinG are required for colony formation in Inv mutants. Appropriate dilutions of overnight cultures at 37°C in MM (OD 1.0–1.5) were plated on MM and LB plates, which were incubated at 37°C. Unmarked positions on the left of (A) (InvA), (B) (InvBE) and (C) (InvABE) are data points for Inv mutants that express all helicases. White boxes: colony forming units (cfu)/ml on MM plates after 48 h incubation; dark grey boxes: cfu/ml on LB plates after 16–24 h incubation; light grey boxes: cfu/ml on LB plates after 48 h of incubation. The hatched box indicates cfu/ml on MM after 3 days incubation. The results are also presented in Supplementary Table S2.
dinG inactivation did not affect the formation of Inv mutant colonies on MM; however, colony formation on LB was strongly impaired for InvA dinG and delayed for InvBE dinG (Figure 2A and B). To test whether transcription is responsible for the LB sensitivity of InvA dinG and InvBE dinG mutants, we used the rpoCΔ215−220 mutation (called rpoC* thereafter). By mimicking the presence of ppGpp, this mutation reduces the stability of transcription elongation complexes (Bartlett et al, 1998, 2000; Trautinger and Lloyd, 2002; Trautinger et al, 2005). rpoC* restored 100% overnight colony formation on LB in both Inv dinG mutants (Figure 2C and D). In InvBE, the oriC-distal att site removes rrnB P1 Fis-binding sites (Supplementary Figure S1A), so that the promoter is weakened about seven-fold during steady-state growth in rich medium, but remains growth-rate regulated (Appleman et al, 1998; Hirvonen et al, 2001). Specifically, deleting the highly expressed rrnE operon in the InvBE dinG mutant fully restored the plating efficiency on LB (Supplementary Table S2). We conclude that DinG is required for efficient colony formation on rich medium when a highly expressed rrn operon is inverted on the chromosome, and that the growth defect observed in Inv dinG mutants is completely overcome by reducing the transcription level (growth on MM or inversion of only rrnB, which is deprived of Fis sites in this construction).
rep inactivation causes cell elongation in rich medium
Most of the rep mutants were constructed in the presence of a conditional Rep+ plasmid (IPTG dependent) that was cured before each experiment, (Supplementary Table S1; Lestini and Michel, 2008). Inactivation of rep in InvA or InvBE mutants did not cause any loss of plating efficiency (Figure 2A and B), although InvBE rep overnight colonies on LB were quite small. The introduction of rpoC* or the deletion of rrnE in InvBE rep suppressed this slow-growth phenotype, again suggesting a deleterious effect of the inverted highly expressed rrn operons (not shown). This idea was confirmed by the use of a strain with a large inverted region carrying the three operons rrnA, rrnB and rrnE (InvABE, 277.3 kb inverted): InvABE rep was sensitive to LB and this defect was fully suppressed by the rpoC* mutation (Figure 2C). In contrast, all Inv uvrD mutants were fully viable on LB as on MM (Figure 2). We conclude that the Rep helicase (and not UvrD) is required for colony formation on LB when at least three highly transcribed rrn operons are oriented opposite to replication. In contrast, a 277 kb inversion does not impair growth of the rep mutant providing that the rrn operons are only moderately expressed (InvABE cells grown in MM) or that the stability of the RNA polymerase is compromised (rpoC* mutant on LB, Figure 2).
Formation of a visible colony requires about 24 generations and to determine whether the rep mutation affects Inv cell growth at early times, Inv rep cells were analysed by differential interference contrast (DIC) microscopy. Both InvA rep and InvBE rep cultures, shifted for 1 h from MM to LB, contained a high percentage of elongated cells, higher than rep or Inv single mutants (Table I). Cell elongation was weaker in MM and was strongly decreased by the rpoC* mutation or the deletion of rrnE and rrnB (from 29 to 3% in InvA rep and from 48 to 5–11% in InvBE rep, Table I), indicating that it is caused by the strong expression of inverted rrn. It was also specific for the rep mutation, as Inv uvrD cells were no more elongated than single uvrD mutants (6–11% elongated cells, Table I), and Inv dinG cells were not (InvBE) or only slightly (InvA) elongated (4 and 16% of elongated cells, respectively, Table I). The contrast between the elongated phenotype of Inv rep cells after a shift to LB and a wild-type efficiency of colony formation overnight on LB plates suggest an early defect followed by a recovery. Conversely, the absence of cell elongation of the Inv dinG mutants after a shift to LB contrasts with their plating defect suggests late, possible cumulative defects. These ideas were tested by analysing micro-colony formation by time-lapse microscopy (Supplementary Figure S2). InvA rep micro-colonies grown for a few hours on LB contained normal-sized cells, owing to the splitting of some elongated cells. Conversely, InvA dinG normal-sized cells growing on an LB agar pad produced micro-colonies composed of non-dividing, mostly elongated cells (Supplementary Figure S2).
Table 1.
Cell elongation after a shift to LB
| Strain | Relevant genotype | MMa | LB 1 hb | ||||||
|---|---|---|---|---|---|---|---|---|---|
| dinG | rep | uvrD | 1 Nc | 2 N | >3 N | 1 N | 2 N | >3 N | |
| JJC3524 | + | + | + | 73 | 27 (20) | 0 | 62 (1) | 36 (20) | 2 |
| InvA strains | |||||||||
| JJC4010/4802 | + | + | + | 58 | 41 (19) | 1 (1) | 61 | 36 (19) | 3(2) |
| JJC4678/4881S | dinG | + | + | 60 | 36 (12) | 4 (1) | 40 | 44 (12) | 16 (8) |
| JJC4408 | + | rep | + | 51 | 39 (8) | 10 (9) | 23 | 44 (5) | 29 (12) |
| JJC4873 | + | + | uvrD | 68 (1) | 29 (1) | 3 (3) | 42 | 47 (16) | 11 (7) |
| JJC4880 | dinG | + | uvrD | 48 | 39 (1) | 12 (2) | 17 | 26 (2) | 56 (7) |
| JJC4828S | dinG | rep | + | 60 | 29 (3) | 10 (6) | 24 | 30 (1) | 46 (3) |
