Abstract
Purpose
Hypoxia inducible factor-1 (HIF-1) is the central mediator of the cellular response to low oxygen and functions as a transcription factor for a broad range of genes that provide adaptive responses to oxygen deprivation. HIF-1 is over-expressed in cancer and has become an important therapeutic target in solid tumors. In this study, a novel HIF-1α inhibitor was identified and its molecular mechanism was investigated.
Experimental Design
Using a HIF-responsive reporter cell-based assay, a 10,000-membered natural product-like chemical compound library was screened to identify novel HIF-1 inhibitors. This led us to discover KC7F2, a lead compound with a central structure of cystamine. The effects of KC7F2 on HIF-1 transcription, translation and protein degradation processes were analyzed.
Results
KC7F2 markedly inhibited HIF-mediated transcription in cells derived from different tumor types, including glioma, breast and prostate cancers and exhibited enhanced cytotoxicity under hypoxia. KC7F2 prevented the activation of HIF-target genes such as Carbonic Anhydrase IX, Matrix Metalloproteinase 2 (MMP2), Endothelin 1 and Enolase 1. Investigation of the mechanism of action of KC7F2 showed that it worked through the down-regulation of HIF-1α protein synthesis, an effect accompanied by the suppression of the phosphorylation of eukaryotic translation initiation factor 4E binding protein 1 (4EBP1) and p70 S6 kinase (S6K), key regulators of HIF-1α protein synthesis.
Conclusion
These results show that KC7F2 is a potent HIF-1 pathway inhibitor and that its potential as a cancer therapy agent warrants further study.
Introduction
Hypoxia, a reduction in partial oxygen pressure, is a major hindrance to effective solid tumor therapy. The microenvironment of rapidly growing solid tumors shows increased energy demand and diminished vascular supply, resulting in focal areas of prominent hypoxia (1). The hypoxic fraction of tumors is resistant to traditional therapies. Radiotherapy is compromised because of the reduced reaction of oxygen with radiation-induced DNA free radicals (2). Chemotherapy is hampered by the diffusion-limited drug delivery to hypoxic regions from distant vasculature. Also, many anticancer drugs are most effective against rapidly proliferating cells and hypoxia (and deficiencies in other nutrients such as glucose) can cause a reduction in cell proliferation rate (3). This is compounded by the induction of the multi-drug resistance (MDR1) gene product P-glycoprotein in hypoxic tissue (4), further reducing drug efficacy. Hypoxic tumor regions also impede immune responses, and may promote the growth of cancer stem cells (5, 6).
Hypoxia drives malignant tumor progression. Tumor hypoxia increases malignant progression and metastasis by promoting angiogenesis through induction of both pro-angiogenic proteins such as VEGF and metabolic adaptation through elevation of glycolytic enzymes (7, 8). Hypoxia also generates selective pressure for cells to acquire genetic alterations (e.g.TP53, K-ras ) that will circumvent hypoxia-induced apoptosis (9, 10). For all these reasons, it is rational to design novel therapies targeted at the hypoxic fraction in tumors (11).
In response to hypoxia, HIF-1, a heterodimeric transcription factor consisting of α and β subunits, is activated. The HIF-1α subunit is highly regulated by oxygen concentration and serves as a marker of hypoxia. Under normoxic conditions, the HIF-1α subunit is constitutively transcribed and translated, but rapidly degraded, while it is stabilized under hypoxia. In contrast, HIF-1β is constitutively expressed and not affected by O2 levels. The binding of HIF-1α to HIF-1β and co-activators p300/CBP forms an active transcription factor that transactivates a variety of target genes by binding to hypoxia responsive elements (HRE) in their promoters (12). HIF-target genes encode critical factors for the adaptation to low oxygen, including pro-angiogenic factors (VEGF), vasoconstrictors (Endothelin 1), enzymes mediating metabolic adaptation to anaerobic respiration (Enolase 1), glucose transporters (Glut-1), matrix remodeling enzymes (MMP2), regulators of pH (Carbonic Anhydrase IX) and pro-motility factors (7, 13–18).
