Abstract
Background
The composition of the gut microbiome is affected by host phenotype, genotype, immune function, and diet. Here we used the phenotype of RELMβ Knockout (KO) mice to assess the influence of these factors.
Methods and Results
Both wild-type and RELMβ KO mice were lean on a standard chow diet, but upon switching to a high fat diet, wild-type mice became obese while RELMβ KO mice remained comparatively lean. To investigate the influence of diet, genotype, and obesity on microbiome composition we used deep sequencing to characterize 25,790 16S rDNA sequences from uncultured bacterial communities from both genotypes on both diets. We found large alterations associated with switching to the high fat diet, including a decrease in Bacteroidetes and an increase in both Firmicutes and Proteobacteria. This was seen for both genotypes (i.e. in the presence and absence of obesity), indicating that the high fat diet itself, and not the obese state, mainly accounted for the observed changes in the gut microbiota. The RELMβ genotype also modestly influenced microbiome composition independently of diet. Metagenomic analysis of 537,604 sequence reads documented extensive changes in gene content due to a high fat diet, including an increase in transporters and two-component sensor-responders as well as a general decrease in metabolic genes. Unexpectedly, we found a substantial amount of murine DNA in our samples that increased in proportion on a high fat diet.
Conclusions
These results demonstrate the importance of diet as a determinant of gut microbiome composition and suggest the need to control for dietary variation when evaluating the composition of the human gut microbiome.
Introduction
The prevalence of diet-induced obesity is reaching epidemic proportions in industrialized nations. In parallel, there has been a dramatic increase in type 2 diabetes mellitus1. Together, these two diseases are an enormous health and financial burden. To maintain body mass, energy input through food intake and absorption, must match energy expenditure through physical activity, basal metabolism and adaptive thermogenesis. Since triglycerides, stored as fat in white adipose tissue, are the most efficient means of energy storage, alterations in energy balance favoring “energy input” can lead to obesity2.
Growing evidence demonstrates that the normal gut microbiome contributes to the development of diet-induced obesity3-5. The human intestine is home to some 100 trillion microorganisms, representing hundreds and perhaps thousands of species. The density of bacterial cells in the colon has been estimated at 1011 to 1012 per ml, which makes the colon one of the most densely populated microbial habitats known on Earth6, 7. The genome size of this pool of intestinal microbes is estimated to exceed the size of the human nuclear genome by two orders of magnitude7.
The colonization of germfree mice with the normal gut microbiota harvested from conventionally-housed mice leads to an increase in fat mass despite a decrease in food intake8. Furthermore, the gut microbiome of ob/ob mice, which are hyperphagic and become morbidly obese due the absence of the leptin satiety factor, exhibit an altered ratio of abundance in the two dominant bacterial divisions, the Bacteroidetes and the Firmicutes3 similar to that observed in obese humans4. Metagenomic analysis of these communities reveals an increased representation of genes encoding proteins important for the synthesis of short chain fatty acids from the fermentation of dietary carbohydrates3 which can be utilized by the host for hepatic lipogenesis8. In this manner, the microbiome associated with obesity demonstrates an increased capacity to salvage energy from the diet.
We have previously reported the characterization of a colonic goblet cell-specific gene, RELMβ, whose expression is dependent upon the presence of the gut microbiome9. Th2-mediated immune responses strongly activate the expression of RELMβ10. In turn, RELMβ has been shown to be an effector of intestinal immune function11-13. RELMβ protein is secreted apically into the lumen of the bowel by goblet cells, is found at high levels in the stool, and can also be detected in the serum9, 14. A high fat diet induces the expression of RELMβ in the stool14.
Herein, we take advantage of the RELMβ phenotype to investigate the linkage between diet, obesity, and gut microbiome composition. We used deep sequencing of 16S rDNA gene segments and shot-gun metagenomic analysis of fecal bacteria to show that both RELMβ and diet, but not the metabolic phenotype of the host, are independent determinants of the gut microbiome.
Materials and Methods
Animals
RELMβ KO mice were generated on a mixed background, 129Svev/C57BL6, as previously described11. Female RELMβ KO mice were back-bred for five generations onto a C57BL6 background. Wild-type and KO mice were generated from heterozygote parental crosses and were maintained on standard chow (LabDiet 5001) or high fat diet (Research Diets D12451). The composition of these two diets is shown in Supplementary Table 1. Food intake was assessed over 24 hours for a minimum of three consecutive days. All mice had free access to chow and water and were maintained on a twelve-hour light cycle. All experimental protocols were approved by the IACUC committee at the University of Pennsylvania.
We have studied the effect of a high fat diet on the development of diet-induced obesity in several cohorts of RELMβ KO and wild-type mice on a 129Svev/C57BL6 background11 as well as additional cohorts of mice backcrossed five generations to a C57BL/6 strain. Although a divergence in weight was observed in both genders of 129Svev/C57BL6 background mice, this effect was observed only in female mice on the C57B/6 background. Sexual dimorphic effects on diet-induced obesity have been previously described in mice15. Four of these studies showed the divergence of weight on a high fat diet described above, two studies failed to reproduce these findings, indicating incomplete penetrance of the phenotype.
