Abstract
Pre- and neonatal overfeeding programmes a permanent obesity disposition and accompanying diabetic and cardiovascular disorders, by unknown mechanisms. We proposed that early overfeeding may alter DNA methylation patterns of hypothalamic promoter regions of genes critically involved in the lifelong regulation of food intake and body weight. We induced neonatal overfeeding by rearing Wistar rats in small litters (SL) and thereafter mapped the DNA methylation status of CpG dinucleotides of gene promoters from hypothalamic tissue, using bisulfite sequencing. Neonatal overfeeding led to rapid early weight gain, resulting in a metabolic syndrome phenotype, i.e. obesity, hyperleptinaemia, hyperglycaemia, hyperinsulinaemia, and an increased insulin/glucose ratio. Accompanying, without group difference to controls, the promoter of the main orexigenic neurohormone, neuropeptide Y, was methylated at low levels (i.e. < 5%). In contrast, in SL rats the hypothalamic gene promoter of the main anorexigenic neurohormone, proopiomelanocortin (POMC), showed hypermethylation (P < 0.05) of CpG dinucleotides within the two Sp1-related binding sequences (Sp1, NF-κB) which are essential for the mediation of leptin and insulin effects on POMC expression. Consequently, POMC expression lacked upregulation, despite hyperleptinaemia and hyperinsulinaemia. Accordingly, the extent of DNA methylation within Sp1-related binding sequences was inversely correlated to the quotients of POMC expression/leptin (P= 0.02) and POMC expression/insulin (P < 0.001), indicating functionality of acquired epigenomic alterations. These data for the first time demonstrate a nutritionally acquired alteration of the methylation pattern and, consequently, the regulatory ‘set point’ of a gene promoter that is critical for body weight regulation. Our findings reveal overfeeding as an epigenetic risk factor of obesity programming and consecutive diabetic and cardiovascular disorders and diseases, in terms of the metabolic syndrome.
Introduction
Overweight is becoming increasingly prevalent even in childhood, for unclear reasons. Overweight and obesity are probably the most important risk factors for the development of type 2 diabetes, the metabolic syndrome and cardiovascular diseases (Ford et al. 2002). Consequently, even the latter at present applies to children and adolescents (Goodman et al. 2005). Accordingly, obesity epidemics and subsequent morbidity and mortality are among the major health challenges nearly worldwide.
Environmental alterations during ‘critical periods’ of development, especially materno-fetal hyperglycaemia, early overfeeding, and rapid weight gain may permanently increase the risk of overweight and associated diseases (Plagemann et al. 1999a; Boullu-Ciocca et al. 2005; Plagemann, 2005; Stettler et al. 2005; Singhal et al. 2007). For this and other phenomena of early imprinting of fundamental life functions and its potential consequences for the development of chronic diseases, the term ‘perinatal programming’ has been proposed (Dörner, 1975; Plagemann, 2005). Although this concept dates back to the 1970s (Dörner, 1975), it had not been widely recognized until the ‘small-baby-syndrome’ hypothesis was formulated (Hales & Barker, 1992). Obesity and diabetes, with its metabolic and cardiovascular consequences following altered pre- and neonatal conditions, have to be regarded as the longest- and best-investigated examples of perinatal programming (Dörner, 1975; Plagemann et al. 1999a; Plagemann, 2004, 2005; Boullu-Ciocca et al. 2005; Gillman, 2005; Stettler et al. 2005; Singhal et al. 2007).
Rats raised in ‘small litters’ (SL) are an established animal model in this field. Through artificial reduction of the natural litter size to only three pups per nest SL neonates are subjected to early overfeeding, indicated e.g. by early hyperglycaemia, rapid fat accumulation, and obesity. By unknown mechanisms obesity disposition is preserved permanently, accompanied by hyperphagia, hyperinsulinaemia, hyperleptinaemia, diabetic and cardiovascular disturbances throughout later life (Plagemann et al. 1999a; Plagemann, 2005; Boullu-Ciocca et al. 2005).
Neuropeptidergic circuits in the mediobasal hypothalamus play a key role in the regulation of feeding and body weight (Schwartz et al. 2000). Physiologically, circulating leptin and insulin stimulate in particular the expression of the anorexigenic neurohormone proopiomelanocortin (POMC), while inhibiting the orexigenic neuropeptide Y (NPY) (Schwartz et al. 2000; Cone, 2005; Horvath, 2005). Early overfeeding leads to permanent dysregulation of these hypothalamic circuits, including functional resistance to insulin and leptin, which may underlie permanently increased food intake and overweight (Plagemann et al. 1999b; Davidowa & Plagemann, 2000, 2007; Davidowa et al. 2003; Plagemann, 2005). Remarkably, similar findings have recently been obtained in offspring of rat dams with diet-induced obesity during pregnancy (Kirk et al. 2009). Causal molecular mechanisms are unknown.
Hormones play a pivotal role in the functional and structural organization of feeding circuits (Plagemann, 2004, 2005), as recently shown for the melanocortinergic system (Gao et al. 2007). The question remains, however, whether and how such processes of programming might be translated to the genomic level. Epigenetic molecular events leading to permanent alterations in gene expression, such as changes in DNA methylation status (Jirtle & Skinner, 2007), could form a general basis of perinatal programming. For glucocorticoids, it has been shown that environmental influences during neonatal life can induce alterations in central nervous promoter methylation, with lasting consequences for gene expression and consecutive stress response (Weaver et al. 2004; McGowan et al. 2009). Similarly, neonatal overfeeding might lead to altered methylation of promoter regions of neuropeptides at central sites critical for the regulation of body weight and metabolism. This could result e.g. in permanent uncoupling of neuropeptide gene expression from circulating leptin and insulin levels, explaining lasting adipogenic and diabetogenic consequences of neonatal overfeeding. We therefore hypothesized that early overnourishment may lead to acquired epigenetic alterations, in terms of altered promoter methylation, of neurohormonal systems involved in feeding, metabolism and body weight regulation (Plagemann, 2005).
To evaluate our hypothesis we investigated whether rats overfed in SL develop alterations in DNA methylation patterns of promoters of genes encoding orexigenic and anorexigenic hypothalamic neuropeptides, thereby potentially contributing to the development of obesity, diabetes and cardiovascular alterations.
Methods
Animal model
The investigations were performed in offspring, bred in our laboratory, of Wistar rats of an outbred colony (CRL:WI; Charles River, Sulzfeld, Germany). All animal procedures were approved by the local animal welfare committee (G 0093/02; LAGETSI Berlin, Germany) and were in accordance with the European Communities Council Directive (86/609/EEC). Virgin female rats were time-mated with normal males at the age of 3 months. On the first day of life, newborns were distributed among the mothers (n= 13) randomly, for minimizing confounding by differences in maternal care and lactational performance. To induce neonatal overnutrition, the primary litter size was then reduced on the third day of life to only three pups per litter (small litters/nests; SL; n= 8 litters). In normal litters, the size was adjusted to 12 neonates per mother (normal litters; NL; n= 5 litters) (Fiorotto et al. 1991; Plagemann et al. 1999a,b; Davidowa & Plagemann, 2003; Boullu-Ciocca et al. 2005). The time point of the third day of life was chosen because it has been shown that lactation is fully established then (Fiorotto et al. 1991).
All animals were kept under standard conditions with 12 h:12 h inverse light–dark rhythm. They had free access to tap water and standard pellet diet (commercial control diet for rats, energy content: 3.5 kcal g−1, Code 1000; Altromin, Lage, Germany).