| JJC4879Sc | + | rep | uvrD | 11 | 25 | 64 (5) | 4 | 24 | 72 (1) |
| InvA rpoC* | |||||||||
| JJC4962 | dinG | + | + | 75 (5) | 23 (13) | 0 | |||
| JJC4995 | + | rep | + | 64 (3) | 33 (23) | 3 (2) | |||
| JJC4963 | dinG | + | uvrD | 54 (5) | 30 (12) | 17 (10) | |||
| JJC4914S/4919 | dinG | rep | + | 54 (5) | 33 (9) | 13 (1) | |||
| JJC5140S/5143 | + | rep | uvrD | 43 | 39 (5) | 18 (0) | |||
| InvA recA | |||||||||
| JJC4027 | + | + | + | 86 (7) | 11 (3) | 3 (1) | |||
| JJC5040 | + | + | uvrD | 55 (2) | 38 (15) | 8 (3) | |||
| JJC5042 | dinG | + | uvrD | 31 (1) | 52 (9) | 17 (1) | |||
| JJC5053S | + | rep | + | 71 (0) | 24 (5) | 5 (4) | 15 | 36 (2) | 49 (5) |
| InvA lexA | |||||||||
| JJC5096 | + | rep | + | 56 | 38 (12) | 6 (4) | 17 | 55 (4) | 28 (9) |
| InvBE strains | |||||||||
| JJC4349 | + | + | + | 83 (4) | 17 (11) | 1 (1) | 52 (1) | 40 (11) | 7 (5) |
| JJC4920 | dinG | + | + | 77 (1) | 23 (14) | 0 | 50 (1) | 45 (13) | 4 (2) |
| JJC4700S/4978S | + | rep | + | 33 | 50 (9) | 17 (13) | 17 | 35 (3) | 48 (10) |
| JJC4870/4997 | + | + | uvrD | 78 (4) | 22 (14) | 0 | 45 | 48 (11) | 7 (3) |
| JJC4981 | dinG | + | uvrD | 63 (1) | 34 (14) | 3 (2) | 25 | 43 (7) | 27 (10) |
| JJC4746S/5009S | dinG | rep | + | 52 | 34 (3) | 14 (6) | 7 | 37 (1) | 56 (7) |
| InvBE rpoC* | |||||||||
| JJC4987 | + | rep | + | 46 (1) | 49 (23) | 5 (3) | |||
| JJC4966/4979 | dinG | + | uvrD | 55 (2) | 26 (10) | 18 (9) | |||
| JJC4975 | dinG | rep | + | 58 (6) | 36 (19) | 5 (3) | |||
| InvBE ΔrrnE | |||||||||
| JJC4951 | dinG | + | + | 80 (4) | 19 (11) | 0 | |||
| JJC4973 | dinG | + | uvrD | 39 (0) | 48 (11) | 13 (5) | |||
| InvBE ΔrrnE ΔrrnB | |||||||||
| JJC5125 | dinG | + | + | 82 (5) | 19 (11) | 0 | |||
| JJC5154 | + | rep | + | 44 | 45 (18) | 11 (10) | |||
| JJC5158 | dinG | + | uvrD | 39 | 50 (11) | 11 (10) | |||
| JJC5156S | dinG | rep | + | 56 | 41 (14) | 3 (3) | |||
| JJC5157Sc | + | rep | uvrD | 80 (7) | 19 (17) | 1 | 30 | 35 (4) | 34 (10) |
| InvBE recA | |||||||||
| JJC4631 | + | + | + | 69 (7) | 26 (15) | 4 (1) | |||
| JJC5036 | dinG | + | + | 59 (1) | 36 (13) | 4 (2) | |||
| JJC5058S | + | rep | + | 35 | 43 (10) | 22 (8) | 24 | 36 (2) | 40 (5) |
| JJC5034 | + | + | uvrD | 54 (2) | 30 (5) | 15 (7) | |||
| Non-inverted strains | |||||||||
| JJC3424 | + | + | + | 71 | 27 (20) | 0 | 60 (1) | 36 (20) | 2 |
| JJC4400 | dinG | + | + | 62 | 36 (12) | 1 (1) | |||
| JJC4984 | + | rep | + | 35 | 49 (8) | 16 (13) | |||
| JJC4858 | + | + | uvrD | 74 | 25 (16) | 1 | 42 | 52 (19) | 6 (3) |
| JJC4872 | dinG | + | uvrD | 45 | 45 (1) | 10 (5) | |||
| JJC4804S | dinG | rep | + | 83 (6) | 6 (4) | 4 (2) | |||
| JJC4878Sc | + | rep | uvrD | 45 | 44 | 11 (3) | 25 | 39 (2) | 36 (3) |
| Non-inverted rpoC* | |||||||||
| JJC5164S/5165 | + | rep | uvrD | 63 (1) | 26 (15) | 11 (3) | |||
| JJC4629 | recA | 79 (6) | 16 (5) | 2 (1) | |||||
| ‘S': the pAM-rep plasmid was cured before the experiment, the strain number is followed by an ‘S' to indicate that experiment was performed after the plasmid has been segregated. In each medium, the smallest wild-type cells produced by division (baby cells, 1.5 μM in MM and 2.1 μM in LB) were used as cell unit and their size was, as expected, half that of the smallest cells with a detectable septum. Numbers indicate the percentage of cells in each of the following categories: 1 N: cells whose length was from baby wild-type cells to twice as long; 2 N: cells whose length was between twice and three times that of baby wild-type cells; >3 N: cells longer than three times the size of baby wild-type cells. With few exceptions, 150–300 cells were counted. Numbers in parentheses indicate the percentage of cells with a visible septum in formation. Data in bold differ at least three-fold from their parental values (InvA and InvBE single mutants, and non-inverted cells carrying the same helicase or recA mutations); for these mutants, results are the average of two independent experiments. | |||||||||
| aCells in exponential phase in MM. | |||||||||
| bCells in exponential phase shifted for 1 h in LB. | |||||||||
| c JJC4879, JJC5157 and JJC4878 are rep uvrD recF mutants. | |||||||||
The effects of the dinG, rep and uvrD mutations are additive
To analyse whether DinG, Rep and UvrD have independent or overlapping roles, we tested whether the inactivation of two of these three helicases is synergistic. Cells that do not carry a chromosome inversion were tested first, showing that dinG uvrD and dinG rep double mutants are fully viable (Figure 2E). As described earlier (Petit and Ehrlich, 2002), non-inverted rep uvrD cells were (i) nearly lethal on MM (small colonies appeared in 3 days), (ii) lethal on LB and (iii) mainly rescued by recF inactivation (Figure 2E). Therefore, Inv rep uvrD mutants were tested in a recF mutant background. As recF inactivation per se does not affect the growth of Inv strains (Supplementary Table S3, see below) and is beneficial to rep uvrD cells, we consider thereafter that the growth defects of rep uvrD recF mutants carrying an inversion result from the inactivation of the rep and uvrD genes and not from the recF mutation.
All Inv mutants lacking two helicases were sensitive to rich medium as they formed colonies on LB plates with a very low efficiency (Figure 2A and B). As the InvA dinG mutant was already quite sensitive to rich medium, the deleterious effect of inactivating uvrD in this mutant can be deduced from the increased level of elongated cells after only 1 h of propagation in LB (Table I). A high percentage of elongated cells are observed in all Inv mutants lacking two helicases. It is accompanied by a decrease in the number of cells with a visible septum (number in parenthesis in Table I), in agreement with a cell division defect. Therefore, dinG, rep and uvrD mutations are synergistic, indicating overlapping functions.