Antibiotic Treatment of Mice
20 female C57B/6 mice 14 weeks of age were equally divided into 4 groups, standard chow, standard chow with oral antibiotics, high fat diet, and high fat diet with oral antibiotics. The oral antibiotics, which were delivered in drinking water, consisted of ampicilllin (1.0 g/l), neomycin sulfate (1.0 g/l), metronidazole (1.0 g/l), and vancomycin (0.5 g/l) as previously described16. After 1 month, spontaneously voided fecal pellets were collected and the colonic tissue was harvested for RNA isolation.
Measurement of Dietary Fat Absorption
Fats with negligible amounts of behenic acid (e.g., safflower oil) were fed to mice along with the sucrose behenate esters (5% of the total dietary fat). After two days of ad lib ingestion of the diet, approximately 10-30 mg of a fecal sample was analyzed by GC for fatty acids following saponification and methylation17.
Assessment of Body Composition
MRI was performed using Echo MRI 3-in-1™, with fat and lean magnetic resonance analysis (Echo Medical Systems, Houston, TX).
Indirect Calorimetry
Energy expenditure was measured by open-circuit calorimetry (Oxymax system, Columbus Instruments, Columbus OH) and locomotor activity was measured simultaneously by infrared beam interruption (Optovarimax System, Columbus Instruments, OH). Mice were housed individually in calorimetry cages at approximately 22° C and acclimatized for 24 hours. Room air was pumped at a rate of 0.52 liters/min and exhaust air was sampled at 27 min intervals for 24 hours.
Core Body Temperature
Rectal body temperatures were measured at room temperature using a thermistor (YSI Model 4600, YSI Temperature, Dayton, OH).
RNA isolation, qRT-PCR, and RELMβ Immunoblot
RNA was isolated followed by reverse transcription and Syber Green qRT-PCR for RELMβ and GAPDH as previously described10. The method used for the isolation of stool protein and immunoblot detection using a polyclonal donkey anti-rabbit antibody to mRELMβ has been previously described9.
Transitions in gut bacterial populations associated with transition from standard chow to high fat diet
Five female RELMβ KOs and five female wild-type littermate controls were raised on standard chow (LabDiet 5001) for 13 weeks at which time fecal pellets were collected and stored at -80°C for subsequent DNA isolation and protein extraction. Mice were then switched to a high fat diet (Research Diets D12451) for 21 weeks at which time fecal pellets were again collected, stored at -80°C, and used for DNA isolation and protein extraction. Previous studies have documented that pellets contain bacterial populations resembling those present in the lower GI tract and so provide a convenient sample source18. DNA was extracted and bacterial 16S rDNA composition quantified using the 454/Roche GS FLX as in McKenna et al.19 and metagenomic analysis was carried out by shotgun sequencing of pooled DNA from pellets according to the 454/Roche protocol (http://www.454.com/). All sequences will be deposited in GeneBank upon acceptance of the paper for publication. We identified the lowest common taxonomic ancestor of the DNA sequences from BLAST hits (bit score > 50) on the nt database using the MEGAN algorithm20. To study the functional genomics of the samples, we collected BLAST hits (e-value < .00001) to UniProt sequences annotated to cellular process pathways by the Kyoto Encyclopedia for Genes and Genomes21 using annot8r (http://www.nematodes.org/bioinformatics/annot8r/index.shtml). Gene function was further investigated using the metagenomics RAST Server (default settings)22. Statistical analysis was carried out in R.
Results
Experimental Plan
We sought to determine the changes in the gut microbiome associated with a high fat diet in mice, and to determine whether the obese state or the high fat diet itself accounted for any changes observed. We took advantage of the phenotype of RELMβ KO, which in this cohort remained comparatively lean on the high fat diet compared to the wild-type controls (this phenotype is incompletely penetrant but was quite strong in the cohort studied here). Mutant or wild-type mice were maintained on standard chow and pellets were harvested. Then mice were switched to the high fat diet for 21 weeks, and pellets sampled again. Below we first describe the RELMβ KO phenotype in this cohort, and then describe the associated microbiome on the two diets.
A high fat diet enhances the expression of RELMβ dependent upon the presence of the gut microbiome
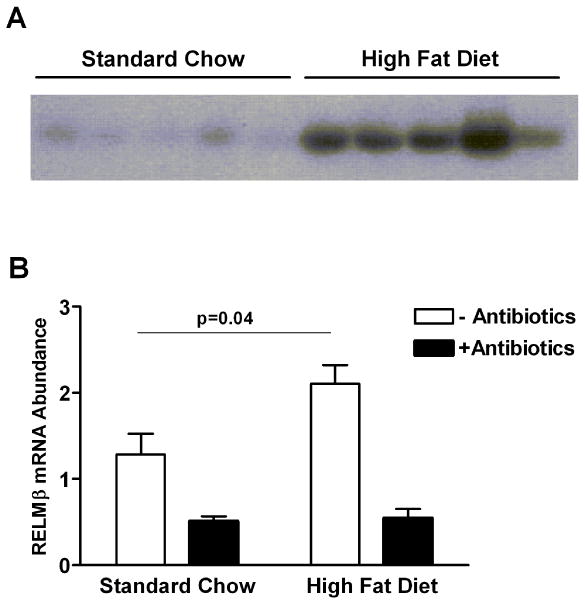
We have previously shown that RELMβ expression23 in the colon and stool9 is dependent upon microbial colonization of the intestinal tract. Alteration in diet also effects RELMβ expression. Mice on a high fat diet and obese db/db mice lacking the leptin receptor have higher levels of colonic RELMβ expression14. Figure 1A shows that C57/B6J mice fed a high fat diet for three months exhibited higher levels of RELMβ protein in the stool. The induction of RELMβ expression by a high fat diet is dependent upon commensal gut microbiota since treatment of mice with orally-delivered antibiotics reduces the mRNA expression of colonic RELMβ similarly in both mice fed a standard chow as well as a high fat diet (Figure 1B).
