Abstract
Locomotion relies on neural networks called central pattern generators (CPGs) that generate periodic motor commands for rhythmic movements1. We have identified a spinal input to the CPG that drives spontaneous locomotion using a combination of intersectional gene expression and optogenetics2 in zebrafish larvae. The photo-stimulation of one specific cell type was sufficient to induce a symmetrical tail beating sequence that mimics spontaneous slow forward swimming. This neuron is the Kolmer-Agduhr (KA) cell3, which extends cilia into the central cerebrospinal fluid containing canal of the spinal cord and has an ipsilateral ascending axon that terminates in a series of consecutive segments4. Genetically silencing KA cells reduced the frequency of spontaneous free swimming, indicating that KA cell activity provides necessary tone for spontaneous forward swimming. KA cells have been known for over 75 years, but their function has been mysterious. Our results reveal that during early development in low vertebrates these cells provide a positive drive to the spinal CPG for spontaneous locomotion.
In vertebrates, the excitatory synaptic drive for inducing the spinal CPG can originate from either supraspinal glutamatergic inputs or from within the spinal cord5,6. We searched for novel spinal neurons which trigger the CPG in the zebrafish larva by using “Intersectional Optogenetics”, a combination of trans-gene expression in specific cell types7 and genetic tools for manipulating neuronal activity with light2. The light-gated channel LiGluR8,9 was selectively expressed in distinct subsets of spinal cord neurons by crossing transgenic animals carrying UAS:LiGluR10 with a series of fish lines11 which express GAL4, the transcription factor that activates the UAS promoter, in distinct cellular patterns. We looked for common behavioral outcomes induced by light stimulation in lines with partially overlapping expression patterns that could be attributed to the activity of a common cell type (Suppl. Fig. 1).
Five day old zebrafish (5dpf) larvae exhibit spontaneous forward slow swims12. These occur in brief bursts, with each burst consisting of a series of symmetrical, dampening left-right oscillations (Fig 1a). We chose a number of Gal4 transgenic lines to drive expression of LiGluR in different subsets of spinal neurons, and asked whether optical activation of these neurons elicits a forward swim-like behavior. We first tested the Gal4s1020t line, which labels a heterogeneous population of ventral spinal neurons. When crossed to UAS:LiGluR, and labeled with the chemical photoswitch MAG18-10, 94% of the double-transgenic animals (n=37) exhibited robust tail oscillations upon stimulation of the caudal spinal cord with a short light pulse (see Methods) (Fig 1b, Suppl. Movie 2). The frequency and initial deflection angle of these oscillations closely resembled the spontaneous slow swim that we observed in free animals (Fig. 1c-f). The optical stimulation had no effect on non-transgenic larvae (n=12) or on LiGluR-expressing larvae not incubated with MAG1 (n=12).