Inv dinG uvrD mutants were fully viable on MM whereas a significant plating defect of both Inv dinG rep mutants on MM indicates replication impairment by moderately expressed rrn in this mutant and suggests overlapping functions of Rep and DinG (MM, Figure 2A and B). Inv rep uvrD recF cells were also impaired on MM; the plating defect was stronger for InvBE than for InvA, suggesting a possible replication impairment also at non-rrn sequences (MM, Figure 2A and B; Inv rep uvrD RecF+ colonies were not obtained).
rrn expression is responsible for the growth defects of helicase mutants on LB
rpoC* and rrn deletion alleles (ΔrrnE and ΔrrnB, Supplementary Figure S1) were used to ascertain the role of rrn in the observed growth defects. rpoC* was first tested in a non-inverted rep uvrD mutant. Importantly, rpoC* rescued colony formation of rep uvrD cells on MM and on LB, regardless of the recF status (Figure 2E). This result indicates that (i) in E. coli the presence of both Rep and UvrD is required because of a high level of transcription and (ii) decreasing transcription by affecting the stability of RNA polymerase bypasses the need for RecFOR inactivation.
In InvA mutants, rpoC* restored the viability of both dinG uvrD and dinG rep cells (although InvA dinG uvrD rpoC* remained slightly impaired on LB) and the InvA rep uvrD rpoC* mutant formed colonies on LB in 2 days (Figure 2C–E; Table I). Therefore, the growth defects of all the three InvA mutants lacking two helicases result from the high level of rrnA expression.
In InvBE dinG rep, introduction of the rpoC* allele improved viability and decreased cell elongation in LB, as observed for the InvA strain (Figure 2D; Table I). Accordingly, deletion of both rrnE and rrnB also fully rescued the InvBE dinG rep mutant, confirming that these highly expressed operons are the only deleterious sequences in this mutant (Figure 2D, Table I). In contrast, rpoC* did not rescue InvBE dinG uvrD, but deletion of both rrnE and rrnB allowed a full recovery of colony formation on LB (Figure 2D; Table I; Supplementary Table S2). These observations allow us to conclude that rrn are also the only deleterious sequences in InvBE dinG uvrD, but that even in the presence of the rpoC* mutation, inverted rrn impair growth of this mutant on rich medium.
In InvBE rep uvrD cells, introduction of rpoC*, deletion of rrnE or of both rrn allowed colony formation on MM but cells remained sensitive to LB, even in a recF context (Figure 2D; Supplementary Table S2). Therefore, the inversion of genes other than rrn is deleterious in rich medium in rep uvrD and rep uvrD recF mutants.
The requirement for UvrD in Inv dinG mutants is not because of its anti-RecF-RecA action
An recF null mutation was used to test whether UvrD is required in Inv dinG mutants to counteract a deleterious DNA binding of RecFOR, and in turn RecA. recF inactivation did not improve the growth of dinG, uvrD or dinG uvrD Inv mutants (Supplementary Table S3). We conclude that in Inv dinG uvrD mutants, the deleterious effect of the absence of UvrD is not because of the lethal binding of RecFOR-RecA to DNA. We propose that the synergistic effects of dinG and uvrD inactivation in cells carrying a highly expressed inverted rrn operon reflect a redundant function of these two helicases.
In agreement with a previous report, we observed that recA inactivation did not affect the viability of InvA and InvBE single mutants (Esnault et al, 2007; Supplementary Table S3). However, recA deletion prevented growth of InvBE dinG and Inv rep mutants on LB (Supplementary Table S3). Furthermore, no plasmid-less colony could be obtained from Inv dinG rep recA; [pAM-rep] cells even on MM, indicating that in both Inv backgrounds the dinG rep recA combination of mutations is lethal (Supplementary Table S3; uvrD recA colonies were slow growing on LB and were not affected by inversion, Supplementary Table S3). This suggests that the lack of Rep and/or DinG in Inv mutants generates ssDNA that renders homologous recombination and/or SOS induction crucial for viability. Notably, in Inv rep mutants the inactivation of the SOS response by a lexAind mutation also delayed (InvA) or prevented (InvBE) colony formation on LB (Supplementary Table S3; Supplementary Figure S2), indicating that the plating defect of Inv rep recA mutants may mainly result from the absence of SOS induction.
The combination of rep uvrD dinG recF mutations is lethal in non-inverted strains and rescued by rpoC*
We attempted to construct a rep uvrD dinG recF mutant by eliminating the pAM-Rep+ plasmid from rep uvrD dinG recF [pAM-Rep+] cells. Small plasmid-less colonies were obtained in 3 days on MM but some failed to grow in overnight cultures and others exhibited variable plating efficiencies, indicating that the simultaneous inactivation of the three helicases Rep, UvrD and DinG is nearly lethal in a recF E. coli mutant (Supplementary Table S2). Therefore, the viability of each helicase double mutant relies on the presence of the third helicase when all genes are in their original orientation. The rpoC* mutation also failed to restore rep uvrD dinG colony formation. Therefore, in cells lacking all three helicases, neither decreasing RNA Pol stability nor preventing RecA binding to blocked forks is sufficient to allow colony formation, even in slow-growth conditions (MM). However, when the stability of the RNA polymerase was compromised by the rpoC* mutation and recF was inactivated, the resulting rep uvrD dinG rpoC* recF mutant formed colonies on MM and on LB in 2 days (Figure 2E; Supplementary Table S2). This result indicates that an E. coli mutant lacking all three helicases is killed by collisions between replication and transcription complexes; in the presence of the rpoC* mutation, the triple helicase mutant is killed by RecFOR-RecA binding to arrested forks.
Pulse field gel electrophoresis analysis of Inv helicase mutants
To investigate the effects of helicase inactivation on the progression of replication forks across inverted sequences, chromosomes of Inv mutants were analysed using two approaches, pulse-field gel electrophoresis (PFGE) and 2D gels.
Y or X structures, that is replication or recombination intermediates, prevent migration of linear DNA fragments in PFG (Azvolinsky et al, 2006 and references therein). Therefore, by measuring the proportion of DNA fragments that remain trapped in the wells after PFGE, we could quantify the formation of abnormal DNA structures in the inverted region. Chromosomes were digested by a rare cutting enzyme (NotI), and a probe specific for the NotI fragment carrying the inverted region (named Inv-fragment below) was used (Figure 3A).
Figure 3.
rep, uvrD and/or dinG mutations prevent Inv-fragment migration in PFGE. (A) InvA dinG rep cells (left panel) or InvBE rep cells (right panel) were propagated in MM or in LB for 1 or 2 h as indicated above each lane. Cells were lysed in plugs, chromosomes were treated with NotI, and restriction fragments were separated by PFGE. As the InvA-fragment is 50 kb and the InvBE-fragment is 138 kb (Figure 1), different migration conditions were used for InvA and InvBE mutants. For each panel, left lanes show the Et Br stained gel, the position of the wells and of the Inv fragment is indicated; right lanes Southern hybridization with a probe that detects the Inv-fragments after DNA transfer to a nylon membrane. (B–E) Percentage of Inv-fragment DNA retained in wells for various mutant strains, quantified after Southern hybridization. Unmarked positions on the left of (A) (InvA) and (B) (InvBE) are data points for Inv mutants that express all helicases. White boxes: percentage of non-migrating Inv fragment in cells grown in MM; dark grey boxes: percentage of non-migrating Inv fragment in cells grown in LB for 1 h; light grey boxes: percentage of non-migrating Inv fragment in cells grown in LB for 2 h. The results are also presented in Supplementary Table S4.