Figure 1.
Induction of RELMβ expression in the stool and colon is dependent upon gut bacteria. A) RELMβ immunoblot using proteins isolated from fecal pellets collected from wild-type mice fed a standard chow diet for 13 weeks and again after 21 weeks on a high fat diet; B) Quantitative RT-PCR of colonic mRNA for RELMβ in mice fed a standard chow diet and a high fat diet with and without the administration of oral antibiotics, Mean±SEM, N=5 mice per group.
RELMβ KO mice remain comparatively lean on a high fat diet
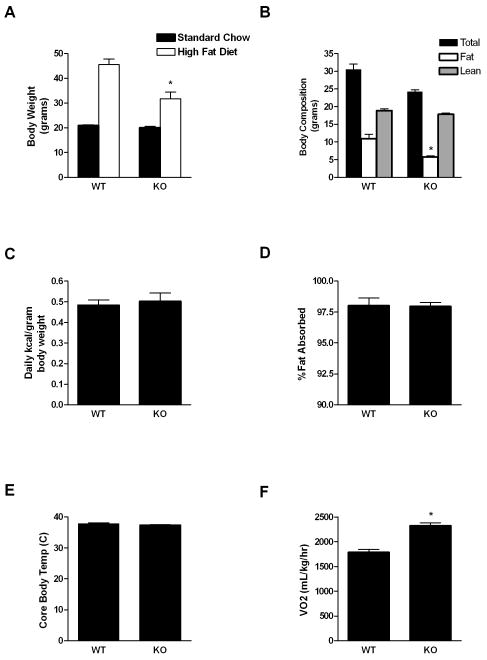
RELMβ KO mice do not exhibit an overt phenotype11 (data not shown), but our cohort showed altered weight gain on the high fat diet. Although RELMβ KO mice weighed the same as wild-type mice at 13 weeks of age on a standard chow diet, after 21 weeks on a high fat diet, RELMβ KO mice exhibit diminished weight gain (Figure 2A) due to decreased accumulation of fat mass relative to wild-type controls (Figure 2B). The reduction in diet-induced obesity in these RELMβ KO mice was not due to an alteration in food intake, fat absorption, or core body temperature (Figures 2C-E) but was rather caused by an increase in energy expenditure, as measured by an increase in oxygen consumption over a period of 4 hours (light cycle) via indirect calorimetry (Figure 2F). Importantly, RELMβ KO did not exhibit any difference in physical activity compared to wild-type controls during this period (data not shown).
Figure 2.
RELMβ KO mice remain comparatively lean on a high fat diet compared to wild-type littermate controls. A) Body weight of female RELMβ wild-type (WT) and Knockout (KO) mice at 13 weeks of age on a standard chow diet or after 21 weeks on a high fat diet, Mean±SEM, N=4-5 mice per group; *p<0.05; B) MRI body composition analysis after 8 weeks on the high fat diet, Mean±SEM, N=4-5 mice per genotype, *p=0.004; C) Daily food intake in RELMβ KO and wild-type mice fed a high fat diet for 4 weeks, Mean±SEM, N=4-5 mice per genotype; D) Percent dietary fat absorbed by RELMβ KO and wild-type mice fed a high fat diet for 5 weeks, Mean±SEM, N=4-5 mice per genotype; E) Rectal temperatures of RELMβ KO and wild-type mice fed a high fat diet for 8 weeks, Mean±SEM, N=4-5 mice per genotype; F) Oxygen consumption (VO2) measured via indirect calorimetry over 4 hours (light cycle) in RELMβ KO and wild-type mice after 21 weeks on a high fat diet, Mean±SEM, N=4 mice per group, *p<0.0001.
Transitions in gut bacterial populations associated with diet
To determine the effect of a high fat diet on the composition of the gut microbiome, spontaneously voided fecal pellets were collected from the five RELMβ KO mice and five wild-type controls at 13 weeks of age, while on a standard chow diet, and again after 21 weeks on a high fat diet. DNA was purified from pellets and samples were analyzed by 16S rDNA profiling and metagenomic analysis using 454/Roche pyrosequencing.
The 16S rDNA PCR primers were chosen based on the published reconstruction studies of Liu et al. to maximize the reliability of community analysis and phylogenetic assignments24. In order to analyze all the 16S rDNA sequences in parallel, samples were amplified using bar coded primers as previously described19, 25 and individual samples sorted after sequencing using the bar code information. A total of 25,790 sequence reads passed quality filters with an average read length of 262 nt. Sequence counts per sample ranged from 617 to 2448 per sample.