Figure 1. Optical stimulation of specific spinal neurons leads to distinct locomotor behaviors.
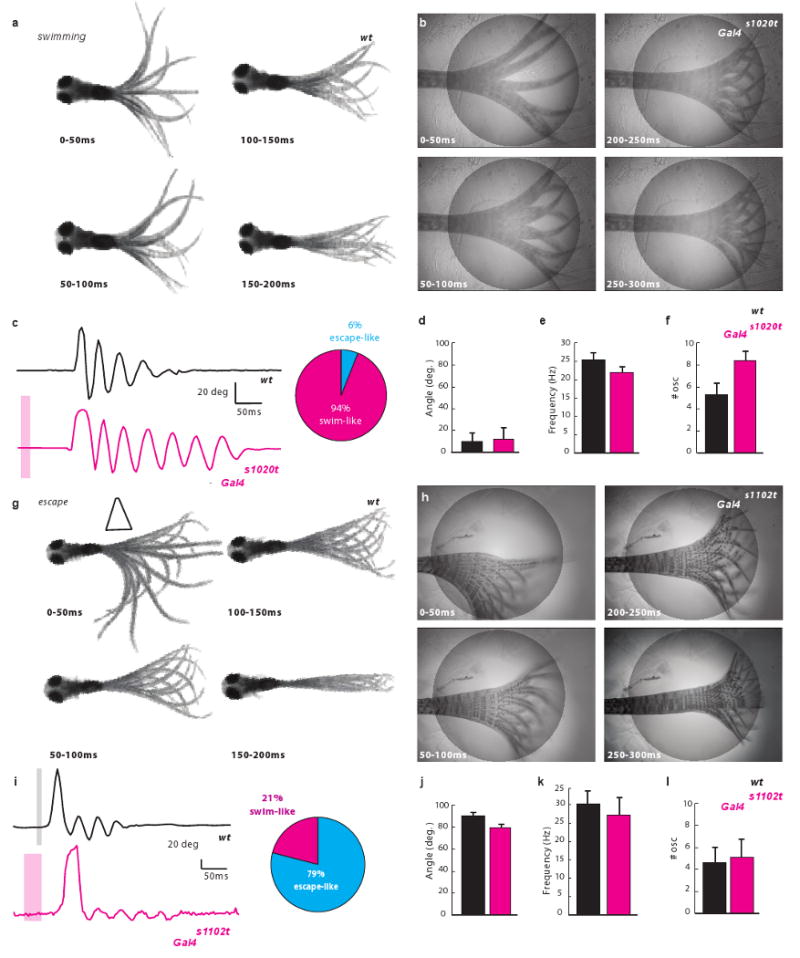
a, Spontaneous swim (superimposed frames). b, Optical stimulation (circle) of Gal4s1020t/UAS:LiGluR evokes a “spontaneous swim”-like behavior. c, Comparison of deflection angle traces corresponding to a) (top, black) and b) (bottom, magenta, bar for stimulation) (inset: 94% of responses were a swim (n=18)). No difference in angle (p > 0.51; n=9) (d), or frequency (f), but more oscillations (e), in light-induced swims. g, Escape elicited by a water jet (trapeze) consists of sharp C-turn away from stimulus followed by a forward swim. h, Light-induced escape induced by stimulation of RB cells in Gal4s1102t/UAS:LiGluR larvae. i, Tail deflection traces corresponding to g) (top) and h) (bottom) (inset: 79% of responses were an escape (n= 11)). j-l, No difference in j) deflection angle (p > 0.13; n=7); k, frequency (p > 0.42; n=7); l, number of oscillations ((p > 0.4101;n=7).
The swim-like response induced by light in Gal4s1020t/UAS:LiGluR larvae differed from the well described touch-escape response13, in which larvae respond to touch on one side of the tail by an initial sharp bend of the tail (”C-bend”) to the opposite side that propels the fish away from the touch (Fig. 1g, Suppl. Movie 3). A C-bend to either the left or right side was elicited by bilateral illumination of the tail in UAS:LiGluR/Gal4s1102t line10 larvae expressing the LiGluR in Rohon-Beard (RB) touch sensing neurons of the tail. A C-bend was evoked in 79% of trials (7 larvae tested 5 times each) (Fig. 1h-l; Suppl. Movie 4), resembling the natural escape of free-swimming fish (see initial one-sided tail bend, the frequency and the number of the ensuing tail beats in Fig. 1c-f). The left/right symmetry of the beating oscillations and small deflection angle seen in the Gal4s1020t line distinguish it from this RB cell-induced asymmetric escape response of the Gal4s1102t line (compare Fig. 1c, e to Fig. 1i, k).
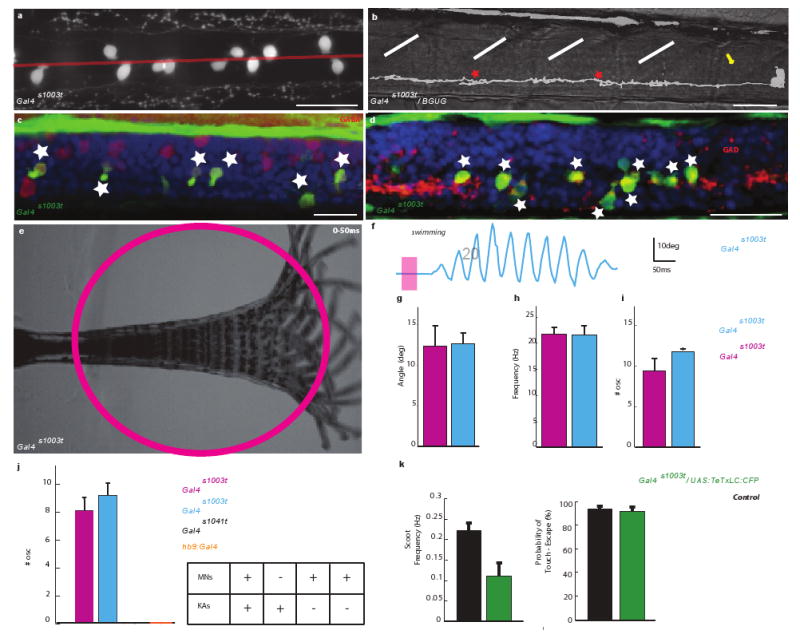
Gal4s1020t drives expression in several cell types in the ventral spinal cord (Fig. 2a). Inverse PCR cloning indicates that the transposon is integrated near the Olig2 gene11. Indeed, the expression pattern of Gal4s1020t is indistinguishable from that seen in an Olig2:GFP transgenic line14 (Suppl. Fig. 2). Using the BGUG expression system to determine which cell types express GAL4 in the Gal4s1020t line (Suppl. Methods), we found that 79% of the 250 cells imaged in 73 fish were motoneurons (26.4% primary motoneurons: Fig. 2b, top panel; 52.4% secondary motoneurons: Fig. 2b, bottom panel). The remaining cells (20.4%) were neurons with a central axon and lacking dendrites (Fig. 2c, d, f). A small number of cells (2 out of 250 cells, i.e. 0.8%) resembling oligodendrocytes