In the InvA background, after a 1 h shift to LB the percentage of Inv-fragment trapped in the wells increased from 3% (InvA) to 41% in a dinG mutant and 52% in a rep mutant, whereas it remained weak in the InvA uvrD mutant (Figure 3B). The level of trapped DNA was still high after 2 h in LB for the dinG mutant, whereas it decreased slightly in the rep mutant, suggesting an adaptation to LB in this mutant. All double helicase mutants exhibited a high level of trapped Inv-fragment after 1 or 2 h of propagation in LB (56–86%, Figure 3B). DNA trapping was abolished (in the rep mutant), or decreased (in uvrD dinG and rep dinG mutants) by the rpoC* mutation (compare Figure 3B and D), indicating that trapping of the Inv-fragment results mainly from the high level of rrnA transcription in LB.
In the InvBE rep mutant the percentage of trapped Inv-fragments increased from 15 to 79% after 1 h in LB and, as in the InvA rep mutant, this increase was transient (Figure 3C). Trapping was only increased after 2 h of propagation in LB in the InvBE dinG mutant (35%), and remained weak in InvBE uvrD (Figure 3C). In contrast, InvBE mutants lacking two helicases exhibited a high level of DNA trapping (64–90% Figure 3C; InvBE rep uvrD recF grew too poorly to be tested). The rpoC* mutation had partial effects (Figure 3D). Deleting rrnE and rrnB suppressed Inv-fragment trapping in dinG (Supplementary Table S4) and dinG uvrD cells, confirming that rrn are the only inverted genes that perturb replication in these mutants (Figure 3D; Supplementary Table S4). Although InvBE rep and InvBE dinG rep lacking both rrnE and rrnB were also fully viable (Figure 2D; Supplementary Table S2), they retained a weak but significant level of DNA trapping when propagated in LB for 1 or 2 h (Figure 3D; Supplementary Table S4). Finally, the LB sensitivity of the InvBE rep uvrD recF ΔrrnE ΔrrnB mutant correlates with a high level of Inv-fragment trapping (Supplementary Table S4), confirming a role for Rep and UvrD at other inverted genes, as well as at rrn.
Interestingly, the recovery of normal DNA migration in Inv rep mutants propagated for 2 h in LB was not observed in a recA or lexAind context, indicating it requires SOS induction (Supplementary Table S4). In other mutants, the proportion of Inv-fragments trapped in wells was only marginally affected by the recA mutation (Supplementary Table S4). The observation that the proportion of non-migrating Inv-fragments is similar in the absence of RecA indicates that these non-linear structures are not recombination intermediates but rather replication intermediates.
As expected, the percentage of DNA trapping was low in non-inverted strains, or when a probe hybridizing with a NotI fragment other than the Inv-fragment was used as a control (Supplementary Table S4). DNA trapping was also low when Inv cells were grown in MM; the presence of abnormal DNA structures only when cells are grown in LB indicates that this non-migrating DNA only forms when cells are propagated at a high growth rate (Supplementary Table S4).
In helicase mutants, replication intermediates accumulate in the inverted rrn operon
As increased DNA trapping correlates with a high level of rrn transcription, we examined replication progression in the rrn operon by 2D gel analysis (Brewer and Fangman, 1987). rrnA and rrnE operons were each analysed after DNA cleavage with two different restriction enzymes and a specific probe just downstream of the rrn operon (Figure 4A). As replication forks move at a speed of about 800 bp/s in E. coli, replication intermediates are not detectable in the chromosome of wild-type cells. Actually, we never detected replication intermediates in control 2D gels performed with an Inv mutant that expresses all helicases (InvA and InvBE single mutants) and with non-inverted strains (wt chromosome) lacking one or two helicases (see for instance non-Inv rep Figure 4C; and data not shown).
Figure 4.
Replication forks are arrested in inverted rrn. 2D gels were used to examine DNA replication in restriction fragments containing a large 3′ region of rrnA in InvA mutants and of rrnE in InvBE mutants. (A) Schematic representation of the restriction fragments used for 2D gels, left InvA, right InvBE. Top line, the position of rrn and of restriction sites are shown; bottom lines, schematic representation of the forked fragments when replication is arrested at the 5′ end of rrn, distances from the restriction sites to the 5′ end of rrn and the relative size of the forked fragments compared with linear fragments are indicated. (B, C) DNA from various InvA (B) and InvBE (C) mutants were digested with the indicated restriction enzyme, analysed by 2D gels and probed for the sequence just downstream of the analysed rrn. The left panel shows a simulation of replication arrest in the entire restriction fragment with an increased arrest in about 500 pb around the rrn transcription terminator sequence. The mutants used are indicated above each panel. In the InvBE rep and rep dinG rpoC* mutants, the signal of increased replication arrest in the transcription termination region was not always observed, independently of the restriction enzyme used, and one example of each situation is shown.
A ‘simple Y' arc corresponding to the accumulation of Y-shaped replication intermediates was clearly detected in all mutants exhibiting more than 40% Inv-fragment trapped in wells in PFGE, indicating replication fork arrest within the transcribed region of the restriction fragment. Interestingly, an intense enlarged spot was observed on the simple Y arc (Figure 4B and C), indicating a specific accumulation of replication intermediates. The spot position moves along the simple Y arc depending on the restriction fragment analysed, allowing us to map this replication arrest zone to the 3′ end of the rrn operon, including the transcription terminator (Figure 4). The spot was of weaker and variable intensity in InvA dinG and InvBE rep mutants. The replication arrest zone was often prolonged 100–300 base pairs downstream of the operon calculated according to computer simulations (Viguera et al, 1998), particularly for rrnE, suggesting a possible impairment of replication by transcription-induced supercoiling or a defect in transcription termination. Replication intermediates were not detected with cells growth in MM and were of weak intensity in the rpoC* context, confirming that replication is strongly impaired only when rrn are highly expressed (Figure 4; and data not shown).
DinG is required to remove R-loops and RNA Pol
In addition to their very high level of expression, a characteristic of rrn operons is the production of non-translated RNA, which favours the formation of R-loops by the annealing of rRNA with its template DNA. Therefore, replication blocks within actively transcribed rrn can result from collisions of replication forks with RNA Pol and/or R-loops. To determine whether R-loop formation has a role in the defects of Inv helicase mutants, we used a multicopy plasmid that carries the rnhA gene encoding RNase H, which degrades R-loops (pEM001, Masse et al, 1997, or an ApR derivative pEM-Ap). Vectors pACYC184 and pBR322 were used as controls. In the InvA dinG mutant, over-expression of RNase H clearly suppressed the plating defect on LB and the trapping of Inv-fragments (Figure 5; RNase H over-expression also suppressed the growth delay of InvBE dinG cells, Supplementary Table S5) . We conclude that the role of DinG is to remove R-loops (or to prevent their formation).