Body weight, body fat, blood glucose, insulin and leptin
Body weight was measured on postnatal days (P) 1, 3, 7, 14 and 21 with an accuracy of 0.1 g (scales, Sartorius MC 1, Laboratory LC 6200). Body length was taken on P21 with an accuracy of 0.5 cm. For determination of body fat, carcass mass was measured after stomach and intestine had been removed. Body fat content was evaluated by drying the carcass to constant weight, followed by whole-body chloroform extraction in a Soxhlet apparatus (Schmidt et al. 2000).
Trunk blood was collected after rapid decapitation on P21 for determining blood glucose, insulin and leptin. Blood glucose was assayed by photometry using the glucose oxidase–peroxidase method (Dr. Lange GmbH, Berlin, Germany). Within one assay for determination of immunoreactive plasma insulin a modified commercial radioimmunoassay was performed (Adaltis, Freiburg, Germany). Rat insulin (Novo Nordisk Biolabs, Copenhagen, Denmark) with a biological potency of 21.3 IU mg−1 was used as standard preparation. The intra-assay coefficient of variation was 4.5–7.4% in a concentration range of 9.1–94.2 mIU l−1. Leptin concentration was measured using a commercial radioimmunoassay (rat leptin RIA kit, Linco). Recombinant rat leptin (Linco) served as the standard preparation. The intra-assay variation was 2.0–4.6% in a concentration range of 1.6–11.6 μg l−1. A total of nine rats from five normal litters and nine rats from eight small litters were randomly assigned to this part of the study.
Tissue preparation
Rats were killed on P21 by rapid decapitation. Brains were quickly removed, frozen on dry ice immediately, and then stored at −70°C until subsequent preparation. Preparation of whole hypothalami was performed on a pre-chilled glass slide within a cryostat at −20°C. The brain was placed with its ventral surface facing up under a stereomicroscope. Transverse cuts through the caudal optic chiasm and immediately posterior to the mamilliary bodies were performed using a pre-chilled razor blade. By a horizontal cut below the anterior commissure and vertical cuts passing through the perihypothalamic sulci the hypothalami were removed en bloc and were subsequently analysed in total (Plagemann et al. 1998, 1999b; McGowan et al. 2009).
DNA extraction and sodium bisulfite modification
Dissected tissues were homogenized in Trizol Reagent (Gibco BRL, Invitrogen, Karlsruhe, Germany). Genomic DNA was isolated according to the manufacturer's instructions.
Sodium bisulfite modification, amplification of modified DNA, ligation and cloning were performed as described by Olek et al. and McGowan et al. with slight modifications (Olek et al. 1996; Harder et al. 2004; McGowan et al. 2009). The method of sodium bisulfite modification was chosen for detecting 5′-methylcytosines because it has been demonstrated to be the most sensitive method of detecting CpG methylation (Clark et al. 2006). While treating fragmented DNA with sodium metabisulfite, unmethylated cytosines are converted to uracil residues by deamination. During PCR both uracil and thymine are amplified as thymine.
Bisulfite modification
The bisulfite reaction was carried out in 3 μg of linear fragmented DNA after digestion with the restriction enzyme Rsa I. Following ethanol-induced DNA precipitation, DNA was denatured by 1 m NaOH and embedded in agarose beads (2% low melting point agarose) (Sigma-Aldrich Chemie GmbH, Steinheim, Germany). Sodium metabisulfite (Sigma-Aldrich Chemie GmbH), to a final concentration of 2.5 m, and hydroquinone (Sigma-Aldrich Chemie GmbH), to a final concentration of 125 mm, pH 5.0, were added to agarose beads and incubated at 50°C for 5 h. Reaction was stopped by 1 × Tris/HCl/ethylenediamine tetra-acetic acid (EDTA); Tris (Merck, Darmstadt, Germany), HCl (Merck), EDTA (Sigma-Aldrich Chemie GmbH). Purification of agarose beads was performed as previously described (Harder et al. 2004).
Selection of NPY promoter and POMC promoter fragments
Using all currently available literature and gene data bank information, we selected for investigations a 246 base-pair fragment of the NPY promoter (Genbank accession no.: M15792). According to the definition by Gardiner-Garden & Frommer (1987), the rat NPY promoter is located in a CpG island (Larhammar et al. 1987). The NPY fragment investigated here (−246 –−1) included the minimal promoter of the NPY gene (Larhammar et al. 1987; Minth & Dixon, 1990; Minth-Worby, 1994). The fragment covered all currently known consensus sequences of major transcription factor binding sites. These include the binding sequences of nerve growth factor (NGF), activator protein 1 (AP-1), activator protein 2α (AP-2α), nerve growth factor-induced gene A (NGFI-A) and specific protein 1 (Sp1) (Minth & Dixon, 1990; Higuchi et al. 1992; Minth-Worby, 1994; Lerchen et al. 1995; Li et al. 2000; Shimizu-Albergine et al. 2001). Analogously, a 404 base-pair fragment of the POMC promoter was investigated (Genbank accession no.: M74296). The rat POMC gene promoter is also located in a CpG island (Newell-Price et al. 2001). The POMC promoter fragment investigated here spans the tissue-specific 5′CpG-island which was described for the human POMC promoter to be differently methylated in various tissue types (Newell-Price et al. 2001) and includes all DNA binding sequences of regulatory elements that were characterized to be relevant for transcriptional expression of the POMC gene. This included the consensus sequences of nuclear factor κB (NF-κB), Sp1, signal transducer and activator of transcription 3 (STAT 3), negative glucocorticoid receptor element (nGRE) and forkhead box O1 (FoxO1) (Therrien & Drouin, 1991; Drouin et al. 1993; Jin et al. 1994; Liu & Mortrud, 1995; Mynard et al. 2002; Kitamura et al. 2006).
Amplification of promoter regions, ligation, cloning and sequencing
Bisulfite-modified DNA embedded in agarose beads was amplified either with the forward primer NPY (−290 bp/−264 bp): 5′-TTT TTG TTT TAT TTT TTT TTT TGG TAG-3′ and the reverse primer NPY (+83 bp/+105 bp): 5′-TCC CAA TTA ATC CTA ACA CTC AC-3′ (annealing 54°C, 45 cycles) or with the forward primer POMC (−540 bp/−515 bp): 5′-GTT TTG GGT TGT TAT GAT TTT TGA T-3′ and the reverse primer POMC (−1 bp/−23 bp): 5′-AAT CCC TAT CAC TCT TCT CTC TT-3′ (annealing 58°C, 40 cycles). Genomic fragments of NPY promoter of 395 bp and of POMC promoter of 540 bp were amplified and purified with QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions.
Amplified and purified DNA was ligated into the vector pCR2.1-TOPO (Invitrogen, Karlsruhe, Germany) and transformed into chemically competent cells E. coli TOP10F′ (Invitrogen), according to the manufacturer's instructions of TOPO TA Cloning Kit (Invitrogen). Plasmids were plated on media containing ampicillin (Sigma-Aldrich Chemie GmbH), X-gal (Bio Vectra, Charlottetown, Canada) and IPTG (Sigma-Aldrich Chemie GmbH) and were incubated overnight. Transformants were assayed for the presence of recombinant inserts by the blue/white colony phenotype and plasmids were extracted with QIAprep Spin Miniprep Kit (Qiagen) according to the manufacturer's instructions. Inserts were controlled with restriction enzyme Bstx I (Promega, Mannheim, Germany). The inserts in plasmids were amplified with the forward primer pUC/M13: 5′-GTT TTC CCA GTC ACG AC-3′ and the reverse primer pUC/M13: 5′-CAG GAA ACA GCT ATG AC-3′ (annealing 50°C, 35 cycles). Purification was performed with the QIAquick PCR Purification Kit (Qiagen) according to the manufacturer's instructions. The NPY fragment included nucleotides −246 to −1, the POMC fragment nucleotides −404 to −1 of their respective primary promoter.