were also GFP-positive (not shown). The neurons with a central axon appeared to represent a single cell type. They are located near the central canal and have an ascending axon that projects ipsilaterally, making terminals in a series of 2-6 consecutive segments (Fig. 2c, d, f). Instead of dendrites these cells have a brush of cilia emanating from the somata, which appear to contact the cerebrospinal fluid (CSF), as shown by the alignment of the cilia with the central canal (Fig. 2e). Antibody staining showed that these neurons are GABA and GAD65/67-positive (Fig. 2f, h; Suppl. Fig. 3) as well as somatostatin-positive (Suppl. Fig. 4). Combined, these features are consistent with these neurons being Kolmer-Agduhr (KA) cells 3,15.
Figure 2. The Gal4s1020t line drives expression in motoneurons and KA neurons.

a, Expression in ventral cells including motoneurons projecting out of cord (arrows) (lateral view). b-e, Random labeling in Gal4s1020t/BGUG identifies solely two cell types: b, primary (top) and secondary (bottom) motoneurons. Dorsal (c) and lateral (d-f) views of neuron with a central ipsilateral ascending axon. Note contact feet (red stars in c-d) and a “toothbrush” morphology (e) (cilia, yellow arrows) characteristic of KAs. In c), larva was slightly tilted to show enlarged contacts on axon (midline, red line and segment, white lines). e, Dense BGUG pattern with multiple KAs shows the alignment of the brush of cilia (arrows) with central canal. f, The ascending axon runs near the ventral edge of the spinal cord before aiming dorsally. g,h, KAs at 5dpf in Gal4s1020t/BGUG (green) are GABAergic neurons (anti-GAD (g) and anti-GABA (h) immunostaining in red). Scale bars = 25μm.
To find out whether the KA neurons are responsible for triggering the swim-like behavior in the Gal4s1020t/UAS:LiGluR fish, we screened more Gal4 lines and found one line, Gal4s1003t, in which expression in the spinal cord is restricted to KAs. These cells shared morphology, cell body position, and marker expression with the sensory neuron labeled in Gal4s1020t (Fig. 3a-c; Suppl. Fig. 5). As in Gal4s1020t/UAS:LiGluR, the light-induced response in Gal4s1003t/UAS:LiGluR consisted of an alternating symmetrical tail beat at the slow swim frequency (Fig. 3e-i; Suppl. Movie 5), confirming that KAs are indeed able to trigger the CPG. The properties of the light-induced swim Gal4s1003t/UAS:LiGluR were indistinguishable from those of Gal4s1020t/UAS:LiGluR (see Fig. 3), suggesting that at the intensities applied, motoneurons are not activated in Gal4s1020t/UAS:LiGluR. Indeed, calcium imaging in tetrodotoxin (to block action potentials and confine activity to the optically stimulated cells) revealed that the light pulses used in the behavioral experiments were strong enough to activate the narrow region surrounding the central canal where KA cells are located, but not elsewhere in the ventral spinal cord where the majority of motorneurons are situated (Suppl. Fig. 6).