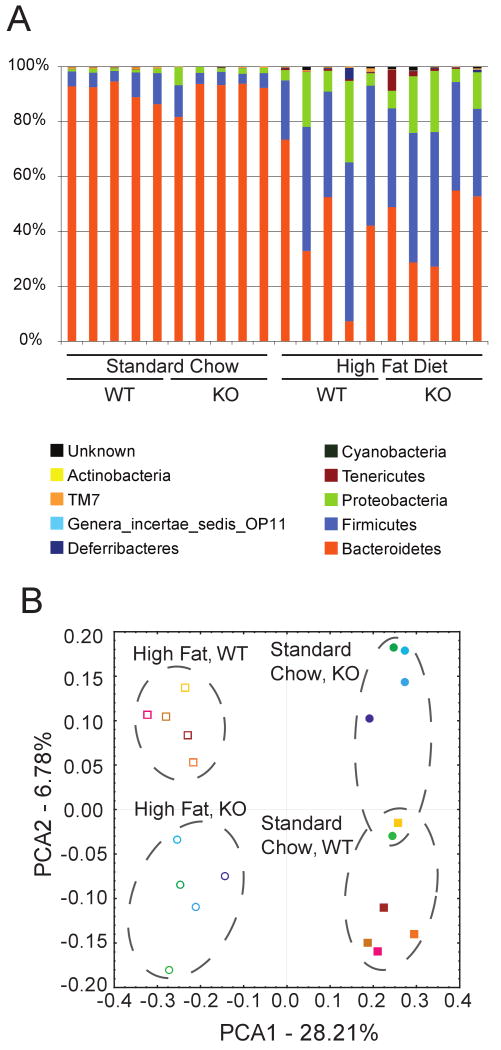
Sequence reads were aligned using NAST and the GreenGenes database and phylogenetic placements were determined using ARB's parsimony insertion tool and the Hugenholtz tree26, 27. Taxonomic assignments were then extracted from the phylogenetic tree. (Figure 3A). Communities from both wild-type and RELMβ KO mice on standard chow diet were relatively similar in composition among the ten samples. Each was dominated by gram-negative anaerobes of the Bacteriodetes phylum. The next most abundant group was Firmicutes, particularly of the anaerobic genus Clostridia. Less abundant but detectable were bacteria of the Proteobacteria, Tenericutes, and TM7 phyla (Figure 3A).
Figure 3.
Analysis of gut bacterial communities by 16S rDNA analysis from mice on the standard chow and high fat diets. A) The figure shows the percentages of each community contributed by the indicated Phyla. Diet and genotype are indicated below the figure; B) UniFrac analysis of the bacterial communities studied. Each point corresponds to a community from a single mouse. Samples from the Standard chow and High Fat Diet are from the same mouse, as indicated by the color code. Open symbols indicate high fat diet, closed symbols the standard chow. Circles indicate the knockouts, squares the wild-type controls. Colors indicated individual mice.
Samples from the wild-type mice after three months on the high fat diet resulted in a drastic change in the detectable 16S rDNA sequences (Figure 3A). In all ten samples the Firmicutes class Clostridiales was greatly expanded at the expense of the Bacteroidetes Class Bacteriodales. The Delta-Proteobacteria were also greatly expanded. Within the Bacteriodetes more than thirty different lineages were reduced in abundance. Orders affected included Bacteriodaceae, Prevotellaceae, and Rickenellaceae. The increase in Firmicutes was due to an increase in Clostridiaceae. The bloom of Proteobacteria was largely accounted for by the Desulfovibrionaceae. A previous study of high fat diet in mice reported a bloom of Mollicutes5, and we saw an increase in Mollicutes as well, but numerically these were comparatively modest in number in our study. To confirm the taxonomic placements of the main lineages detected, 184 near full-length 16S rDNA sequences were determined from these communities. Analysis indicated that these yielded phylogenetic placements consistent with the pyrosequence data (Supplementary Data Table 2).
The high fat diet, and not the obese state, accounts for the altered microbial communities
The changes seen between diets for the wild-type community could have been due either to the obese state resulting from the high fat diet, or direct effects of the diet on bacterial populations. The RELMβ KO mice remained comparatively lean on the high fat diet, allowing us to distinguish between these models. As can be seen in Figure 3A, the general changes in the composition of the gut microbiome were similar between wild-type and KO mice, indicating that effects of diet dominated.
Figure 3B presents a global analysis of the communities based on the pairwise phylogenetic distances calculated using UniFrac28, 29. Pairs of communities were marked on a common phylogenetic tree, and then the fraction of the branch length unique to each community determined. This provides a measure of the distance between communities in terms of their shared evolutionary history. Distances for all pairs of communities were than calculated and principal coordinate analysis used to generate the scatter plot in Figure 3B. The first principal coordinate (x-axis), which explained 28% of the variance, separated the communities by the type of diet. The second principal coordinate (y-axis), which accounted for 7% of the variance, separated the communities by genotype within each diet. The final position of the two genotypes differed within diets, indicating an interaction between diet and genotype. Thus the difference in diet had the predominant effect on community structure, but effects of REMLβ genotype were detectable.