Figure 5.
Inv dinG mutants are rescued by Rnase H overproduction, but not other mutants. (A) Plating efficiencies of InvA mutants [pEM-Ap] overnight cultures determined as in Figure 2. (B) Percentage of InvA-fragment retained in PFG wells in pEM-Ap containing cells determined as in Figure 3.
This result indicates that Rep and UvrD (present in Inv dinG mutants) do not efficiently remove R-loops formed at rrn in vivo. This conclusion is strengthened by the observation that the plasmids pEM001 and pEM-Ap had no effect in Inv rep mutants (Supplementary Table S5) and did not suppress the defects conferred by dinG inactivation in Inv dinG uvrD double mutants (Figure 5; Supplementary Table S5; and data not shown; the plasmid could not be introduced, even on MM, in Inv dinG rep and Inv rep uvrD recF). If we assume that the only possible obstacles to replication progression in oppositely oriented rrn are R-loops and RNA Pol, then the growth defect and the high level of non-migrating Inv-fragment in InvA dinG uvrD and InvBE dinG uvrD mutants that overproduce RNaseH can logically be interpreted as the occurrence of collisions of replication forks with RNA Pol. In these mutants, replication impairment is observed on inactivation of both the dinG and uvrD genes, but not when only dinG or only uvrD is inactivated, suggesting that the UvrD and DinG proteins share a common function. Consequently, this reasoning leads us to suggest that DinG and UvrD are both participating in RNA Pol removal. Similarly, our observation that RNaseH overproduction does not decrease the high level of replication intermediates and cell elongation in InvA rep and InvBE rep cells (Figure 5B; Supplementary Table S5) can be interpreted as an increased level of replication-RNA Pol collisions in these mutants. We suggest that Rep is also involved in RNA Pol removal.
The helicases do not prevent replisome disassembly
In E. coli, restart of inactivated replication forks involves the reloading of the replication machinery by ‘replication restart' proteins. The main restart pathway is catalysed by PriA and its partners (Sandler, 2000). We constructed InvA priA, InvBE priA and InvABE priA mutants. priA mutants devoid of chromosome inversion were used as a control (Supplementary Table S1). All mutants were constructed in the presence of a PriA+ IPTG-dependent plasmid, pAM-priA, which can be cured by growing cells in the absence of IPTG (Grompone et al, 2004). No plasmid-less colony could be recovered by growing Inv priA mutants in MM devoid of IPTG, whereas plasmid-less priA colonies were obtained in the non-inverted strains as expected (Supplementary Table S2). Therefore, the PriA pathway is essential for viability in Inv mutants. Importantly, this result indicates that DinG, Rep and UvrD do not prevent replication arrest. We propose that these helicases act after replisome disassembly and allow PriA-dependent restarted forks to replicate across the obstacle created by the inversion.
Discussion
When replication and transcription proceed in opposite directions, the DnaB helicase collides on the lagging strand with RNA Pol and, as shown here and elsewhere, replication progression is hampered. In this work, we show that the three E. coli helicases DinG, Rep and UvrD are recruited to the replication fork to allow replication across oppositely oriented highly transcribed ribosomal operons. Furthermore, these helicases are also crucial in wild-type E. coli, where replication and rrn transcription are co-directional. Although DinG, Rep and UvrD helicases have overlapping functions, our results show that they do not act on exactly the same molecular substrate, and do not act at exactly the same time.
DinG removes R-loops
Defects conferred by the single dinG mutation in cells that carry an oppositely oriented rrn are suppressed by RNase H over-expression and by the rpoC* mutation. These findings indicate that R-loops form within highly expressed rrn operons and block replication, and that DinG is the only helicase that removes them in vivo. The identification of R-loops as an in vivo target for DinG is in full agreement with the in vitro properties of the purified protein (Voloshin and Camerini-Otero, 2007), and with the deleterious phenotype of a dinG mutation in an rnh background, attributed to an excess of R-loops (Yasuda et al, 1996). The stronger defects of the InvA mutant compared with InvBE suggest that R-loops are more prone to form in rrnA than in rrnE. Either R-loops may form more often when rrn are facing the direction of replication, or they may form at a similar efficiency in Inv and wild-type cells but they may be deleterious only when replication and transcription move through the operon in the opposite orientation. However, because our data indicate that neither Rep nor UvrD act on R-loops in Inv mutants, the synergistic effects of inactivating dinG in rep or uvrD mutants indicate that R-loops are not the only target of DinG.
Rep, DinG and UvrD participate in RNA Pol removal
Several data indicate that these three helicases share a common function. First, in Inv Rep+ cells the combination of uvrD and dinG mutations leads to the accumulation of replication intermediates at early times after a shift to LB and prevents colony formation. This rich medium sensitivity persists when RNase H is overproduced and is only observed when the inversion carries a highly expressed rrn, indicating that together with Rep either UvrD or DinG is required for the dislodging of RNA Pol from inverted rrn operons but not from other genes. Second, in Inv rep mutants both DinG and UvrD must be present for colony formation, even on MM. We conclude that replication across inverted rrn requires the presence of two out of these three helicases. It is noteworthy that RecA and RecF, which are required for replication fork progression across DNA lesions (Courcelle and Hanawalt, 2003) are not required for replication across inverted highly transcribed sequences (Esnault et al, 2007, Supplementary Table S3).
Rep acts early after a shift to rich medium
In a strain that lacks Rep and carries an inverted rrn, a shift to LB induces cell elongation and the accumulation of replication intermediates. As the obstacles to replication are not R-loops (because they are not abolished by RNase H overproduction) they are most likely RNA Pols. Interestingly, Inv rep mutants spontaneously recover and eventually form 100% colonies on LB. The rescue of InvA rep and InvBE rep mutants requires SOS induction and the presence of both DinG and UvrD (it is abolished in recA, lexAind, dinG and uvrD contexts). Actually, the SOS response is induced by replication impairment, caused by DNA lesions or various replication defects (Sassanfar and Roberts, 1990; Lestini and Michel, 2007). We propose that, after replication blockage, Inv rep mutants are rescued by the SOS-induced DinG and UvrD helicases. It is tempting to speculate that the Rep helicase acts early owing to an efficient targeting to blocked replication forks, whereas in its absence UvrD and/or DinG may be efficient at unblocking forks only when they are at a high concentration, that is SOS induced.