Figure 3. Optical stimulation of KAs of Gal4s1003t line induces a forward swim.
a, Expression in spinal cord is confined to cells close to the midline (dorsal view). b, Lateral view with BGUG shows ventral neurons with a central axon forming contact feet (red stars), characteristic “toothbrush” morphology (arrow) of KAs. c, d, These cells (green) are GABAergic (anti-GAD and anti-GABA staining in red). e-f, Optical stimulation induces a swim-like response. e, Superimposed images. f, Deflection angle trace. g-i, No difference in deflection angle (g), frequency (h) and number of oscillations (i) between Gal4s1003t (blue) to Gal4s1020t (magenta) (respectively p>0.85; p>0.98, p>0.36, n=9). j, Side-to-side comparison of number of oscillations evoked by a 100ms pulse of light shows that only lines expressing in KAs show a swim-like response while line with motoneurons (MNs) and no KAs (Gal4s1041t, black; Hb9:Gal4, orange), do not. k, Reduction of the spontaneous swimming frequency in Gal4s1003t/UAS:TeTxLC-CFP (p> 0.0075; n=10) but no change in the probability of touch-response (p> 0.45; n=12).
To rule out the possibility that motoneuron activation in Gal4s1020t/UAS:LiGluR fish elicits swimming movements, we tested two additional Gal4 lines with motoneuron expression but no expression in KA cells: Gal4s1041t and Hb9:Gal416 (Suppl. Fig. 7). Light pulses that reliably triggered swim-like behavior in animals expressing LiGluR in the Gal4s1020t and Gal4s1003t lines produced no effect in either of these motoneuron lines (6 LiGluR larvae tested for each line; Fig. 3j). However, increasing the intensity or duration of illumination by ≥10-fold evoked contraction on the illuminated side of the tail, which were distinct from the forward swim-like behavior (Suppl. Fig. 8). The requirement for stronger illumination to evoke the contraction is consistent with the larger size and lower input resistance of motorneurons17. Altogether, these observations show that the forward swim can be attributed specifically to the activation of the KA cells.
KA cells are GABAergic. To test the role of GABAergic transmission in the light-induced response of Gal4s1020t/UAS:LiGluR we injected the GABA-A antagonist bicuculline into the spinal cord. This treatment greatly reduced the number of oscillations evoked by light in Gal4s1020t/UAS:LiGluR fish (p < 10-6, t(7)= 11.2950; Suppl. Fig. 9), abolishing the light-induced response entirely in 4 out of 8 larvae. These experiments indicate that the optical stimulation of KAs is sufficient to initiate a swim-like behavior by a GABA dependent process.
Having shown that activation of KAs is sufficient for inducing a swim-like behavior, we next asked whether they are also necessary for spontaneous swimming by blocking synaptic transmission from KA cells via a targeted expression of the tetanus toxin light chain (TeTxLC) fused to cyan fluorescent protein (CFP) (UAS:TeTxLC-CFP18 crossed with Gal4s1020t and Gal4s1003t). Three to five day old larvae expressing TeTxLC-CFP were easily identified by their CFP fluorescence (Methods). We compared the swimming behavior of CFP-positive larvae to that of siblings that did not have CFP fluorescence (i.e. did not express TeTxLC). Gal4s1020t/UAS:TeTxLC-CFP larvae expressing the TeTxLC were paralyzed at five days, as expected for expression of GAL4 in motoneurons. On the other hand, Gal4s1003t/UAS:TeTxLC-CFP larvae, which lack motoneuron expression, were not paralyzed, enabling behavioral assays. Gal4s1003t/UAS:TeTxLC-CFP exhibited spontaneous burst-swimming, but the frequency of the swims was greatly reduced (p< 0.0075; t(9)= - 3.4278) (Fig. 3k). These results indicate that KAs provide a positive drive to spontaneous swimming. It should be noted that only half of the KAs express the UAS transgene in the Gal4s1003t line (Suppl. Fig. 3), suggesting that block of synaptic transmission in all of the KAs could have an even more profound effect on spontaneous swimming. Strikingly, we found that Gal4s1003t/UAS:TeTxLC-CFP still respond to touch by a touch-escape (p<0.45; t(11)= 0.7863), indicating that KAs do not play a significant role in initiating touch-escape.