Bacterial lineages affected by RELMβ
Analysis of lineages indicated that much of the difference between RELMβ KO and wild-type mice was due to changes in abundance of relatively low-level lineages. Fifteen Bacteriodetes lineages, and one lineage of Proteobacteria, changed in abundance between genotypes, whereas fifteen Firmicutes lineages changed in abundance. Though detectable, these changes were small compared to those associated with diet.
Metagenomic analysis of gut bacterial populations from mice on standard chow or high fat diets
To investigate the changes in bacterial gene content associated with diets in more detail, we carried out a metagenomic analysis of pellets from wild-type mice on the standard chow and high fat diets. Bacterial cells are known to exchange DNA sequences by a wealth of mechanisms30, so the types of bacteria present, as inferred from 16S rDNA sequence reads, may not be reflective of the gene content of the bacterial communities. The DNA samples from the gut communities were combined from mice on either standard chow or mice on the high fat diet, then samples were sheared, ligated to linkers, and subjected to 454 pyrosequencing. The average length sequence read returned was ∼260 nt. A total of 239,905 sequences were recovered for the standard chow pool and 297,699 for the high fat diet pool (Figure 4).
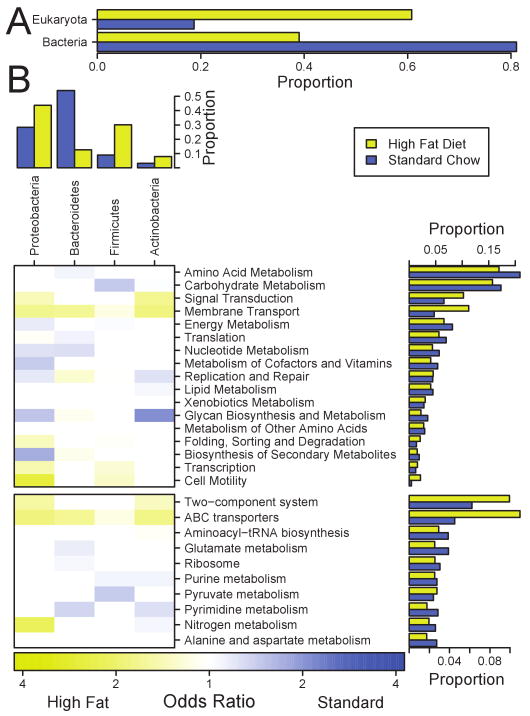
Figure 4.
Metagenomic analysis of community composition. Samples were pooled for wild-type mice from standard chow (blue) and high fat (yellow) diets; A) Comparison of contributions of bacterial and murine DNA for the two communities. Groups were assigned using MEGAN. The great majority of sequences from “Eukaryota” were murine; B) Metagenomic analysis of bacterial taxa and gene types. The bacterial phyla are indicated along the top of the figure for the most abundant four bacterial phyla (95% of total), the functional categories in the column to the right. The colored tiles in the body of the figure indicate the changes in gene content within each Phylum (that is, changes in gene types are compared between diets considering only sequences from each Phylum). Functional classes were assigned using KEGG and SEED annotation. The color scale (bottom) reports the odds ratio (set to the conservative edge of the confidence interval; if the interval included 1 the value was set to 1).
MEGAN was used to identify the taxonomic origin of the DNA sequences assignable with the BLAST cut-off used, revealing unexpected differences between the two diets (Figure 4A). Taxa were assigned for 41,980 of the standard chow reads and 79,257 reads from the high fat diet. On the standard chow diet, 81% of the sequence reads were annotated to within the Bacteria domain and 19% to the Eukaryote domain. Of the Eukaryotic reads, the vast majority were further classified as murine. In samples from the high fat diet, the murine assignments were the most abundant, accounting for 61% of reads, while bacterial sequences only accounted for 39%. This was further supported by using a quantitative PCR assay for bacterial 16S rDNA sequences, which showed the proportion of bacterial sequences was greater for the standard chow than the high fat diet samples (data not shown). One possible explanation is that the high fat diet led to greater shedding of intestinal epithelial cells into the lumen leading to the accumulation of mouse DNA in fecal pellets.
The bacterial phyla identified using MEGAN included Proteobacteria, Bacteriodetes, Firmicutes, and Actinobacteria, paralleling the 16S rDNA analysis. After the switch to the high fat diet, the proportions of Proteobacteria, Firmicutes and Actinobacteria increased and Bacteriodetes decreased, paralleling results with 16S rDNA sequencing. A few minor groups did differ between the metagenomic and 16S rDNA data. For example the Deferribacteres was not identified in the metagenomic data, probably because there are no full genome sequences for this group in the database to serve as a target for alignment.
Shotgun DNA sequences were aligned to the UniProt database using BLAST, and assigned to functional categories using SEED and KEGG annotation. KEGG pathways were assigned for 33,267 reads from the standard chow reads and 33,465 from the high fat diet.
The proportions of gene types were then compared between the two diets (Figure 4B, column to the right of the figure). Multiple categories showed changes in abundance, many of which are involved in metabolism. Genes for amino acid metabolism and carbohydrate metabolism decreased in abundance upon the switch to the high fat diet, while genes for signal transduction and membrane transport increased. Finer level analysis showed that most of the genes involved in signal transduction were two-component response-regulator systems, and the membrane transport molecules were mostly ABC transporters (bottom right, Figure 4B).