Model for helicase action
The lethality of Inv priA mutants implies that in Inv cells replication forks are arrested and disassembled. Therefore, the three helicases act after fork arrest, either on naked replication forks, or in conjunction with a reassembled, restarting replisome. As DinG migrates in the 5′–3′ direction on DNA it is conceivable that it acts on the lagging strand template whereas Rep and UvrD, which migrate in the 3′–5′ direction, progress on the leading strand template (Figure 6). In Inv cells inactivating only the uvrD has no deleterious effects and we propose that RNA Pols are dislodged by the concerted action of Rep and DinG (Figure 6A). If Rep is lacking, it is replaced by UvrD and the replication restart is then delayed because UvrD needs to be SOS induced to be efficient (Figure 6B). If DinG is lacking, the fork will recruit UvrD in addition to Rep and because these two helicases progress on the same strand, they will efficiently remove a series of RNA Pol provided that no R-loop forms (R-loops form on the other strand) (Figure 6C). Rep alone (DinG and UvrD absent) and UvrD alone (DinG and Rep absent) can only progress through co-directional highly expressed rrn genes or through moderately expressed inverted rrn, on which less RNA Pol travel and RNA Pol stability is compromised by ppGpp or by a mutation that mimics its presence (Figure 6D and E). Finally, DinG alone does not allow normal replication progression across an inverted rrn or across other inverted sequences (Inv rep uvrD recF mutants), whereas it is sufficient for the growth of non-inverted E. coli cells, provided that the stability of RNA polymerase is compromised (rep uvrD rpoC* cells, Figure 6F).
Figure 6.
Rescue of transcription-blocked replication forks by helicases. Schematic representation of a replication fork blocked by a transcription unit. Top, a replication fork encounters an oppositely oriented highly expressed rrn operon (Inv mutant in LB): (a) in cells proficient for all helicases and in a uvrD single mutant, Rep translocating towards the transcription unit on the leading strand template and DinG on the lagging strand template act in concert; (b) in a rep mutant both UvrD and DinG are required, UvrD translocating on the leading strand template and DinG on the lagging strand template act in concert; (c) in a dinG mutant, both Rep and UvrD are required, they both translocate towards the transcription unit on the leading strand template; because R-loops form on the lagging strand template (not shown) where no helicase is present, R-loops are deleterious. Middle, a replication fork encounters an oppositely oriented moderately expressed rrn operon (Inv mutant in MM): (d) Rep only (dinG uvrD mutant) or (e) UvrD only (dinG rep mutant)is sufficient for replication. Bottom, a replication fork encounters a normally oriented (co-directional) moderately expressed rrn operon (wild-type chromosome in MM): this is the only condition in which DinG alone (rep uvrD recF mutant) allows full viability. Full lines: template DNA; dashed lines: newly synthesized DNA; oval: replisome; yellow circles: DnaB helicase; green indented circles: RNA Pol; pink lines: rRNA. Helicases are shown as grey indented circles: hatched DinG, light grey Rep, dark grey UvrD.
Replication arrest at inverted rRNA operons
The analysis of mutants in which both inverted rrn have been deleted shows that rrn are the main obstacle to replication in dinG uvrD and (to a lesser extent) in rep dinG mutants, and one of the obstacles to replication in the rep uvrD recF mutant. To get insight into the nature of the elements that slow down replication in the rrn operon we analysed replication intermediates by 2D gels. Replication forks are slowed down in the transcribed region in rrn. Replication intermediates are similar in the dinG and in the rep mutant, although they result from the encounter of R-loops in the former and RNA Pol in the latter. Interestingly, we observed a strong accumulation of replication intermediates at the very end of the operon, including the transcription terminator, suggesting the encounter of replication forks with a highly stable nucleoprotein complex. To our knowledge such complexes have not been described so far in the rrn transcription termination region, even though RNA Pols may accumulate at the terminator and replication arrest was described at rrn terminators in E. coli plasmids during co-directional collisions (Mirkin et al, 2006).
Suppression of the LB sensitivity of the rep uvrD mutant by rpoC* indicates that in wild-type E.coli these two helicases are essential for replication across highly transcribed regions. On chromosome inversion, Rep and UvrD are also required to replicate across genes other than rrn, as the InvBE rep uvrD recF cells lacking rrnE and rrnB remain sensitive to LB. It is noteworthy that the InvBE inversion also carries genes encoding ribosomal proteins. Future work will tell whether replication is hampered by these or by other specific sequences, and/or by the high number of genes in an inverted orientation.
RNA Pol removal in other contexts
In E. coli, removal of RNA Pol from damaged DNA is performed during transcription coupled repair by the Mfd helicase, which also attracts the nucleotide excision repair machinery (reviewed in Selby and Sancar, 1994). Mfd translocates on DNA behind the RNA Pol in the direction of transcription, pushing a blocked RNA Pol forward, which causes its dissociation from DNA when its advance is prevented by a lesion or a DNA-bound protein (Park et al, 2002). Mfd action on a series of RNA Pol is likely to be prevented by steric hindrance; however, it would be interesting to test whether Mfd participates in the dislodging of single RNA Pol from replication forks. It is noteworthy that Mfd would only dislodge RNA Pol as long as the replisome prevents its forward movement.
The presence in numerous organisms of DinG and UvrD homologues underlines the importance of these proteins. In several Gram-positive bacteria, rep and uvrD are a single, essential gene, pcrA (Petit and Ehrlich, 2002 and references therein). Interestingly, in a two-hybrid assay PcrA interacts with the Bacillus subtilis RNA polymerase (Noirot-Gros et al, 2002). DinG in B. subtilis carries an N-terminal exonuclease domain, raising the possibility that DinG in E. coli functions in conjunction with an RNase (Moser et al, 1997). DinG is homologous to Rad3 in S. cerevisiae and XPD (ERCC2) in humans, which act in nucleotide excision repair, a function fulfilled by UvrD in E. coli. However, the helicases that remove RNA Pol and other DNA-bound proteins from the path of replication forks in S. cerevisiae are not the DinG, Rep or UvrD homologues but rather two related SF1 helicases, Rrm3 and to a lesser extent Pif1. Helicases of the Pif1 family are conserved from yeast to humans (reviewed in Boule and Zakian, 2006). Mutants lacking these enzymes have been extensively studied in vivo and although Rrm3 is clearly the closest functional homologue to DinG, Rep and UvrD, it travels with the fork and may act before replisome dissociation (Azvolinsky et al, 2006). Owing to the difficulty of purifying these helicases, only the action of Pif1 could be analysed in vitro on model substrates showing that Pif1 unwinds DNA–RNA hybrids, similar to DinG and UvrD (Matson, 1989; Boule and Zakian, 2007; Voloshin and Camerini-Otero, 2007), and recognizes fork substrates, similar to DinG and Rep (Lahaye et al, 1993; Heller and Marians, 2007; Voloshin and Camerini-Otero, 2007).
The physiological role of DinG, Rep and UvrD helicases could only be deduced from detailed in vivo analyses. By revealing the relevant physiological substrates for these helicases, this study paves the way for future experiments aimed at understanding the molecular mechanism of action of enzymes that displace RNA Pol from the path of replication forks.