In addition to its post-translational regulation by cellular oxygen content, HIF-1α is also controlled at the protein synthesis level, through oncogenic signal transduction activated by growth factors. For example, growth factors, such as insulin, insulin-like growth factor 1 and 2, angiotensin II, thrombin and platelet-derived growth factor, promote the accumulation of HIF-1α protein in a cell-type-specific manner even under normoxia (19–24). There are two major signaling pathways involved in the regulation of HIF-1α protein levels and function, the phosphatidylinositol 3-kinase (PI3K)-Akt and the mitogen-activated protein kinase (MAPK) pathways (22, 25, 26). Signaling from the PI3K pathway increases HIF-1α stabilization through the mammalian target of rapamycin protein complex (mTOR), likely by enhancing HIF-1α translation. The MAPK pathway may stimulate the transactivation function of HIF-1α through direct phosphorylation of HIF-1α (27) or by up-regulating its cofactor p300 (28).
In many human cancers, deregulation of the HIF-1 pathway results in its over-expression, and associates with poor patient prognosis (29, 30). Therefore, various small molecules targeting the HIF pathway have been developed (11, 31–37). Many of these molecules have an indirect mechanism of action, display pleiotropic effects suggesting a multiplicity of targets, show unwanted side effects, or have poor pharmacological properties necessitating the development of further HIF inhibitors (38, 39). Recent studies show that up-regulation of mTOR signaling increases HIF-1α protein expression through translation initiation factors such as 4E-BP1 and p70 S6 kinase (S6K) (40–42), which are particularly interesting targets for HIF-1α inhibitor development.
We previously screened a 10,000-membered natural product-like chemical compound library using a cell line stably expressing an HRE-dependent reporter and identified several “hit” structures, including 103D5R, which inhibits HIF-1α protein accumulation (31). In this study, we report a new chemical compound derived from this screen, KC7F2, as a notable inhibitor of HIF-1α protein synthesis and investigated its mechanism of action. The identification and development of novel HIF-1 pathway inhibitors may lead to the development of a new type of treatment for cancer, potentially applicable to many solid malignancies.
Materials and Methods
Cell lines and culture conditions
Human cancer cell lines LN229, LNZ308, U251MG, MCF7, PC3, D54MG, U87MGD and the human fibroblast cell line HFF-1 were maintained in DMEM media (Mediatech, Herndon, VA) supplemented with 10% FBS, sodium pyruvate (1 mM), penicillin (100 IU/ml) and streptomycin (100 μg/ml). A549 cells were kept in RPMI 1640 medium (Mediatech, Herndon, VA) with the same supplements. Primary cultures of human dermal microvascular endothelial cells (HDMVEC) were obtained from the Dermatology Department Core Facility at Emory University and maintained in Medium 131 with attachment factor (Invitrogen, CA). Primary cultures of mouse neurons were maintained in Neurobasal medium (Invitrogen, CA) with 5% FBS, 2% B-27 (Invitrogen), 0.6% dextrose, sodium pyruvate (1 mM), l-glutamine (0.5 mM), penicillin (25 IU/ml) and streptomycin (25 μg/ml). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2 and 21% O2 (normoxia) or 1% O2 (hypoxia) in a hypoxia workstation (InVivo1,000, Ruskinn). The LN229-HRE-AP reporter cell line for HIF transcriptional activity was created by stably transfecting LN229 cells with the pACN188 plasmid, which contains an alkaline phosphatase gene driven by six HREs derived from the VEGF gene (31, 43).
Cycloheximide (CHX), an inhibitor of protein synthesis, was used to test whether KC7F2 affects the protein degradation rate of HIF-1α. Cells were plated on 60 mm dishes and incubated under hypoxic conditions for 4 hrs to induce HIF-1α protein. At time zero, CHX and KC7F2 were added at a final concentration of 100 μM and 40 μM, respectively. Then cells were incubated under normoxia or hypoxia and harvested at different time points.