We further examined the KA induced-swim and the RB-induced escape behaviors by performing local photo-activation. Since mechanical activation on one side of the larva elicits, as part of the escape response, a C-turn on the opposite side (Fig. 1g), we predicted that one-sided optical activation of RBs would have the same effect. We tested this by confining the illumination to a small portion of the spinal cord with a Digital Light Processing (DLP) array (Fig. 4a). One-sided optical stimulation of RB cells in the Gal4s1102t line triggered a reliable large-angle contralateral bend (n=9 out of 9; Fig. 4c), resembling the C-bend induced by one-sided mechanical stimulation (Fig. 1g). In contrast, one-sided optical stimulation of Gal4s1020t elicited a symmetrical forward swim-like behavior (Fig. 4b), closely resembling the response to bilateral optical stimulation (Fig. 1c).
Figure 4. Dissection of the light-evoked responses in Gal4s1020t and Gal4s1102t by unilateral stimulation and lesion studies.

a-c, Patterned illumination for stimulation. a, semi-restrained Gal4s1020t larva aimed bilaterally (cartoon and fluorescence image of Kaede expression in three segments, top) and on left (L) or right (R) side (bottom panels, Scale bar = 25μm). b-c, Deflection angle traces and mean values induced by L (green) and R (red) stimulation; b) L and R activations induce similar symmetric oscillations of tail in Gal4s1020t line (n=5). c, L and R activations induce large and opposite directed C-bends in Gal4s1102t (n=9). d-f, Effect induced by isolation of the spine. d, Pattern in Gal4s1020t pre (top) and post (below) lesion. e-f, No reduction of the light-induced swim behavior in Gal4s1020t ((e), n=7) but abolition of the light-induced escape behavior in Gal4s1102t ((f), n=4) (pre and post-lesion, top and bottom).
To test the involvement of supraspinal inputs in the KA-elicited swim-like behavior, we performed hindbrain lesions that ablated the connections between the brain and the spinal cord (Fig. 4d). Tactile stimuli sensed by RB cells are known to be transmitted to the hindbrain13 where the command for escape is relayed back to the tail, and ablation of the hindbrain was shown earlier to suppress the fast contralateral C-bend that begins the escape13. Consistent with this, the C-bend component of the response to optical activation of RB cells in the Gal4s1102t/UAS:LiGluR was abolished by the hindbrain lesion (n = 4 out of 4; Fig. 4d, f). In contrast, the light-evoked swim-like behavior in Gal4s1020t remained intact after the lesion (n=7; Fig. 4e), demonstrating that intra-spinal activation of Gal4s1020t-positive neurons is sufficient to drive the swim-like behavior.
Prior work in vertebrates has implicated specific classes of spinal interneurons in regulating locomotion speed19,20 or movement strength and their activity was associated with specific states of the spinal networks recorded during fictive locomotion16,21. These studies were based on either loss of function19 or on correlation of the activity of neurons with specific phases of ventral root activity during fictive locomotion 16,20. We show that it is possible to combine genetic targeting of light-gated channels with a simple behavioral assay as an alternative way for identifying neurons that are necessary for a behavior and that, at the same time, can establish sufficiency (Suppl. Fig1).
Previous studies in the lamprey showed that the GABAergic system is a strong modulator of fictive swimming22, but there are many types of spinal GABAergic neurons and the neuronal basis for the observed modulation was not known. We demonstrate here that a single GABAergic cell, the KA neuron, is a major modulator of locomotion in the awake behaving animal. Although KA neurons were first described decades ago3, their role in spinal circuits remained enigmatic. We show that KAs are necessary for the normal frequency of spontaneous swimming and sufficient to drive the CPG in early development, when GABAergic transmission is excitatory23.
The KA neurons of zebrafish resemble the CSF contacting cells of lamprey and other vertebrates, including mammals24,25, in that they express GABA and the transcription factor olig226 (Suppl. Fig. 2), are located next to the spinal canal and project a brush of stereocilia into the CSF and have axons that run longitudinally25. It remains to be determined whether the KA-like cells of mammals affect the spinal locomotory CPG.
While we have determined that KAs provide a positive drive to the CPG in larval fish, in adult fish, and in postnatal mammals, GABAergic transmission is inhibitory and blocking GABA receptor in the adult lamprey enhances swim frequency22. This suggests that KA activity may suppress swimming in adult zebrafish. The “liquor-contacting” cilia of the KA neurons in the spinal canal3,15,22 could permit them to sense mechanical deformation of the spine or chemical signals in the central canal, such as low pH, as proposed recently for mammalian CSF contacting neurons27. The natural drive to KA cells and their function later in life remain to be defined.