Eleven to eighteen clones per animal were sequenced for each promoter (Genetic Analyzer 3100; Applied Biosystems, Darmstadt, Germany) using the pUC/M13 reverse primer and the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit v2.0 (Applied Biosystems) according to the manufacturer's instructions. A total of eight rats from five normal litters and eight rats from eight small litters were randomly assigned to this part of the study.
RNA extraction, cDNA synthesis and RT-PCR
In hypothalami prepared as described above, the arcuate hypothalamic nucleus (ARC) was dissected (Plagemann et al. 1999b) and mRNA was isolated by TRIZOL Reagent (GibcoBRL, Invitrogen) according to the manufacturer's instructions. RNA (3 μg) was reverse transcribed into complementary DNA (cDNA) using the reagents provided in the SuperScript First-Strand Synthesis System (Invitrogen) for RT-PCR according to the manufacturer's instructions. Primers were designed spanning an intron, POMC forward (5′-GAG ATT CTG CTA CAG TCG CTC-3′) and POMC reverse (5′-TTG ATG ATG GCG TTC TTG AA′) (Millington et al. 1999). The cDNA was amplified by an annealing temperature of 59°C and 30 cycles. To get a measure of POMC expression relative to the individual leptin or insulin level of each animal, POMC expression was divided by leptin and insulin level, respectively, and presented as relative POMC expression (log a.u. (arbitrary units)). A total of eight rats from five normal litters and eight rats from eight small litters were randomly assigned to this part of the study (same animals as for the methylation part).
Immunocytochemistry
After rapid decapitation brains were removed and fixed in Bouin's solution for 48 h. After embedding, 5 μm-thick serial coronal sections were cut through the hypothalamus at planes 24–32, corresponding to bregma levels −1.40 to −3.60 mm (Paxinos & Watson, 1986). Alternate slides were stained with cresyl violet (Nissl+), or immunostained for POMC, using the avidin–biotin–peroxidase complex (ABC) method (Vectastain Kit; Vector Laboratories, Burlingame, CA, USA). For immunocytochemistry, slides were pretreated with Triton X-100 (Ferak, Berlin, Germany) followed by incubation in methanol containing 0.3% hydrogen peroxide and rinsed with 2% normal horse serum (Vector). Incubation with a rabbit antibody to rat POMC (concentration 1:5000; Phoenix, Belmont, CA, USA) was performed for 48 h in a humid chamber at 4°C. After washing, slides were treated with biotinylated anti-mouse IgG (1:500; Vector) and then incubated with ABC for 2 h. Sections were exposed to a 0.05% solution of 3,3′-diaminobenzidine tetrahydrochloride (Sigma) for 20 min. Specificity of the labelling procedure was verified by the absence of immunocytochemical reaction in sections in which the primary antibody was omitted or was substituted by normal serum. In each animal, the complete cranio-caudal extention of the ARC was covered by stained sections.
Neuromorphometry
Morphometric analysis within the ARC was performed using an image analysing system (KS 400 V.3.0; Zeiss, Jena, Germany) connected by a video camera (DXC-390P; Sony, Japan) to a light microscope (Axioscope; Zeiss). Using successive serial sections, percentage immunopositivity of POMC+ neurons was analysed within the ARC at a final magnification of ×1700 in both hemispheres of the brain by one investigator, without knowledge of the experimental group. Only neurons with a distinct nucleolus and soma appearance were included in the measurement (Plagemann et al. 1998, 1999a,b; Fahrenkrog et al. 2004; Franke et al. 2005). A total of five rats from five normal litters and six rats from six small litters were used for this part of the study.
Data analysis
For data analysis of sequences the software SeqManII (DNASTAR, Madison, WI, USA) was used. Semiquantitative detection of mRNA concentration was performed using the fluorescence-imager Typhoon 8600 (Amersham Biosciences Europe GmbH, Freiburg, Germany) and the software Image Quant (Amersham Biosciences Europe GmbH).
Methylation status was analysed for every single cytosine position of the NPY promoter and the POMC promoter, respectively, using the approach suggested by Siegmund & Laird (2002) and Meaney and colleagues (Weaver et al. 2004; McGowan et al. 2009): The number of methylated cytosines for each cytosine residue was summed up and divided by the total number of clones. This procedure was repeated for each animal. The resulting proportion of methylated cytosines per position is given as a percentage of all cytosine residues in all animals (Siegmund & Laird, 2002; Weaver et al. 2004; McGowan et al. 2009). For statistical analyses, in all variables distribution was tested for normality. In the case of significant non-normality (insulin levels), the values were log-transformed for statistical tests. Group differences were compared using Student's t test. To further evaluate the robustness of the key results, two additional statistical approaches were applied to the methylation data. First, we corrected for multiple testing using the Bonferroni correction. Second, to account for the hierarchical structure of the experiment, i.e. the fact that pups are nested within a litter and therefore do not represent independent measurements (although randomly assigned), additionally mixed-model (multilevel) analyses were performed using litter as the higher-ranking hierarchical level. For correlation Spearman's rank correlation test was used. Data analysis was performed by SPSS (Statistical Package for the Social Sciences for Windows 11.0; SPSS Inc. Munich, Germany).
Results
Effect of neonatal overnutrition on body weight, body fat content and metabolic parameters
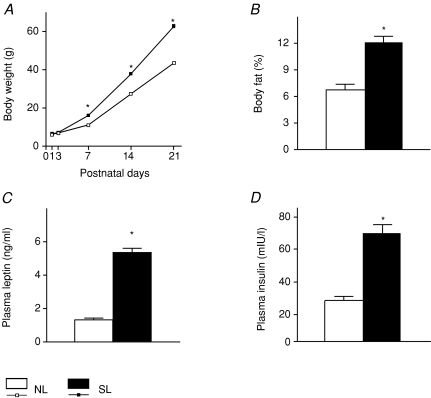
Rats exposed to overfeeding by rearing in SL rapidly developed overweight from P7 onwards (P < 0.001; Fig. 1A). At P21, body weight was highly significantly increased in SL rats, as compared to controls (P < 0.001; Fig. 1A). Analysis of body composition showed that this was due to a nearly doubled percentage of body fat in SL rats (P < 0.001; Fig. 1B). Obesity was accompanied by hyperleptinaemia (P < 0.001), hyperglycaemia (7.5 ± 0.27 mmol l−1vs. 8.3 ± 0.17 mmol l−1; P < 0.05) and hyperinsulinaemia (P < 0.001; Fig. 1C and D). Furthermore, SL rats had a significantly increased insulin/glucose ratio as an indicator of insulin resistance (3.7 ± 0.26 IU mol−1vs. 8.3 ± 0.72 IU mol−1; P < 0.001). Thus, characteristic features of the metabolic syndrome occurred already at weaning in infantile SL animals.
Figure 1.
Body weight development (A), and body fat (B), plasma leptin (C) and insulin (D) in rats raised in small litters (SL) or normal litters (NL; controls) on P21 (n= 9 animals/group). *P < 0.001 (unpaired t test). Data are means ±s.e.m.