MG-132 (Calbiochem, La Jolla, CA), a proteasome inhibitor, was used to investigate whether KC7F2 affects the protein synthesis of HIF-1α. MG132 (10 μM) was added to cell media (time zero) and, 30 min later, KC7F2 was added at a final concentration of 40 μM. Cells were incubated either under normoxia (21% O2) or hypoxia (1% O2), then harvested at zero, one, two and four hrs with RIPA buffer. The lysed cell extracts were subjected to western blot analysis.
Alkaline phosphatase (AP) assay
LN229-HRE-AP cells (4×104 per well) were plated onto 96-well plates and incubated with or without chemicals for 24 hrs under hypoxic or normoxic condition. Cells were washed with PBS and p-nitrophenyl phosphate (Sigma, St. Louis, MO) was added into each well. The plates were incubated at 37°C for 30 min to detect and quantify AP enzymatic activity by measuring optical density (OD) values at 405 nm in a spectrophotometer as described (31).
Sulforhodamine B (SRB) assay
Cells were seeded onto 96-well plates (4×103 per well) and cultured under normoxic (21% O2) and hypoxic (1% O2) conditions with different concentrations of KC7F2 for 72 hrs or treated for various times with 20 μM KC7F2. For proliferation analysis, cells were fixed with 50% Trichloroacetic acid for one hour at 4°C, followed by staining with 0.4% SRB (Sigma) dissolved in 1% acetic acid for 30 min at room temperature. Plates were washed five times with 1% acetic acid to remove unbound dye. Bound dye was dissolved by adding 10 mM unbuffered Tris base. Cell proliferation was calculated by measuring OD values at 564 nm using a spectrophotometer.
Clonogenic assay
D54MG cancer cells (200 cells/well) or HFF-1 immortalized human fibroblast cells (500 cells/well) were seeded onto 6-well plates. After 16 hrs, KC7F2 was added to final concentrations of 0, 5, 10 μM and the cells treated for 72 hrs under hypoxia (1% O2) or normoxia (21% O2). Therafter, the cells were returned to normoxic conditions. After 11 days, the cells were fixed and stained with crystal violet (0.9 %). Cell colonies were counted and percent of survival was calculated by comparing to colonies obtained in untreated plates.
Western blot analysis
Cells were washed twice with cold PBS and lysed with radio-immunoprecipitation assay (RIPA) buffer consisting of 50 mM Tris, 150 mM NaCl, 1% NP40, 2 mM EDTA, 1 mM fluoride/iodide/pyrophosphate/orthovanadate. The buffer was supplemented with 1 mM DTT, 1 mM PMSF and protease inhibitor cocktail (Roche) before cell lysis. The cells were harvested by mechanical scraping, then subjected to centrifugation at 15,000 rpm for 1 min to remove cell debris. The collected proteins were electrophoresed on 7.5–12.5% Tris-HCl gels (BioRad, Richmond, CA), then electroblotted to nitrocellulose membranes. The membranes were subjected to immunoblot analysis using anti-HIF-1α (1: 600 dilution, BD Bioscience, San Diego, CA), anti-HIF-1β (1:1,000 dilution, BD Bioscience, San Diego, CA), anti-Carbonic Anhydrase IX (1:1,000 dilution, Novus Biologicals), anti-Enolase 1 (1:500 dilution, Novus Biologicals), anti-MMP2 (1:1,000 dilution, Novus Biologicals), anti-β-actin (1:1,000 dilution, Santa Cruz Biotechnologies), anti-α-tubulin (1:1,000 dilution, Santa Cruz Biotechnologies) antibodies. Antibodies specific for Akt, p-Akt, mTOR, p-mTOR, S6K, p-S6K, 4EBP1, p-4EBP1 (Cell Signaling Technology, Beverly, MA) were used at a dilution of 1:1,000 (total form) or 1:500 (phosphorylated form). Corresponding secondary horseradish peroxide-conjugated antibodies were used to detect the target proteins.