Methods Summary
To determine which cell types express in a GAL4 line and to quantify their abundance, we used the BGUG transgene (short for Brn3c:Gal4; UAS: mGFP) which labels a small random subset of the GAL4-expressing cells due to variegated expression of the gene encoding a membrane-targeted GFP11,28. The transgenic line UAS:LiGluR, as well as all Gal4 lines from the enhancer trap screen were published previously (see10,11). To make the Hb9:Gal4 transgenic construct, 3Kb of genomic sequence upstream the hb9 coding sequence 18,29 was amplified by PCR and inserted upstream of the GAL4 coding sequence30. Synthesis of MAG-1 was carried out as described8-10. Five days old larvae were bathed in 200 μM MAG-1, 4% DMSO for 45 min at 28.5°C in the dark. The fast photoswitching light source was coupled to an upright Zeiss epifluorescence microscope. Patterned illumination was accomplished using a Digital Mirror Devices. Motion of the tail was monitored at 250fps using a high speed camera. Lesions were performed on anesthetized larvae bathed in Evans solution using a fine tungsten needle. Embryos at appropriate stages were fixed in 4% PFA in PBS and processed for immunohystochemistry according to published protocols28. Injection of the calcium dye was performed 30-60 minutes after MAG-1 labeling. Fluorometric Ca++ measurements were performed using a confocal Olympus laser scanning microscope equipped with a UV SIM scanner. Tracking of the tail position as well as calcium imaging analysis were performed using a custom made script written in Matlab 2007 (Mathworks, MA, USA).
Supplementary Material
Acknowledgments
We thank M. Volgraf for MAG-1 synthesis, K. Kawakami for the UAS:TeTxLC-CFP line, B. Appel for the Olig2-DsRed line, W. Staub for animal care, D. Li for help with screening of BGUG larvae, B. Vigh, C. Girit, E. Brustein, P. Drapeau and S. Hugel for helpful discussion; P.G. de Gennes and Noam Sobel for moral support, and O. Wyart for aesthetic input. We are grateful to K. Best and P. Tavormina (Molecular Devices), H. Aaron (Molecular Imaging Center, UC Berkeley) and R. Ayer (Sutter Instruments, CA), B. Nowak (Photonic Instruments) and M. Ulbrich for helpful advice on the design of the photostimulation setup. Support for the work was from the Marie Curie Outgoing International Fellowship (with the CNRS - UMR5020 “Neurosciences Sensorielles, Comportement Cognition” laboratory, Lyon, France) (C.W.), the Human Frontier Science Program Long-term Postdoctoral Fellowship (F.D.B), the National Institutes of Health Nanomedicine Development Center for the Optical Control of Biological Function (5PN2EY018241) (E.Y.I., D.T. and H.B.), the Human Frontiers Science Program (RGP23-2005) (E.Y.I. and D.T.), the Lawrence Berkeley National Laboratory LDRD (E.Y.I. and D.T.), R01 NS053358 (H.B.), and a Sandler Opportunity Award (H.B.).
Footnotes
Author Contributions: C.W., F.D.B, H.B and E.Y.I. made critical primary contributions to this study. C.W. built the photostimulation setup, performed behavioral experiments, lesions, pharmacology, calcium imaging, imaging of the immunolabeled larvae, anatomical analysis based on BGuG imaging and wrote the matlab scripts for analyzing both behavior and imaging. F.D.B. generated the transgenic lines UAS:LiGluR10 and Hb9:Gal4, as well as performed the immunochemistry experiments. E.W. participated in the anatomical analysis of BGuG. E.K.S. and H.B. generated the enhancer trap gal4 screen which made the “intersectional optogenetic” approach possible11. E.Y.I. and D.T. developed chemical optogenetics with LiGluR8. C.W. and E.Y.I. wrote the manuscript with helpful feedback from H.B. and F.D.B. H.B. and E.Y.I. supervised C.W. and F.D.B. and contributed to the planning of all aspects of this project.