DNA methylation pattern of the hypothalamic NPY gene promoter in neonatally overfed rats
Against the background of this acquired phenotype, using bisulfite sequencing as the ‘gold-standard’ technique we mapped the methylation status of cytosines within CpG dinucleotides of the NPY promoter in hypothalamic tissue. The NPY promoter spans over 246 base pairs including 17 CpG dinucleotides (Larhammar et al. 1987). Among the five transcription factor binding sites which were covered by the sequence (NGF-RE, Sp1, AP-1, AP-2, NGFI-A), only those for NGF-RE, AP-2 and NGFI-A included CpG dinucleotides and were therefore potentially accessible to DNA methylation (Fig. 2A). Analysis of single CpG residues showed that the NPY promoter was methylated at low levels in both SL and NL rats: in none of the positions were more than 5% of CpGs found to be methylated (Fig. 2B). The analysis of the three CpG-containing transcription factor binding sites showed no significant group differences (Fig. 2C).
Figure 2.
A, sequence map of the neuropeptide Y (NPY) gene promoter region including functionally regulatory elements (boxed) and CpG dinucleotides (longer lines). *The binding sequences of Sp1 and AP-1 do not contain CpG dinucleotides and are therefore not accessible to DNA methylation. B, methylation analyses of the hypothalamic NPY promoter from small litter (SL) and normal litter (NL; controls) rats. C, percentage of CpG residues that were methylated within the CpG-containing binding sequences for nerve growth factor regulatory element (NGF-RE; −78 to −36), nerve growth factor-induced gene A (NGFI-A; −62) and activator protein 2 (AP-2; −62) within the NPY promoter region. Group differences were not statistically significant (P > 0.05 t test with Bonferroni correction; P > 0.05 after adjustment for litter origin by multilevel analysis). Data are means ±s.e.m. (11–18 clones sequenced per animal; n= 8 animals/group).
DNA methylation pattern of the hypothalamic POMC gene promoter in neonatally overfed rats
Using the same technique, we then mapped the methylation status of CpG dinucleotides within the hypothalamic POMC promoter. The POMC promoter spans over 404 base pairs including 21 CpG dinucleotides (Newell-Price et al. 2001). The sequence covers four activating transcription factor binding sites (FoxO1, STAT3, Sp1, NF-κB) and one inhibiting transcription factor binding site (nGRE). However, only those for NF-κB and nGRE contain CpG dinucleotides and are therefore accessible for methylation. Whereas the Sp1 binding site itself does not contain CpGs, it is known that methylation of the region upstream of this sequence decisively influences Sp1-induced gene transcription (Zhu et al. 2003). Therefore, we additionally investigated this region (Fig. 3A).
Figure 3.
A, sequence map of the proopiomelanocortin (POMC) gene promoter region including functionally regulatory elements (boxed) and CpG dinucleotides (longer lines). *The binding sequences of FoxO1, STAT3 and Sp1 do not contain CpG dinucleotides and are therefore not accessible to DNA methylation. B, methylation analyses of the hypothalamic POMC promoter from small litter (SL) and normal litter (NL; controls) rats. CpG at position –368 was methylated at 84% in both groups (NL: 84 ± 4.3%vs. SL: 84 ± 2.6%) and is therefore not displayed to keep the scale of the y-axis. C, percentage of CpG residues that were methylated within the CpG-containing binding sequences for Sp1 (upstream Sp1; −156 to −15), nuclear factor-κB (NF-κB; −152) and negative glucocorticoid receptor element (nGRE; −62) within the POMC promoter region. *P < 0.05 (t test with Bonferroni correction; P < 0.01 after adjustment for litter origin by multilevel analysis). Data are means ±s.e.m. (11–18 clones sequenced/animal; n= 8 animals per group).
Compared to the NPY promoter, the POMC promoter showed considerably higher levels of DNA methylation. Analysis of single dinucleotides showed that upstream parts of the promoter generally were more strongly methylated in SL rats, as compared to control animals, while, to the contrary, more downstream CpGs tended to be less methylated in SL (Fig. 3B). To quantify these results and to evaluate a potential impact of these group differences on POMC gene expression, we calculated the levels of DNA methylation within the three CpG-containing transcription factor binding sites and regions. Binding sequences of the activating transcription factor NF-κB (P < 0.05) and the region upstream of the Sp1 binding site showed significantly increased methylation in SL rats, as compared to controls (P < 0.05; Fig. 3C). In contrast, the inhibiting nGRE binding sequence showed a non-significantly decreased methylation in SL rats (Fig. 3C). The significance of increased methylation of the NF-κB/SP1 region was confirmed after Bonferroni correction (P < 0.05). The level of significance was even slightly higher for both regions when litter origin was accounted for by multilevel analysis (P < 0.01).
Relations between metabolic parameters and POMC promoter methylation
To evaluate the hypothesis that metabolic alterations due to neonatal overfeeding could be responsible for increased promoter methylation in SL rats, we performed correlation analyses between metabolic parameters and POMC promoter methylation. Remarkably, blood glucose levels showed a significant positive correlation to the degree of POMC promoter methylation (r= 0.63; P= 0.01; Fig. 4A). The absence of significant correlations to leptin (r= 0.12; P= 0.65) and insulin (r= 0.32; P= 0.24) indicated that this was not an unspecific finding caused by group differences in the respective parameters.
Figure 4.
Hypothalamic POMC promoter methylation, presented as a function of neonatal blood glucose levels (A), and hypothalamic POMC expression calculated per corresponding level of leptin (B), and insulin (C), presented as a function of the percentage of CpG residues that are methylated upstream of the SP1 binding sequence of the POMC promoter. Correlation coefficients and significances derived from Spearman rank correlation test.
Consequences of increased POMC promoter methylation for leptin- and insulin-dependent hypothalamic POMC gene expression
We then investigated whether increased methylation of activating transcription factor binding sites might have had consequences for hypothalamic POMC gene expression. Despite 2- to 3-fold increased leptin and insulin levels in SL rats (Fig. 1), which normally would lead to a strong increase in POMC expression (Brown et al. 2006; Sahu, 2008), hypothalamic POMC expression in SL showed no increase, as compared to NL rats (NL: 0.64 ± 0.03 a.u. vs. SL: 0.55 ± 0.05 a.u.; P= 0.13). Additionally, we replicated this finding of a lack of upregulation of POMC on the protein level, showing by immunocytochemistry that POMC immunopositivity did not differ quantitatively between hyperleptinaemic/hyperinsulinaemic SL rats and normoleptinaemic/normoinsulinaemic NL rats (NL: 362 ± 37 POMC+ mm−2vs. SL: 395 ± 6.8 POMC+ mm−2; P= 0.42).
Because of the well-known dependency of POMC expression on circulating leptin and insulin (Schwartz et al. 2000; Cone, 2005), we additionally calculated the quotient of POMC expression per unit of leptin and insulin, respectively. In SL rats, POMC expression was clearly decreased per corresponding leptin ((131 ± 29) × 103 a.u. vs. (35 ± 3) × 103 a.u.; P= 0.01) and insulin ((47 ± 5.8) × 102 a.u. vs. (27 ± 3.2) × 102 a.u.; P= 0.01).
Given that POMC expression is regulated decisively by leptin and insulin, it is not reasonable to correlate the extent of promoter methylation to POMC expression without considering the actual levels of leptin and insulin. We therefore finally used the above-described integrative parameters ‘POMC expression per unit leptin’ and ‘POMC expression per unit insulin’ to evaluate whether the extent by which hypothalamic POMC expression is regulated by leptin and/or insulin might have depended on the level of neonatally acquired POMC promoter methylation. To do so, we correlated the individual percentage of methylation upstream of the activating Sp1 binding site to POMC expression per unit of leptin and insulin, respectively. This analysis revealed significant inverse correlations between the extent of DNA methylation at this promoter site and POMC expression per unit of leptin (Fig. 4B) and insulin (Fig. 4C). That is, the higher the degree of methylation was, the smaller the leptin- and insulin-induced upregulation of hypothalamic POMC expression.