Materials and methods
Strains and plasmids
All E. coli strains are derivatives of MG1655. Plasmids and strains are described in Supplementary Table S1. MM is M9 (Miller, 1992) complemented with 0.04% glucose. Standard transformation and transduction procedures were as described earlier (Miller, 1992). Chromosome inversions were made as described earlier (Valens et al, 2004). Briefly, strains were first P1 transduced for attR (linked to a kanR marker) and attL (linked to a cmR marker) to construct the non-inverted parental strain. For inversion, the attR attL carrying strain was transformed at 30°C with the plasmid pTSA29-CXI (ts replication, cI857 –PR-(xisλ-intλ), ApR); a transformant propagated at 30°C in exponential phase was shifted to 37°C for 10 min (a control culture was not shifted) and then incubated at 30°C for 1 h. Appropriate dilutions were plated on MM Ap X-gal plates and incubated for 3 days at 30°C, only the cultures that were shifted to 37°C gave rise to blue colonies (Lac+, about 50%). Blue colonies were streaked on MM and then cultured at 37°C with a 4 h shift to 42°C to cure pTSA29-CXI. The inversion was verified by PCR using the oligonucleotides shown in Supplementary Table S6. uvrD mutants were tested for UV sensitivity and mutator phenotype (100-fold excess of RifR clones in overnight cultures). AttL2 insertions and gene inactivation were verified using the oligonucleotides listed in Supplementary Table S6. Oligonucleotides used to synthesize PCR DNA fragments for strain construction, or probes, are listed in Supplementary Table S6. pAM-rep and pAM-priA plasmids were segregated before each experiment as published earlier (Grompone et al, 2004). Briefly, overnight cultures propagated in the presence of 500 μg/ml IPTG and 100 μg/ml Ap were diluted 1000-fold and propagated for 7–8 h in MM at 37°C. Appropriate dilutions were then plated on MM and MM containing IPTG and Ap. Routinely, the number of colonies on MM was 10-fold higher than the number of clones on IPTG/Ap MM plates and more than 90% of the clones obtained on MM were sensitive to Ap.
Measures of plating efficiencies
Overnight cultures (OD650 1.0–1.5) were diluted and plated on MM or LB plates, incubated at 37°C. LB plates were counted after 24 and 48 h of incubation. MM plates were counted after 48 h incubation.
Microscopy
Cells were grown in MM to OD650 0.05–0.1, centrifuged, grown for 1 or 2 h in LB or MM to reach OD650 0.15–0.3 and then observed with a Zeiss microscope by DIC. Photographs were acquired with the Metamorph software; cell lengths were measured by hand under Image J software.
PFGE and 2D gels
Cells grown for microscopy analysis were lysed in plugs as described earlier (Seigneur et al, 1998). Chromosomes embedded in plugs were treated with the appropriate restriction enzyme for 6 h at 37°C according to the instructions of the suppliers.
PFGE was performed in 1% agarose gels, TEB 0.5 × , at 14°C, 6 V/cm, angle 120 deg, in a CHEF DRIII apparatus (Bio-Rad). InvA gels: 11 h, switch time 1–6 s. InvBE gels 19 h, switch time 5–30 s.
2D gel migration was in 1 × TEB and as follows: InvBE/BmgB1 or BssHII and InvA/AflIII: 1st dimension 0.4% agarose, 0.9 V/cm, 22 h at room temperature (RT); 2nd dimension 1% agarose, ethidium bromide (Et Br) 0.5 μg/ml, 5 V/cm, 9 h at 4°C. InvA/BstEII: 1st dimension 0.35% agarose, 0.9 V/cm, 39 h RT; 2nd dimension 0.8% agarose, Et Br 0.5 μg/ml, 1.7 V/cm, 27 h at 4°C.
DNA was transferred from PFG or 2D gels to a nylon membrane and hybridized by the classical Southern technique. Storage phosphor imaging was performed with a Typhoon; image analysis was performed with ImageQuant software.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank F Boccard, S Duigou, R Gourse, CJ Herbert and M-A Petit for very helpful reading of the manuscript, and S Zeytouni and E Long for experiments realized during their undergraduate internship. We are very grateful for all members of the Boccard laboratory for strains, plasmids, and help in the realization of certain experiments. This work was financed by the following grants: ANR-05-BLAN-0204-01, ANR-08-BLAN-0230-01 and Prix ‘coup d'élan' from the foundation Bettencourt-Schueller. ALS was financed by the French Ministry of Research. EV acknowledges the support of the Acciones Integradas' project (Ref. HF2006-0154), Spanish Ministry of Science and Innovation grant BFU2007-64153, a grant from the Junta de Andalucía, and an Egide ‘Picasso' grant.
Footnotes
The authors declare that they have no conflict of interest.
References
- Appleman JA, Ross W, Salomon J, Gourse RL (1998) Activation of Escherichia coli rRNA transcription by FIS during a growth cycle. J Bacteriol 180: 1525–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA (2006) The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev 20: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett MS, Gaal T, Ross W, Gourse RL (1998) RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol 279: 331–345 [DOI] [PubMed] [Google Scholar]
- Bartlett MS, Gaal T, Ross W, Gourse RL (2000) Regulation of rRNA transcription is remarkably robust: FIS compensates for altered nucleoside triphosphate sensing by mutant RNA polymerases at Escherichia coli rrn P1 promoters. J Bacteriol 182: 1969–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P, Hochstrasser M, Jongeneel CV, Alberts BM (1983) Properties of the T4 bacteriophage DNA replication apparatus: the T4 dda DNA helicase is required to pass a bound RNA polymerase molecule. Cell 34: 115–123 [DOI] [PubMed] [Google Scholar]
- Bidnenko V, Lestini R, Michel B (2006) The Escherichia coli UvrD helicase is essential for Tus removal during recombination-dependent replication restart from Ter sites. Mol Microbiol 62: 382–396 [DOI] [PubMed] [Google Scholar]
- Boule JB, Zakian VA (2006) Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res 34: 4147–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule JB, Zakian VA (2007) The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res 35: 5809–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2007) Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amst) 6: 994–1003 [DOI] [PubMed] [Google Scholar]
- Brewer BJ (1988) When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53: 679–686 [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Lockshon D, Fangman WL (1992) The Arrest of Replication Forks in the rDNA of Yeast Occurs Independently of Transcription. Cell 71: 267–276 [DOI] [PubMed] [Google Scholar]
- Condon C, Philips J, Fu ZY, Squires C, Squires CL (1992) Comparison of the expression of the seven ribosomal RNA operons in Escherichia coli. EMBO J 11: 4175–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Squires C, Squires CL (1995) Control of rRNA transcription in Escherichia coli. Microbiol Rev 59: 623–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin RW, Paliy O, Yang F, Shabanowitz J, Platt M, Lyons CE Jr, Root K, McAuliffe J, Jordan MI, Kustu S, Soupene E, Hunt DF (2003) Toward a protein profile of Escherichia coli: comparison to its transcription profile. Proc Natl Acad Sci USA 100: 9232–9237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC (2003) RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet 37: 611–646 [DOI] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC (2001) Comparative gene expression profiles following UV exposure in wild- type and SOS-deficient Escherichia coli. Genetics 158: 41–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS (1996) DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Esnault E, Valens M, Espeli O, Boccard F (2007) Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet 3: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores MJ, Sanchez N, Michel B (2005) A fork-clearing role for UvrD. Mol Microbiol 57: 1664–1675 [DOI] [PubMed] [Google Scholar]
- French S (1992) Consequences of replication fork movement through transcription units in vivo. Science 258: 1362–1365 [DOI] [PubMed] [Google Scholar]