References
- 1.Grillner Sten. Neuron. 2006;52(5):751. doi: 10.1016/j.neuron.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Luo L, Callaway EM, Svoboda K. Neuron. 2008;57(5):634. doi: 10.1016/j.neuron.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agduhr E. In: Cytology and cellular pathology of the nervous system. Penfield W, editor. Vol. 2. Hoeber; New York: 1932. p. 536. [Google Scholar]
- 4.Higashijima S, Mandel G, Fetcho JR. Journal of Comparative Neurology. 2004;480(1):1. doi: 10.1002/cne.20278. [DOI] [PubMed] [Google Scholar]
- 5.Fenaux F, Corio M, Palisses R, et al. Experimental Brain Research. 1991;86(2):393. doi: 10.1007/BF00228963. [DOI] [PubMed] [Google Scholar]
- 6.Kiehn O, Quinlan KA, Restrepo CE, et al. Brain Research Reviews. 2008;57(1):56. doi: 10.1016/j.brainresrev.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Douglas JR, Noga BR, Dai X, et al. Journal of Neuroscience. 1993;13(3):990. doi: 10.1523/JNEUROSCI.13-03-00990.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volgraf Matthew, Gorostiza Pau, Numano Rika, et al. Nature Chemical Biology. 2006;2(1):47. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorostiza Pau, Volgraf Matthew, Numano Rika, et al. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(26):10865. doi: 10.1073/pnas.0701274104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szobota S, Gorostiza P, Del Bene F, et al. Neuron. 2007;54(4):535. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Scott EK, Mason L, Arrenberg AB, et al. Nature Methods. 2007;4(4):323. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- 12.Budick SA, O'Malley DM. Journal of Experimental Biology. 2000;203(17):2565. doi: 10.1242/jeb.203.17.2565. [DOI] [PubMed] [Google Scholar]
- 13.Liu KS, Fetcho JR. Neuron. 1999;23(2):325. doi: 10.1016/s0896-6273(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 14.Shin Jimann, Park Hae-Chul, Topczewska Jolanta M, et al. Methods in Cell Science: An Official Journal of the Society for In Vitro Biology. 2003;25(1-2):7. doi: 10.1023/B:MICS.0000006847.09037.3a. [DOI] [PubMed] [Google Scholar]
- 15.Dale N, Roberts A, Ottersen OP, et al. Proceedings of the Royal Society of London Series B-Biological Sciences. 1987;232(1267):193. doi: 10.1098/rspb.1987.0068. [DOI] [PubMed] [Google Scholar]
- 16.Liao JC, Fetcho JR. Journal of Neuroscience. 2008;28(48):12982. doi: 10.1523/JNEUROSCI.3330-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drapeau P, Ali DW, Buss RR, et al. Journal of Neuroscience Methods. 1999;88(1):1. doi: 10.1016/s0165-0270(99)00008-4. [DOI] [PubMed] [Google Scholar]
- 18.Asakawa K, Suster ML, Mizusawa K, et al. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(4):1255. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosgnach S, Lanuza GM, Butt SJB, et al. Nature. 2006;440(7081):215. doi: 10.1038/nature04545. [DOI] [PubMed] [Google Scholar]
- 20.McLean DL, Masino MA, Koh IYY, et al. Nature Neuroscience. 2008;11(12):1419. doi: 10.1038/nn.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritter DA, Bhatt DH, Fetcho JR. Journal of Neuroscience. 2001;21(22):8956. doi: 10.1523/JNEUROSCI.21-22-08956.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alford S, Sigvardt KA, Williams TL. Brain Research. 1990;506(2):303. doi: 10.1016/0006-8993(90)91267-k. [DOI] [PubMed] [Google Scholar]
- 23.Brustein E, Drapeau P. Journal of Neuroscience. 2005;25(46):10607. doi: 10.1523/JNEUROSCI.2017-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vigh B, Vigh-Teichmann I. Microscopy Research and Technique. 1998;41(1):57. doi: 10.1002/(SICI)1097-0029(19980401)41:1<57::AID-JEMT6>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 25.Stoeckel ME, Uhl-Bronner S, Hugel S, et al. Journal of Comparative Neurology. 2003;457(2):159. doi: 10.1002/cne.10565. [DOI] [PubMed] [Google Scholar]
- 26.Masahira N, Takebayashi H, Ono K, et al. Developmental Biology. 2006;293(2):358. doi: 10.1016/j.ydbio.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 27.Huang AL, Chen XK, Hoon MA, et al. Nature. 2006;442(7105):934. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao T, Baier H. Nature Neuroscience. 2007;10(12):1529. doi: 10.1038/nn2002. [DOI] [PubMed] [Google Scholar]
- 29.Flanagan-Steet H, Fox MA, Meyer D, et al. Development. 2005;132(20):4471. doi: 10.1242/dev.02044. [DOI] [PubMed] [Google Scholar]
- 30.Koster RW, Fraser SE. Developmental Biology. 2001;233(2):329. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