The changes in gene complement are color-coded as a heat map in the table in Figure 4B to show the changes in gene representation within each of the bacterial Phyla in the two diets. Thus, within the Proteobacteria, lineages with more genes for signal transduction, cell motility and membrane transport increased in abundance on the high fat diet, while lineages with more genes for energy metabolism and nucleotide metabolism decreased. For Bacteriodetes, lineages rich in genes for amino acid metabolism, translation, and nucleotide metabolism decreased, while those with more genes for membrane transport and replication and repair increased. For the Firmicutes, lineages with more genes for membrane transport, transcription and cell motility increased in abundance on the high fat diet, while those with more genes for carbohydrate metabolism and energy metabolism decreased. Collectively these data document a community-wide change in metabolism that was distinctive for each Phylum accompanying the switch to the high fat diet.
Under the high fat diet, a collection of genes for ABC transporters increased in abundance. Analysis indicated that these transported a variety of nutrients including lipids, sugars and peptides and metals. This may be a reflection of the different nutrients available in the high fat diet or reduced precursor synthesis, so that lineages with increased numbers of transporters were favored for growth. ABC transporters that act as efflux pumps for antibiotics were also increased in abundance in the high fat diet, suggesting the possibility that high fat diet may diminish the sensitivity of some bacterial groups to antibiotics31.
A collection of genes involved in import and assimilation of sugars were more abundant in samples from the high fat diet, paralleling data of Turnbaugh et al.5. Genes for utilization of fructose, raffinose, D-ribose, sucrose, and mannitol all increased in abundance. Genes for phosphorous metabolism were also increased on the high fat diet, notably phosphotransferase systems active during the uptake and assimilation of sugars.
Several classes of genes changed in abundance as expected from the reduction in Gram-negative Bacteriodetes on the high fat diet. Genes responsible for synthesis of the Gram-negative cell wall were decreased in abundance in the high fat diet and genes for Gram-positive cell wall synthesis increased. Sequences matching a Bacteriodetes conjugative transposon declined sharply in abundance. Genes for respiration increased in the high fat diet with the decline in anaerobic Bacteriodetes. A collection of genes involved in bacterial motility increased in abundance on the high fat diet, and these were associated with the expanded Proteobacteria and Firmicutes Phyla, indicating that the more motile members of these lineages expanded preferentially.
Our findings differed from those of Turnbaugh et al.5 regarding the relationship of bacterial motility in the two diets. We found a collection of gene groups linked to bacterial motility increased on the high fat diet, including “flagellar motility” and “motility and chemotaxis”. In contrast, Turnbaugh et al. found that categories related to flagellar assembly, motility, and chemotaxis decreased in abundance on a Western (high fat) diet.
In summary, the metagenomic analysis documented a community-wide transition in gene content that accompanied the switch to the high fat diet and specified gene classes affected within each lineage.
Discussion
We find, as have others4, 5, that distinctive community-wide changes in the gut microbiome accompany a switch from a standard chow to a high fat diet. This could be a result of either 1) the obese state altering the gut microbiome, or 2) the high fat diet causing changes in the microbiome directly. Here we distinguished between these two models by taking advantage of the RELMβ KO phenotype. The RELMβ KO mice remained comparatively lean when subjected to the high fat diet in our cohort, thereby allowing us to assay the effects of the high fat diet in the absence of obesity. We found that consistent and dramatic changes in microbial communities could be seen upon switching to the high fat diet for both wild-type and RELMβ KO mice, establishing that the high fat diet itself, and not the obese state, was responsible for the altered microbiota.
In the cohort studied here, we found that RELMβ KO reduces diet-induced obesity without altering food intake or fat absorption by increasing energy expenditure. This occurred only on a high fat diet. This finding is similar to other murine model systems where alterations in energy expenditure have been shown to alter the development of obesity only on a high fat diet32, 33. Thus our studies further confirm that relatively modest alterations in energy expenditure can lead to dramatic differences in the accumulation of fat mass over time.
We initially hypothesized that RELMβ might regulate the composition of the gut microbiome because 1) RELMβ is expressed at high levels in the stool of both mice and humans9 dependent upon the presence of the gut microbiome9, 23 and 2) other immune effector molecules secreted by the intestinal mucosa34 (and also intestinal inflammation35) lead to alterations in gut microbiome composition. We and others have previously shown that RELMβ can function as an activator of innate immune cell populations where it can regulate intestinal inflammation11-13. Here we report that RELMβ has a modest but significant effect on the composition of the gut microbiome, thereby specifying another host cell gene that regulates the microbiome.
Our results differ from those of Turnbaugh et al.5 in several ways. Rather than observing a bloom of Mollicutes on a high fat diet, we observed a bloom of Clostridia and Proteobacteria (Figure 3). The major group of Proteobacteria increased in abundance were the Phylum Delta-Proteobacteria, Order Desulfovibrio. This group contains sulfate-reducing bacteria, and genes for sulfate reduction were also detected as increased on high fat in the metagenomic analysis. However, even within Bacteriodetes, the lower level groups present differed considerably between the two diets, emphasizing the extent of the community changes between diets. The reason for the differences between our data and those of Turnbaugh et al. are unclear--the sample types analyzed differed, and it is also possible that the starting gut microbiota in mice from the two groups differed, so that different bacterial lineages bloomed upon switching to the high fat diet.