Discussion
This is the first study to demonstrate an epigenetic ‘programming’ effect of an altered nutritional milieu on a central nervous system regulating feeding and body weight control, affecting overweight disposition and thereby the risk of subsequent diabetic and cardiovascular disorders and diseases in terms of the metabolic syndrome.
The methylation pattern of the promoter of NPY is characterized here for the first time in any species and tissue, in general. NPY, known to be the most potent orexigenic neurohormone, particularly acts within the arcuate-paraventricular hypothalamic axis. This hypothalamic system consists of neurons expressing NPY in the arcuate nucleus which project to the paraventricular nucleus. The expression and release of NPY is regulated by circulating insulin and leptin (Schwartz et al. 2000). In a previous study, we observed that the hypothalamic expression of NPY is not decreased in SL rats at weaning, despite largely increased levels of insulin and leptin (Plagemann et al. 1999b), whereas another group even observed a paradoxical increase of hypothalamic NPY expression in SL rats (Lopez et al. 2005). However, in contrast to our expectations, we did not observe a significant group difference in DNA methylation within the hypothalamic NPY promoter here. Rather, the analysis revealed global hypomethylation in all animals, i.e. independent of the mode of early nutrition. Our data might therefore suggest a genuine resistance of the NPY promoter to environmentally induced modifications. This seems plausible, considering NPY as the most powerful orexigenic neuropeptide, which is vitally important for the individuum and therefore highly conserved in different species (Larhammar et al. 1987). Thus, one may speculate that an epigenomic ‘fine-tuning’ of food intake and body weight regulation is rather realized via anorexigenic than orexigenic mechanisms. This would make sense, even from an evolutionary perspective, to ensure individual survival by adequate fuel supply as a primary and overall purpose.
The anorexigenic melanocortinergic system has been suggested to be the best-characterized hypothalamic circuit involved in the regulation of energy balance (Cone, 2005). In the hypothalamus, POMC, the precursor molecule of the post-translational cleavage peptide α−melanocyte-stimulating hormone which acts via the melanocortin-4 receptor in the paraventricular nucleus, is synthesized exclusively in the ARC (Cone, 2005). An increase in POMC expression in the ARC reduces food intake and decreases body weight, while the expression, synthesis and release of POMC are physiologically increased in the presence of elevated leptin and insulin, which thereby act as circulating satiety signals (Schwartz et al. 2000; Cone, 2005). In this context, it is important to note that POMC expression has been shown to be strongly influenced by promoter methylation (Newell-Price et al. 2001). Our study is the first one to investigate POMC promoter methylation in the hypothalamus. A comparison with the literature reveals that methylation levels within the hypothalamus are lower than in non-POMC-expressing tissues, such as lung, but higher and more variable than in other POMC-expressing organs, such as pituitary (Newell-Price et al. 2001).
We show here that both of the two CpG-containing binding sites which activate hypothalamic POMC expression become hypermethylated through overfeeding. Sp1 is a major activator of POMC transcription (Therrien & Drouin, 1991). It has recently been shown that activation of the Sp1 binding site in the POMC promoter is essential for the mediation of leptin effects on POMC expression (Yang et al. 2009). The expression of Sp1 is physiologically stimulated by insulin (Pan et al. 2001). It is important to note that instead of methylation of the Sp1 binding site itself, methylation upstream of this sequence has been shown to inhibit Sp1 binding (Zhu et al. 2003). Hypermethylation upstream of the Sp1 binding sequence, as observed here, may therefore impair POMC expression in hyperinsulinaemic SL rats in terms of acquired ‘silencing’ at this site. On the other hand, Sp1 can also bind to the NF-κB site. However, this partly overlapping binding sequence also became significantly hypermethylated in SL rats. Moreover, simultaneously the only inhibitory regulator of POMC expression, the nGRE binding site (Drouin et al. 1993), was not normally or hyper-methylated but, on the contrary, was found to be even less methylated, thereby potentially enhancing inhibition of POMC transcription. This complex of specific alterations with varying directions strongly suggests functionality of acquired methylation pattern.
We therefore tested whether increased methylation of activating transcription factor binding sites had consequences for gene expression. It is important to consider that the hypothalamic POMC expression is highly dynamically influenced by the circulating levels of leptin and insulin. In the presence of high leptin and insulin, POMC expression increases (Brown et al. 2006; Sahu, 2008). In neonatally overnourished SL rats, however, marked hyperinsulinaemia and hyperleptinaemia were not accompanied by an increased expression of POMC, as one would expect from the physiology of regulation (Schwartz et al. 2000; Cone, 2005; Brown et al. 2006; Sahu, 2008). Rather, neither POMC mRNA expression nor POMC immunopositivity in the ARC, i.e. the hypothalamic expression locus of POMC (Cone, 2005), differed between overfed and control rats. This lack of upregulation of the major anorexigenic neurohormone despite the presence of hyperleptinaemia and hyperinsulinaemia clearly indicates a neonatally acquired leptin and insulin resistance, as it was functionally demonstrated during previous studies (Davidowa & Plagemann, 2000, 2007).
In our study, methylation patterns were obtained using bisulfite sequencing. Although this method is regarded to be the ‘gold standard’ technique for DNA methylation analysis (Clark et al. 2006), one has to consider the potential limitations of this approach. One of them could be PCR bias. This term is used in the literature to describe a potential pitfall during the conduct of methylation analysis whereby certain sequences may be amplified preferentially during PCR, leading to an inaccurate estimate of methylation (Warnecke et al. 1997). To control for PCR bias, particular attention must be given to the design of PCR primers for the methylation study. Recently, Wojdacz et al. (2008) have suggested a list of five criteria of PCR primer design which have to be met to avoid PCR bias, e.g. melting temperature limits and conduct of specific control PCRs. All of these criteria were met in our study in both cases of promoter analysis, thereby strongly limiting the possibility of PCR bias.
To evaluate the potential limitations of our study regarding the number of clones and animals used, we made two calculations of its statistical power. We first calculated the power of our study to detect a significant result at the level of P < 0.05. Given eight animals in each group (NL, SL) and standard deviations which are 50% of the mean, our study had a power of 91.5% to detect at least a doubling of the methylation level. Secondly, we aimed to calculate how many clones in each animal would be necessary to achieve a stable estimate of methylation for a single animal. Given the assumption that the between-animal variation in methylation within a group would not be larger than the between-group variation in methylation, i.e. fivefold in the case of the NF-κB site, one could go down in number of clones picked in each animal to seven clones from which significant differences in methylation between animals from the same group at the level of P < 0.05 (power: 80%) could be shown. To summarize, these calculations indicate that our study had a great enough statistical power to reveal significant as well as stable results.
Another potential methodological limitation of our study relates to the fact that our hypothalamic tissue samples used for methylation analysis contained various cell types. Therefore, the observed differences in methylation pattern could theoretically also be caused by a shift in the spatial/number distribution of different cell types which could be differentially methylated. However, while DNA methylation pattern is regarded to be organ-specific, it is not established that methylation differences exist between different cell types within one single organ. Accordingly, in none of the most prominent studies on DNA methylation in models of perinatal programming was the specific type of cells from which the DNA probes came considered (Waterland & Jirtle, 2003; Weaver et al. 2004; Bogdarina et al. 2007; Park et al. 2008; Lillycrop et al. 2008; McGowan et al. 2009).