- Grompone G, Sanchez N, Dusko Ehrlich S, Michel B (2004) Requirement for RecFOR-mediated recombination in priA mutant. Mol Microbiol 52: 551–562 [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ (2005) Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J Biol Chem 280: 34143–34151 [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ (2007) Non-replicative helicases at the replication fork. DNA Repair (Amst) 6: 945–952 [DOI] [PubMed] [Google Scholar]
- Hirvonen CA, Ross W, Wozniak CE, Marasco E, Anthony JR, Aiyar SE, Newburn VH, Gourse RL (2001) Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J Bacteriol 183: 6305–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV (1993) Escherichia coli dinG gene encodes a putative DNA helicase related to a group of eukaryotic helicases including Rad3 protein. Nucleic Acids Res 21: 1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye A, Leterme S, Foury F (1993) PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J Biol Chem 268: 26155–26161 [PubMed] [Google Scholar]
- Lane HE, Denhardt DT (1975) The rep mutation. IV. Slower movement of replication forks in Escherichia coli rep strains. J Mol Biol 97: 99–112 [DOI] [PubMed] [Google Scholar]
- Lecointe F, Serena C, Velten M, Costes A, McGovern S, Meile JC, Errington J, Ehrlich SD, Noirot P, Polard P (2007) Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J 26: 4239–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestini R, Michel B (2007) UvrD controls the access of recombination proteins to blocked replication forks. EMBO J 26: 3804–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestini R, Michel B (2008) UvrD and UvrD252 counteract RecQ, RecJ, and RecFOR in a rep mutant of Escherichia coli. J Bacteriol 190: 5995–6001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LK, Jenkins ME, Mount DW (1992) Isolation of DNA damage-inducible promoters in Escherichia-Coli—regulation of polB (dinA), dinG, and dinH by LexA repressor. J Bacteriol 174: 3377–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF (2008) Structure of the DNA repair helicase XPD. Cell 133: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Campistrous A, Semchuk P, Burke L, Palmer-Stone T, Brokx SJ, Broderick G, Bottorff D, Bolch S, Weiner JH, Ellison MJ (2005) Localization, annotation, and comparison of the Escherichia coli K-12 proteome under two states of growth. Mol Cell Proteomics 4: 1205–1209 [DOI] [PubMed] [Google Scholar]
- Masse E, Phoenix P, Drolet M (1997) DNA topoisomerases regulate R-loop formation during transcription of the rrnB operon in Escherichia coli. J Biol Chem 272: 12816–12823 [DOI] [PubMed] [Google Scholar]
- Matson SW (1989) Escherichia coli DNA helicase II (uvrD gene product) catalyzes the unwinding of DNA. RNA hybrids in vitro. Proc Natl Acad Sci USA 86: 4430–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH (1992) A Short Course in Bacterial Genetic. Cold Spring Harbor Press: Cold Spring Harbor, NY [Google Scholar]
- Mirkin EV, Castro Roa D, Nudler E, Mirkin SM (2006) Transcription regulatory elements are punctuation marks for DNA replication. Proc Natl Acad Sci USA 103: 7276–7281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM (2005) Mechanisms of transcription-replication collisions in bacteria. Mol Cell Biol 25: 888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin EV, Mirkin SM (2007) Replication fork stalling at natural impediments. Microbiol Mol Biol Rev 71: 13–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser MJ, Holley WR, Chatterjee A, Mian IS (1997) The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res 25: 5110–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noirot-Gros MF, Dervyn E, Wu LJ, Mervelet P, Errington J, Ehrlich SD, Noirot P (2002) An expanded view of bacterial DNA replication. Proc Natl Acad Sci USA 99: 8342–8347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JS, Marr MT, Roberts JW (2002) E. coli Transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Cell 109: 757–767 [DOI] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL (2004) rRNA transcription in Escherichia coli. Annu Rev Genet 38: 749–770 [DOI] [PubMed] [Google Scholar]
- Petit MA, Ehrlich D (2002) Essential bacterial helicases that counteract the toxicity of recombination proteins. EMBO J 21: 3137–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz RT, O'Donnell M (2008) The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456: 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A (2005) Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 24: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EP, Danchin A (2003) Gene essentiality determines chromosome organisation in bacteria. Nucleic Acids Res 31: 6570–6577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J, Makrantoni V, Ingledew WJ, Stark MJ, White MF (2006) The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol Cell 23: 801–808 [DOI] [PubMed] [Google Scholar]
- Sandler SJ (2000) Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics 155: 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M, Roberts JW (1990) Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol 212: 79–96 [DOI] [PubMed] [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B (1998) RuvAB acts at arrested replication forks. Cell 95: 419–430 [DOI] [PubMed] [Google Scholar]
- Selby CP, Sancar A (1994) Mechanisms of transcription-repair coupling and mutation frequency decline. Microbiol Rev 58: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T (2003) Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JZ, Schnakenberg SL, Zakian VA (2004) Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Mol Cell Biol 24: 3198–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourriere H, Pasero P (2007) Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 6: 900–913 [DOI] [PubMed] [Google Scholar]
- Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG (2005) RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell 19: 247–258 [DOI] [PubMed] [Google Scholar]
- Trautinger BW, Lloyd RG (2002) Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J 21: 6944–6953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, Cornet F, Boccard F (2004) Macrodomain organization of the Escherichia coli chromosome. EMBO J 23: 4330–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA (2005) UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J 24: 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viguera E, Rodriguez A, Hernandez P, Krimer DB, Trellez O, Schvartzman JB (1998) A computer model for the analysis of DNA replication intermediates by two-dimensional agarose gel electrophoresis. Gene 217: 41–49 [DOI] [PubMed] [Google Scholar]
- Vilette D, Ehrlich SD, Michel B (1995) Transcription-induced deletions in Escherichia coli plasmids. Mol Microbiol 17: 493–504 [DOI] [PubMed] [Google Scholar]
- Voloshin ON, Camerini-Otero RD (2007) The DinG protein from Escherichia coli is a structure-specific helicase. J Biol Chem 282: 18437–18447 [DOI] [PubMed] [Google Scholar]
- Voloshin ON, Vanevski F, Khil PP, Camerini-Otero RD (2003) Characterization of the DNA damage-inducible helicase DinG from Escherichia coli. J Biol Chem 278: 28284–28293 [DOI] [PubMed] [Google Scholar]
- Wang JD, Berkmen MB, Grossman AD (2007) Genome-wide coorientation of replication and transcription reduces adverse effects on replication in Bacillus subtilis. Proc Natl Acad Sci USA 104: 5608–5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey-Wrona JE, Matson SW (1992) Bound Lac repressor protein differentially inhibits the unwinding reactions catalyzed by DNA helicases. Nucleic Acids Res 20: 6713–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Nagata T, Ohmori H (1996) Multicopy suppressors of the cold-sensitive phenotype of the pcsA68 (dinD68) mutation in Escherichia coli. J Bacteriol 178: 3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File