Northern blot analysis
Cells were harvested by Trizol reagent (Invitrogen, Carlsbad, CA) and total RNA was extracted according to the manufacturer’s instructions. Equal amounts of total RNA samples were loaded into 1% agarose-formaldehyde gels, separated by electrophoresis, and transferred to nylon membranes. HIF-1α and Enolase 1 cDNA probes were labeled with [α-32P] dCTP (Amersham Biosciences, Piscataway, NY) using Prime-It II Random Primer Labeling Kit (Stratagene, Ceder Creek, TX) and hybridized to the membrane. The hybridization was done with ULTRAhyb buffer (Ambion, Austin, TX) at 42°C overnight. The membrane was washed twice with buffer A containing 2×SSC and 0.2%×SDS, followed by twice with buffer B containing 0.2× SSC and 0.2%×SDS at 42°C. Stripping of the membrane before hybridization with another probe was done in a microwave for 3 min with stripping buffer containing 1 mM Tris (pH 8.0), 1 mM EDTA, and 0.1% SDS.
Results
Identification of KC7F2 as a HIF-1α inhibitor using a cell-based screening assay
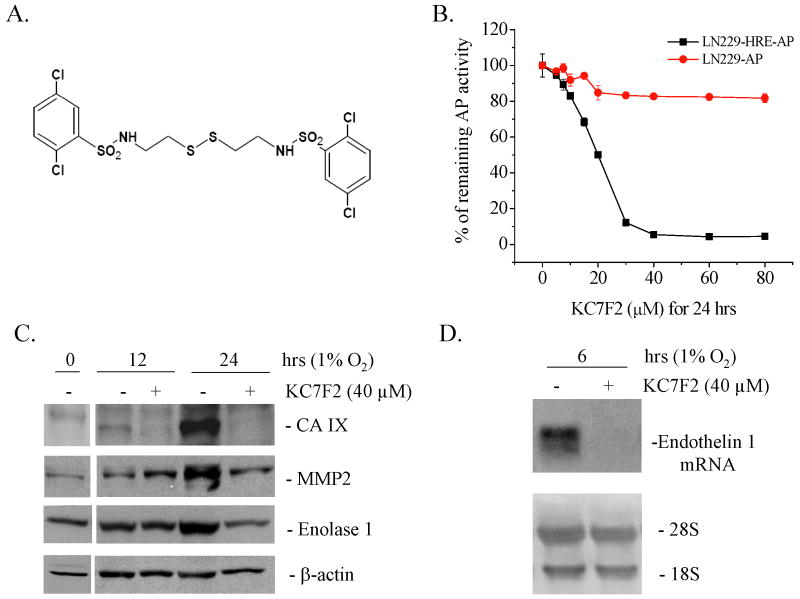
To identify novel small molecules that have the potential of inhibiting HIF-1α biological activity, we previously designed a bioassay using a HIF-reporter cell line, LN229-HRE-AP (31). These cells contain a stably integrated reporter plasmid constituted of a HIF-1 responsive promoter containing six copies of VEGF gene-derived HREs driving the expression of an alkaline phosphatase (AP) gene. The inhibitory effects of tested compounds are measured by a reduction of the AP enzymatic activity present in hypoxic cells. Using this bioassay, we screened a 10,000-membered natural product-like combinatorial library, and identified a novel class of compounds, which have a cystamine group as their central structure. These analogues were subjected to further screening, leading us to identify a small family of compounds for which we determined the IC50 by a dose-response in our bioassay (Fig. 1A and supplementary Fig. S1) (44). The two most potent compounds were KC7F2 and KC7F3, and had an IC50 of 20 and 15 μM, respectively.
Fig. 1. KC7F2 inhibits HRE-driven transcription.