We found an indication that the specifically altered methylation pattern acquired due to overfeeding is functionally relevant for subsequent mRNA expression. Usually, it could be expected that increased promoter methylation leads to decreased absolute mRNA levels of the respective gene. This scenario is likely in cases where the expression of the respective gene is relatively stable, as in tumour suppressor genes, but does not largely depend on factors which dynamically regulate gene expression. However, it has to be considered that hypothalamic POMC gene expression decisively depends on circulating leptin and insulin. Both signals from the periphery are known to regulate gene expression of POMC in a dose-dependent manner, especially by activating the SP1 binding site of the promoter (Yang et al. 2009). Therefore, from a physiological point of view the level of promoter methylation cannot be directly related to gene transcription, but must be put into the actual, cybernetic context of the given leptin and insulin levels of the individual animal. This has to be considered necessarily, because together with the level of methylation of the transcription factor binding sites the actual concentrations of leptin and insulin produce the ‘net effect’ of POMC gene transcription. We therefore calculated a measure of POMC expression in relation to leptin and insulin to account for this. The degree of methylation upstream of the Sp1 binding site in the POMC promoter was significantly inversely correlated to POMC expression per unit of leptin and insulin, respectively. Although this correlation does not necessarily indicate causality, the results show that altered promoter methylation is related to a functional ‘uncoupling’ of POMC expression from actual circulating leptin and insulin, indicated by failure to upregulate POMC. Thus, overnutrition appears to result in an epigenetically acquired alteration of the regulatory ‘set point’ of POMC, with ‘normal’ expression occurring only in the presence of hyperinsulinaemia and hyperleptinaemia, indicating an acquired malprogramming.
The question remains how overfeeding may induce these alterations. Three potential mechanisms should be proposed here. Firstly, hyperinsulinism, as occurring due to overnutrition, induces peripheral as well as central nervous insulin resistance (Plagemann et al. 1999b; Davidowa et al. 2003; Plagemann, 2005; Davidowa & Plagemann, 2007), which leads to decreased Sp1 levels and therefore increased accessibility of the Sp1 binding site to methyltransferases, possibly leading to increased methylation. This idea is supported by positive correlations between the insulin/glucose ratio, an indicator of insulin resistance, and the percentage of methylated CpG residues upstream of the Sp1 binding site (r= 0.62; P= 0.01) as well as the NF-κB binding site (r= 0.52; P= 0.04). Secondly, hyperglycaemia has recently been shown to increase DNA-methyltransferase activity and subsequent DNA methylation, leading to glucose-dependent hypermethylation (Chiang et al. 2008). Accordingly, in our study glucose levels were found to be significantly positively correlated to the percentage of methylated POMC promoter cytosines. This even occurred for the constitutionally hypomethylated NPY promoter (r= 0.59; P= 0.02), strongly supporting causality of the relationships. Since central insulin resistance as well as hyperglycaemia, both typically accompanying overfeeding, are crucial features of the metabolic syndrome, a combination of these two alterations seems to be a particularly plausible and potentially even additive mechanism of acquired epigenetic alterations. Thirdly and finally, in overfed SL pups corticosterone has been shown to be increased (Boullu-Ciocca et al. 2005). Subsequently, increased binding of activated glucocorticoid receptors at the nGRE binding site may therefore decrease its accessibility during early development, consequently resulting in hypomethylation. This mechanism seems the more plausible as glucocorticoid-depending alteration of DNA methylation is one of the best investigated mechanisms of epigenetic programming, in general (Weaver et al. 2004; McGowan et al. 2009). Finally, it should be noted that the mechanisms proposed here may even explain specificity of the acquired alterations observed.
Summarizing, this is the first study revealing nutritionally induced alterations of the methylation pattern within a gene promoter that is critical for body weight regulation and, hence, may be directly relevant for the programming and pathogenesis of obesity and its diabetic and cardiovascular endpoints. Alterations were induced during a critical period of postnatal development, i.e. far beyond the established period of epigenomic reprogramming in early embryonic stages. Specificity of affected transcription factor binding sites, varying direction of altered methylation pattern (i.e. hyper- or hypomethylation) as well as impaired gene expression demonstrate functionality of the acquired alterations.
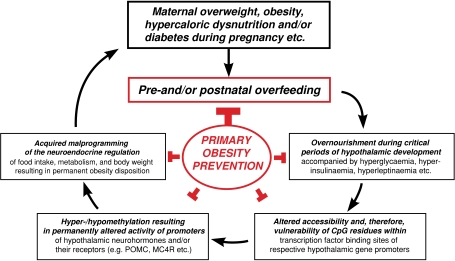
In conclusion, we would like to postulate here a general new aetiological concept of nutritional ‘epigenetic obesity programming’ (Fig. 5). Overfeeding during critical periods of hypothalamic development, especially in the case of maternal gestational overweight/diabetes or neonatal overfeeding, as shown here exemplarily for the SL model, is proposed to lead to an altered accessibility or ‘vulnerability’ of CpG dinucleotides within promoters of hypothalamic genes involved in the regulation of food intake and body weight. The resulting alterations of the methylation pattern of the respective promoters will ultimately change their activity and consecutive gene transcription. Consequently, the respective regulatory system will become malprogrammed, leading to an increased disposition towards obesity and the metabolic syndrome. If female offspring affected in that way enter reproductive age and become pregnant, by being obese and/or diabetic during pregnancy they expose their offspring in a similar way to a hypercaloric/hyperglycaemic perinatal environment that they were exposed to themselves, thereby closing an epigenetic ‘vicious intergenerative circle’. This mechanism might contribute substantially to the epidemics of obesity and ‘diabesity’ but, most of all, seems to be preventable by normalization of food supply in early life.
Figure 5.
Proposal of a general concept of an intergenerative ‘epigenetic vicious circle’ of obesity programming during critical periods of pre- and early postnatal development, caused by permanently altered ‘set points’ of neurohormone and/or receptor expression in neuro-hormonal circuits regulating food intake, metabolism and body weight. Prevention appears to be possible by normalization of food supply in early life.
Acknowledgments
The authors wish to thank Tamas Horvath, New Haven, for critical comments on an earlier draft of the manuscript. This study was supported by funding from the German Research Foundation (PL 241/4-1, 4-2; GRK 1208) to A.P., T.H. and J.W.D.
Glossary
Abbreviations
- ARC
arcuate hypothalamic nucleus
- NF-κB
nuclear factor κB
- NL
normal litters
- NPY
neuropeptide Y
- P
postnatal day
- POMC
proopiomelanocortin
- SL
small litters
- Sp1
specific protein 1
Author contributions
A.P., T.H. and J.W.D. designed the experiments; A.P., T.H., M.B., A.H., K.R., M.W.-S., T.Z., K.S., E.R. and K.M. performed the experiments and analysed the data; A.P., T.H., M.B., A.H., K.R. and M.W.-S. wrote the paper.