The metagenomic analysis showed, unexpectedly, that the high fat diet led to an increase in the proportion of murine DNA in pellets. It is estimated that, in humans, the self-renewal process of the intestinal epithelium results in the release of over 1010 cells into the gut lumen each day36. The proportional increase of murine DNA on a high fat diet may, therefore, be a result of the enhanced intestinal epithelial proliferative response37. We note that another possible explanation for the data is that the absolute amount of bacteria was diminished in the gut on the high fat diet--if the amount of murine DNA stayed the same it would have increased as a proportion. Going forward, it will be of great interest to investigate the generality of this observation in other models and in humans, to assess whether the level intestinal epithelial shedding is increased, and determine whether this can be linked to pathogenic effects of the high fat diet.
The reduction of carbohydrates in the high fat diet may have resulted in a state of nutrient stress on the gut microbiome. This notion is supported by the general decrease in a broad number of metabolic genes under the high fat condition, a response to nutrient deficiency that has been observed in vitro38. Notable exceptions to this trend are the high fat induced ABC transporters, genes of the two-component system, and genes associated with movement. Bacterial ABC transporters, which are ATP-dependent transmembrane proteins that play a role in the import of nutrient substrates31, may be induced to enhance nutrient uptake in an environment of limiting substrates. Genes for ABC transporters are enriched by a high fat diet amongst the most predominant bacterial taxa in the gut microbiome. Two-component systems are the principal signal transduction mechanisms in bacteria allowing prokaryotic organisms to adapt to new environmental conditions under selective pressure39. Therefore, bacteria that have an enriched representation of this system may be better suited for adaptation to gut environment on a high fat diet. Indeed, a two-component system that couples glycan sensing to carbohydrate metabolism has been shown to be an important adaptation of Bacteroides thetaiotaomicron, a major gut commensal40. Finally, a high fat diet enhances the representation of genes for bacterial chemotaxis and flagellar assembly. These genes are principally associated with the bloom of Proteobacteria suggesting that bacterial movement may provide bacteria within this phylum a growth advantage.
There are several limitations of our study. First, we did not examine a separate cohort of mice that would serve as controls to evaluate the effect of aging on the composition of the gut microbiome. The effect of aging on the composition of the adult gut microbiome has not been extensively characterized. However, in human adults, the gut microbiome can remain relatively stable over months42,43, though variations with changes in diet can be extreme. Together with the similarities of our results with those previously reported to be associated with diet-induced obesity5, we believe that the impact of aging on our results would be comparatively minor. Second, we only measured the effect of an altered diet, so the importance of the different components is unknown. The amount of fat in the high fat diet is increased by approximately 4-fold whereas the amount of carbohydrate is reduced by less than 2-fold compared to the normal chow diet. Protein composition in the two diets remained roughly the same. We suggest that it is most likely that the increase in fat is the primary determinant of the observed effects because work of the Gordon lab showed that a high fat and high carbohydrate diet leads to similar alterations in the composition of the gut microbiome5. Third, it is important note that metagenomic studies report only proportional differences gene in content, not absolute values. In addition, it is currently unknown to what degree the alterations bacterial gene representation, identified by metagenomics, correlate with either RNA abundance, protein expression, or biological function. Finally, additional studies with larger cohorts of mice will be required to fully characterize the effects of RELMβ on host metabolism.
Our results suggest that the substantial inter-subject variability in the composition of the human gut microbome18, 41 may be due, in part, to variation in diet. This notion is supported by the relatively modest amount of variability in the composition of the gut microbiome in our study of mice, which are fed a defined diet, compared to a large amount of microbiome variability reported in macaques, that consume a much more varied diet19. Unless human studies are performed on subjects consuming a defined diet or methods are developed to adjust for dietary variation, it may be challenging if not impossible, to make definitive associations between disease state in humans and alterations in the composition of the gut microbiome35, 41.
Supplementary Material
Composition of standard chow and high fat diets.
Full length 16S rDNA sequence reads.
Acknowledgments
This work was supported by NIH grants AI39368 (GDW), DK062348 (RSH), DK078669 (RK), and the Molecular Biology and Mouse Physiology Cores of NIH/NIDDK Center Grants DK50306 and DK019525, respectively. This work was also supported in part by the Penn Genome Frontiers Institute and a grant with the Pennsylvania Department of Health (FDB and GDW). The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions. We are grateful to the University of Pennsylvania Penn Genomic Frontiers Institute for support and NIH instrument grant S10RR024525 as well as to Elizabeth Costello and Catherine Lozupone for their helpful comments on this manuscript.