A, Chemical structure of KC7F2. B, KC7F2 inhibits HRE-mediated alkaline phosphatase (AP) activity under hypoxia (1% O2). LN229-HRE-AP cells were incubated with different concentrations of KC7F2 for 24 hrs. Cells expressing AP constitutively (LN229-AP) were used as a control. The relative remaining AP activities were calculated as the ratios of AP levels in cells treated with KC7F2 versus untreated cells at each concentration. KC7F2 inhibits the transcription of defined HIF-1α target genes in LN229 cells as detected by western blot (C) or northern blot (D).
To verify that the inhibition of AP reporter activity by these two compounds was specific to HRE hypoxia response, we performed the same assay in a control cell line (LN229-AP), which constitutively expresses AP under a CMV promoter. LN229-AP cells retained over 80% of AP activity when exposed to concentrations >25 μM of KC7F2 (Fig. 1B), whereas LN229-HRE-AP showed less than 10% of AP activity at the same concentrations, indicating that the reduction of AP activity observed in LN229 cells was independent of KC7F2-mediated non-specific cytotoxicity or direct inhibition of AP enzyme activity (Fig. 1B). In contrast, KC7F3 showed more pronounced inhibitory activity on control LN229-AP cells (data not shown) and was, therefore, not studied further.
To test whether KC7F2 inhibits the expression of the endogenous HIF-1 target genes, we performed microarray analyses at 24 hrs on LNZ308 human glioma cells grown under hypoxic conditions with or without treatment. The results showed that a panel of well-known HIF target genes are inhibited by KC7F2, such as Carbonic Anhydrase IX (CA IX), Matrix Metalloproteinase 2 (MMP2), Enolase 1 and Endothelin 1 (Supplementary Table). To independently confirm these results we performed western blotting, and showed that the levels of the respective proteins were reduced by 24 hrs post KC7F2 treatment (Fig. 1C). Due to the lack of reliable antibodies for Endothelin 1, we verified the inhibition of its mRNA by northern blot and confirmed that it was reduced at 6 hrs post-treatment (Fig. 1D). These results show that KC7F2 can antagonize the hypoxia-induced expression of endogenous HIF target genes.
Treatment with KC7F2 is cytotoxic to cancer cells and this effect is more severe under hypoxic conditions
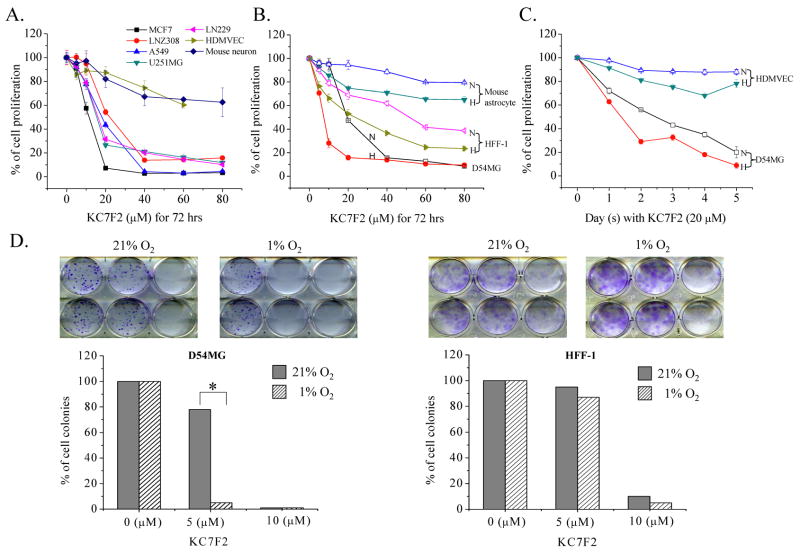
To determine whether HIF inhibition by KC7F2 has inhibitory effects on cell growth and survival, we performed SRB and clonogenic assays under both normoxia and hypoxia. Treatment of a variety of cancer cell lines with KC7F2 for 72 hrs demonstrated a clear dose-response cytotoxicity with an IC50 of approximately 15–25 μM, depending on the cell lines. Remarkably, non-tumoral cells (HDMEV and mouse neurons) showed much less susceptibility to KC7F2 (Fig. 2A). The cytotoxicity of KC7F2 was also increased under hypoxia in the SRB experiments whether in dose-response (Fig. 2B) or in long-term time course response (Fig. 2C). Consistent with SRB experiments, KC7F2 inhibited colony formation of D54MG glioma cells in a dose-dependent manner and this effect was more significant under hypoxia, implying that the cytotoxicity of KC7F2 is pejorated under hypoxia. In contrast, the immortalized fibroblast cell line HFF-1 was more resistant to KC7F2 treatment in colony formation under both normoxia and hypoxia (Fig. 2D). These results suggest that KC7F2 is a lead structure that has potential towards development of a therapeutic agent for the treatment of cancers that depend upon HIF expression for their survival.