References
- Bogdarina I, Welham S, King PJ, Burns SP, Clark AJL. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ Res. 2007;100:520–526. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boullu-Ciocca S, Dutour A, Guillaume V, Achard V, Oliver C, Grino M. Postnatal diet-induced obesity in rats upregulates systemic and adipose tissue glucocorticoid metabolism during development and in adulthood: its relationship with the metabolic syndrome. Diabetes. 2005;54:197–203. doi: 10.2337/diabetes.54.1.197. [DOI] [PubMed] [Google Scholar]
- Brown LM, Clegg DJ, Benoit SC, Woods SC. Intraventricular insulin and leptin reduce food intake and body weight in C57BL/6J mice. Physiol Behav. 2006;89:687–691. doi: 10.1016/j.physbeh.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Chiang EPI, Wang YC, Chen WW, Tang FY. Effects of insulin and glucose on cellular metabolic fluxes in homocysteine transsulfuration, remethylation, S-adenosylmethionine synthesis and global DNA methylation. J Clin Endocrinol Metab. 2008;94:1017–1025. doi: 10.1210/jc.2008-2038. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Statham A, Stirzaker C, Molloy PL, Frommer M. DNA methylation: bisulphite modification and analysis. Nat Protoc. 2006;1:2353–2364. doi: 10.1038/nprot.2006.324. [DOI] [PubMed] [Google Scholar]
- Cone RD. Anatomy and regulation of the central melanocortinergic system. Nat Neurosci. 2005;8:571–578. doi: 10.1038/nn1455. [DOI] [PubMed] [Google Scholar]
- Davidowa H, Li Y, Plagemann A. Altered responses to orexigenic (AGRP, MCH) and anorexigenic (α-MSH, CART) neuropeptides of paraventricular hypothalamic neurons in early postnatally overfed rats. Eur J Neurosci. 2003;18:613–621. doi: 10.1046/j.1460-9568.2003.02789.x. [DOI] [PubMed] [Google Scholar]
- Davidowa H, Plagemann A. Decreased inhibition by leptin of hypothalamic arcuate neurons in neonatally overfed young rats. Neuroreport. 2000;12:2795–2798. doi: 10.1097/00001756-200008210-00037. [DOI] [PubMed] [Google Scholar]
- Davidowa H, Plagemann A. Insulin resistance of hypothalamic arcuate neurons in neonatally overfed rats. Neuroreport. 2007;18:521–524. doi: 10.1097/WNR.0b013e32805dfb93. [DOI] [PubMed] [Google Scholar]
- Dörner G. Perinatal hormone levels and brain organization. In: Stumpf W, Grant LD, editors. Anatomical Neuroendocrinology. Basel: Karger; 1975. pp. 245–252. [Google Scholar]
- Drouin J, Sun YL, Chamberland M, Gauthier Y, De Lean A, Nemer M, Schmidt TJ. Novel glucocorticoid receptor complex with DNA element of the hormonerepressed POMC gene. EMBO J. 1993;12:145–156. doi: 10.1002/j.1460-2075.1993.tb05640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog S, Harder T, Stolaczyk E, Melchior K, Franke K, Dudenhausen JW, Plagemann A. Effects of crossfostering to diabetic rat dams on early development of hypothalamic nuclei regulating food intake, body weight and metabolism. J Nutr. 2004;134:648–654. doi: 10.1093/jn/134.3.648. [DOI] [PubMed] [Google Scholar]
- Fiorotto ML, Burrin DG, Perez M, Reeds PJ. Intake and use of milk nutrients by rat pups suckled in small, medium, or large litters. Am J Physiol Regul Integr Comp Physiol. 1991;260:R1104–R1113. doi: 10.1152/ajpregu.1991.260.6.R1104. [DOI] [PubMed] [Google Scholar]
- Ford ES, Giles WH, Dietz WH. Prevalence of the metabolic syndrome among US adults. Findings from the Third National Health and Nutrition Examination Survey. JAMA. 2002;287:356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- Franke K, Harder T, Aerts L, Melchior K, Fahrenkrog S, Rodekamp E, Ziska T, Van Assche FA, Dudenhausen JW, Plagemann A. Programming of orexigenic and anorexigenic hypothalamic neurons in offspring of treated and untreated diabetic mother rats. Brain Res. 2005;1031:276–283. doi: 10.1016/j.brainres.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Gao Q, Mezei G, Nie Y, Rao Y, Choi CS, Bechmann I, Leranth C, Toran-Allerand D, Priest CA, Roberts JL. Anorectic estrogen mimics leptin's effect on the rewiring of melanocortin cells and Stat3 signalling in obese animals. Nat Med. 2007;13:89–94. doi: 10.1038/nm1525. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gillman MW. Developmental origins of health and disease. N Engl J Med. 2005;353:1848–1850. doi: 10.1056/NEJMe058187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman E, Dolan LM, Morrison JA, Daniels SR. Factor analysis of clustered cardiovascular risks in adolescence: obesity is the predominant correlate of risk among youth. Circulation. 2005;111:1970–1977. doi: 10.1161/01.CIR.0000161957.34198.2B. [DOI] [PubMed] [Google Scholar]
- Hales CN, Barker DJ. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- Harder A, Rosche M, Reuss DE, Holtkamp N, Uhlmann K, Friedrich R, Mautner VF, von Deimling A. Methylation analysis of the neurofibromatosis type 1 (NF1) promoter in peripheral nerve sheath tumours. Eur J Cancer. 2004;40:2820–2828. doi: 10.1016/j.ejca.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Higuchi H, Nakano K, Miki N. Identification of NGF-response element in the rat neuropeptide Y gene and induction of the binding proteins. Biochem Biophys Res Commun. 1992;189:1553–1560. doi: 10.1016/0006-291x(92)90253-h. [DOI] [PubMed] [Google Scholar]
- Horvath TL. The hardship of obesity: a soft-wired hypothalamus. Nat Neurosci. 2005;8:561–565. doi: 10.1038/nn1453. [DOI] [PubMed] [Google Scholar]
- Jin WD, Boutillier AL, Glucksman MJ, Salton SR, Loeffler JP, Roberts JL. Characterization of a corticotropin-releasing hormone-responsive element in the rat proopiomelanocortin gene promoter and molecular cloning of its binding protein. Mol Endocrinol. 1994;8:1377–1388. doi: 10.1210/mend.8.10.7854355. [DOI] [PubMed] [Google Scholar]
- Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk SL, Samuelsson AM, Argenton M, Dhonye H, Kalamatianos T, Poston L, Taylor PD, Coen CW. Maternal obesity induced by diet in rats permanently influences central processes regulating food intake in offspring. PloS One. 2009;4:e5870. doi: 10.1371/journal.pone.0005870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Feng Y, Kitamura YI, Chua SC, Xu AW, Barsh GS, Rossetti L, Accili D. Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat Med. 2006;12:534–540. doi: 10.1038/nm1392. [DOI] [PubMed] [Google Scholar]
- Larhammar D, Ericsson A, Persson H. Structure and expression of the rat neuropeptide Y gene. Proc Natl Acad Sci U S A. 1987;84:2068–2072. doi: 10.1073/pnas.84.7.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerchen RA, Yum DY, Krajcik R, Minth-Worby CA. Transcriptional vs. posttranscriptional control of neuropeptide Y gene expression. Endocrinology. 1995;136:833–841. doi: 10.1210/endo.136.3.7867591. [DOI] [PubMed] [Google Scholar]
- Li BS, Kramer PR, Zhao W, Ma W, Stenger DA, Zhang L. Molecular cloning, expression, and characterization of rat homolog of human AP-2α that stimulates neuropeptide Y transcription activity in response to nerve growth factor. Mol Endocrinol. 2000;14:837–847. doi: 10.1210/mend.14.6.0468. [DOI] [PubMed] [Google Scholar]
- Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPARα promoter of the offspring. Br J Nutr. 2008;100:278–282. doi: 10.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Mortrud M. Low MJ: DNA elements with AT-rich core sequences direct pituitary cell-specific expression of the pro-opiomelanocortin gene in transgenic mice. Biochem J. 1995;312:827–832. doi: 10.1042/bj3120827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez M, Seoane LM, Tovar S, Garcia MC, Nogueiras R, Dieguez C, Senarsi RM. A possible role of neuropeptide Y, agouti-related protein and leptin receptor isoforms in hypothalamic programming by perinatal feeding in the rat. Diabetologia. 2005;48:140–148. doi: 10.1007/s00125-004-1596-z. [DOI] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D’Alessio ACD, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millington WR, Rosenthal DW, Unal CB, Nyquist-Battie C. Localization of pro-opiomelanocortin mRNA transcripts and peptide immunoreactivity in rat heart. Cardiovasc Res. 1999;43:107–116. doi: 10.1016/s0008-6363(99)00076-0. [DOI] [PubMed] [Google Scholar]
- Minth CD, Dixon JE. Expression of the human neuropeptide Y gene. J Biol Chem. 1990;265:12933–12939. [PubMed] [Google Scholar]
- Minth-Worby CA. Transcriptional regulation of the human neuropeptide Y gene by nerve growth factor. J Biol Chem. 1994;269:15460–15468. [PubMed] [Google Scholar]
- Mynard V, Guignat L, Devin-Leclerc J, Bertagna X, Catelli MG. Different mechanisms for leukemia inhibitory factor-dependent activation of two proopiomelanocortin promoter regions. Endocrinology. 2002;143:3916–3924. doi: 10.1210/en.2002-220323. [DOI] [PubMed] [Google Scholar]
- Newell-Price J, King P, Clark AJ. The CpG island promoter of the human proopiomelanocortin gene is methylated in nonexpressing normal tissue and tumors and represses expression. Mol Endocrinol. 2001;15:338–348. doi: 10.1210/mend.15.2.0599. [DOI] [PubMed] [Google Scholar]
- Olek A, Oswald J, Walter J. A modified and improved method for bisulphite based cytosine methylation analysis. Nucleic Acids Res. 1996;24:5064–5066. doi: 10.1093/nar/24.24.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Solomon SS, Borromeo DM, Martinez-Hernandez A, Raghow R. Insulin deprivation leads to deficiency of Sp1 transcription factor in H-411E hepatoma cells and in streptozotocin-induced diabetic ketoacidosis in the rat. Endocrinology. 2001;142:1635–1642. doi: 10.1210/endo.142.4.8083. [DOI] [PubMed] [Google Scholar]
- Park JH, Stoffers DA, Nicholls RD, Simmons RA. Development of type 2 diabetes following intrauterine growth retardation in rats is associated with progressive epigenetic silencing of Pdx1. J Clin Invest. 2008;118:2316–2324. doi: 10.1172/JCI33655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. San Diego: Academic Press; 1986. [Google Scholar]
- Plagemann A. ‘Fetal programming’ and ‘functional teratogenesis’: on epigenetic mechanisms and prevention of perinatally acquired lasting health risks. J Perinat Med. 2004;32:297–305. doi: 10.1515/JPM.2004.055. [DOI] [PubMed] [Google Scholar]
- Plagemann A. Perinatal programming and functional teratogenesis: impact on body weight regulation and obesity. Physiol Behav. 2005;86:661–668. doi: 10.1016/j.physbeh.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Plagemann A, Harder T, Rake A, Melchior K, Rittel F, Rohde W, Dörner G. Hypothalamic insulin and neuropeptide Y in the offspring of gestational diabetic mother rats. Neuroreport. 1998;9:4069–4073. doi: 10.1097/00001756-199812210-00012. [DOI] [PubMed] [Google Scholar]
- Plagemann A, Harder T, Rake A, Voits M, Fink H, Rohde W, Dörner G. Perinatal increase of hypothalamic insulin, acquired malformation of hypothalamic galaninergic neurons, and syndrome X-like alterations in adulthood of neonatally overfed rats. Brain Res. 1999a;836:146–155. doi: 10.1016/s0006-8993(99)01662-5. [DOI] [PubMed] [Google Scholar]
- Plagemann A, Harder T, Rake A, Waas T, Melchior K, Ziska T, Rohde W, Dörner G. Observations on the orexigenic hypothalamic neuropeptide Y-system in neonatally overfed weanling rats. J Neuroendocrinol. 1999b;11:541–546. doi: 10.1046/j.1365-2826.1999.00357.x. [DOI] [PubMed] [Google Scholar]
- Sahu A. Effects of chronic central leptin infusion on proopiomelanocortin and neurotensin gene expression in the rat hypothalamus. Neurosci Lett. 2008;440:125–129. doi: 10.1016/j.neulet.2008.05.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt I, Schoelch C, Ziska T, Schneider D, Simon E, Plagemann A. Interaction of genetic and environmental programming of the leptin system and of obesity disposition. Physiol Genomics. 2000;3:113–120. doi: 10.1152/physiolgenomics.2000.3.2.113. [DOI] [PubMed] [Google Scholar]
- Schwartz MW, Woods SC, Porte D, Seeley RJ, Baskin DG. Central nervous system control of food intake. Nature. 2000;404:661–671. doi: 10.1038/35007534. [DOI] [PubMed] [Google Scholar]
- Shimizu-Albergine M, Ippolito DL, Beavo JA. Downregulation of fasting-induced cAMP response element-mediated gene induction by leptin in neuropeptide Y neurons of the arcuate nucleus. J Neurosci. 2001;21:1238–1246. doi: 10.1523/JNEUROSCI.21-04-01238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Laird PW. Analysis of complex methylation data. Methods. 2002;27:170–178. doi: 10.1016/s1046-2023(02)00071-3. [DOI] [PubMed] [Google Scholar]
- Singhal A, Cole TJ, Fewtrell M, Kennedy K, Stephenson T, Elias-Jones A, Lucas A. Promotion of faster weight gain in infants born small for gestational age: is there an adverse effect on later blood pressure? Circulation. 2007;115:213–220. doi: 10.1161/CIRCULATIONAHA.106.617811. [DOI] [PubMed] [Google Scholar]
- Stettler N, Stallings VA, Troxel AB, Zhao J, Schinnar R, Nelson SE, Ziegler EE, Strom BL. Weight gain in the first week of life and overweight in adulthood: a cohort study of European American subjects fed infant formula. Circulation. 2005;111:1897–1903. doi: 10.1161/01.CIR.0000161797.67671.A7. [DOI] [PubMed] [Google Scholar]
- Therrien M, Drouin J. Pituitary pro-opiomelanocortin gene expression requires synergistic interactions of several regulatory elements. Mol Cell Biol. 1991;11:3492–3503. doi: 10.1128/mcb.11.7.3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke PM, Stirzaker C, Melki JR, Millar DS, Paul CL, Clark SJ. Detection and measurement of PCR bias in quantitative methylation analysis of bisulfite-treated DNA. Nucl Acids Res. 1997;25:4422–4426. doi: 10.1093/nar/25.21.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behaviour. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wojdacz TK, Hansen LL, Dobrovic A. A new approach to primer design for the control of PCR bias in methylation studies. BMC Res Notes. 2008;1:54. doi: 10.1186/1756-0500-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Lim CY, Li C, Xiao X, Radda GK, Li C, Cao X, Han W. FOXO1 inhibits leptin regulation of POMC promoter activity by blocking STAT3 interaction with specific protein 1. J Biol Chem. 2009;284:3719–3727. doi: 10.1074/jbc.M804965200. [DOI] [PubMed] [Google Scholar]
- Zhu WG, Srinivasan K, Dai Z, Duan W, Druhan LJ, Ding H, Yee L, Villalona-Calero MA, Plass C, Otterson GA. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol Cell Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]