Footnotes
No conflicts of interest exist.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Study concept and design (RSA, FB, GDW); acquisition of data (MAH, CH, SAK, YYC); analysis and interpretation of data (CH, SAS, MH, RK, RSA, FB, GDW); drafting of the manuscript (All authors); critical revision of the manuscript for important intellectual content (All authors); statistical analysis (MH, RK, FB, GDW); obtained funding (GDW, RSA, FB, RK); study supervision (RSA, RB, GDW)
References
- 1.Saltiel AR. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell. 2001;104:517–29. doi: 10.1016/s0092-8674(01)00239-2. [DOI] [PubMed] [Google Scholar]
- 2.Spiegelman BM, Flier JS. Obesity and the regulation of energy balance. Cell. 2001;104:531–43. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Backhed F, Fulton L, et al. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–23. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci U S A. 1998;95:6578–83. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Backhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He W, Wang ML, Jiang HQ, et al. Bacterial colonization leads to the colonic secretion of RELMbeta/FIZZ2, a novel goblet cell-specific protein. Gastroenterology. 2003;125:1388–97. doi: 10.1016/j.gastro.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Artis D, Wang ML, Keilbaugh SA, et al. RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc Natl Acad Sci U S A. 2004;101:13596–600. doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McVay LD, Keilbaugh SA, Wong TM, et al. Absence of bacterially induced RELMbeta reduces injury in the dextran sodium sulfate model of colitis. J Clin Invest. 2006;116:2914–23. doi: 10.1172/JCI28121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogan SP, Seidu L, Blanchard C, et al. Resistin-like molecule beta regulates innate colonic function: barrier integrity and inflammation susceptibility. J Allergy Clin Immunol. 2006;118:257–68. doi: 10.1016/j.jaci.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nair MG, Guild KJ, Du Y, et al. Goblet cell-derived resistin-like molecule beta augments CD4+ T cell production of IFN-gamma and infection-induced intestinal inflammation. J Immunol. 2008;181:4709–15. doi: 10.4049/jimmunol.181.7.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shojima N, Ogihara T, Inukai K, et al. Serum concentrations of resistin-like molecules beta and gamma are elevated in high-fat-fed and obese db/db mice, with increased production in the intestinal tract and bone marrow. Diabetologia. 2005;48:984–92. doi: 10.1007/s00125-005-1735-1. [DOI] [PubMed] [Google Scholar]
- 15.Brockmann GA, Bevova MR. Using mouse models to dissect the genetics of obesity. Trends Genet. 2002;18:367–76. doi: 10.1016/s0168-9525(02)02703-8. [DOI] [PubMed] [Google Scholar]
- 16.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–41. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Jandacek RJ, Heubi JE, Tso P. A novel, noninvasive method for the measurement of intestinal fat absorption. Gastroenterology. 2004;127:139–44. doi: 10.1053/j.gastro.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna P, Hoffmann C, Minkah N, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huson DH, Auch AF, Qi J, et al. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–86. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–80. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeJongh M, Formsma K, Boillot P, et al. Toward the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinformatics. 2007;8:139. doi: 10.1186/1471-2105-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang ML, Shin ME, Knight PA, et al. Regulation of RELM/FIZZ Isoform Expression by Cdx2 in Response to Innate and Adaptive Immune Stimulation in the Intestine. Am J Physiol Gastrointest Liver Physiol. 2004 doi: 10.1152/ajpgi.00442.2004. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Lozupone C, Hamady M, et al. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamady M, Walker JJ, Harris JK, et al. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–7. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugenholtz P. Exploring prokaryotic diversity in the genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-2-reviews0003. REVIEWS0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeSantis TZ, Jr, Hugenholtz P, Keller K, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34:W394–9. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bushman FD. Lateral DNA Transfer: Mechanisms and Consequences. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 31.Davidson AL, Dassa E, Orelle C, et al. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72:317–64. doi: 10.1128/MMBR.00031-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachman ES, Dhillon H, Zhang CY, et al. betaAR signaling required for diet-induced thermogenesis and obesity resistance. Science. 2002;297:843–5. doi: 10.1126/science.1073160. [DOI] [PubMed] [Google Scholar]
- 33.Watanabe M, Houten SM, Mataki C, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–9. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 34.Peterson DA, McNulty NP, Guruge JL, et al. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potten CS, Booth C, Pritchard DM. The intestinal epithelial stem cell: the mucosal governor. Int J Exp Pathol. 1997;78:219–43. doi: 10.1046/j.1365-2613.1997.280362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bird RP, Stamp D. Effect of a high fat diet on the proliferative indices of murine colonic epithelium. Cancer Lett. 1986;31:61–7. doi: 10.1016/0304-3835(86)90167-9. [DOI] [PubMed] [Google Scholar]
- 38.Paustian ML, May BJ, Kapur V. Transcriptional response of Pasteurella multocida to nutrient limitation. J Bacteriol. 2002;184:3734–9. doi: 10.1128/JB.184.13.3734-3739.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alm E, Huang K, Arkin A. The evolution of two-component systems in bacteria reveals different strategies for niche adaptation. PLoS Comput Biol. 2006;2:e143. doi: 10.1371/journal.pcbi.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonnenburg ED, Sonnenburg JL, Manchester JK, et al. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Natl Acad Sci U S A. 2006;103:8834–9. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–9. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Composition of standard chow and high fat diets.
Full length 16S rDNA sequence reads.