Fig. 2. Cytotoxicity analysis in response to KC7F2 treatment.
A, Normal cells (HDMVEC and mouse neurons) or cancer cells (MCF7, LNZ308, A549, U251MG and LN229) were exposed for 72 hrs to different doses of KC7F2 under normoxia and their proliferation rates were determined by SRB assay. Cytotoxicity of KC7F2 was more pronounced in tumor cell lines as compared to normal cells.
B, Mouse astrocyte, HFF-1 and D54MG cells were exposed to different concentrations of KC7F2 under normoxia or hypoxia for 72 hrs and their proliferation were analyzed by the SRB assay. N stands for normoxia (21% O2, open symbol). H stands for hypoxia (1% O2, filled symbol).
C. HDMVEC and D54MG cells were treated with 20 μM KC7F2 under normoxia or hypoxia. The anti-proliferation effects of KC7F2 at different time points were determined by the SRB assay.
D. Clonogenic assay of KC7F2 on D54MG and HFF-1 cells under normoxia and hypoxia. The lower panel shows the percent of surviving colonies after treatment.
KC7F2 decreases HIF-1α protein levels in a dose-dependent manner
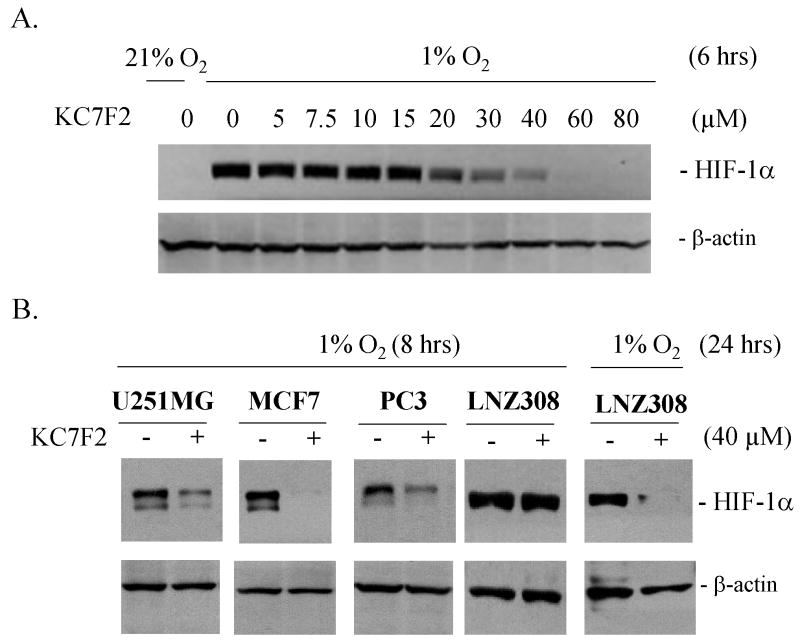
We next examined the molecular mechanism of KC7F2 action. To determine whether the inhibitory effect of KC7F2 was related to HIF-1α protein levels, we performed a western blot analysis on LN229 cells incubated for 6 hrs under hypoxia with KC7F2 at different concentrations (Fig. 3A). KC7F2 specifically reduced the protein levels of HIF-1α in a dose-dependent manner under hypoxic conditions, while the levels of β-actin were largely unaffected. These results suggest that KC7F2 inhibits HIF-1α at the protein level, making it unavailable for HIF-1 mediated transcription.
Fig. 3. KC7F2 reduces the protein levels of HIF-1α in cancer cell lines of different tissue origin and genetic background.
A, LN229 cells were treated with different concentrations of KC7F2 for 6 hrs under hypoxic conditions. Note a strong decrease in HIF-1α levels at concentrations above 20 μM, consistent with the IC50 in the AP reporter assay.
B, U251MG, MCF7, PC3, and LNZ308 cells were treated with 40 μM KC7F2 for 8 or 24 hrs under hypoxia and HIF-1α levels analyzed by western blot.
To examine whether the inhibitory effect of KC7F2 was applicable to a variety of cancer cell lines with different genetic background (wild type or mutated p53, PTEN, p14ARF, CDKN2A) and derived from different organs, we treated U251MG (glioma), PC3 (prostate), MCF7 (breast) and LNZ308 (glioblastoma) cell lines with KC7F2 (Fig. 3B). These cell lines were incubated with or without 40 μM of KC7F2 for 8 or 24 hrs under hypoxia, followed by western blot analysis. HIF-1α protein was strongly suppressed in each cell line by 40 μM of KC7F2 to the same extent as in LN229. Interestingly, LNZ308 cells were resistant to KC7F2 treatment at the early time point and the inhibitory effect became visible only after 24 hrs.
Taken together, these data indicate that KC7F2 suppresses HIF-1 transcription function in a variety of human cancer cells by blocking HIF-1α protein accumulation in response to hypoxia.
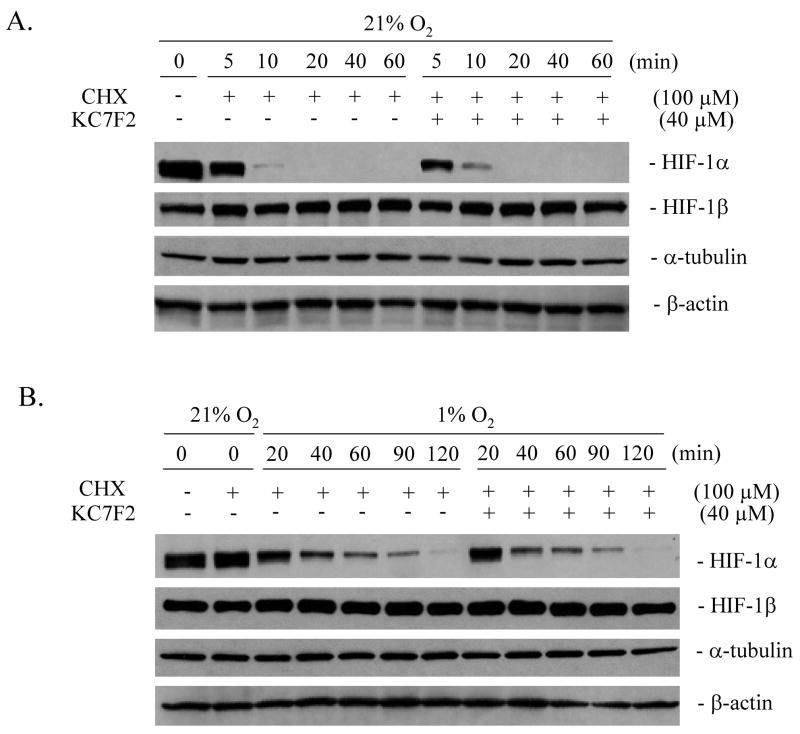
KC7F2 does not affect the rate of HIF-1α protein degradation
To further investigate how KC7F2 inhibits HIF-1α expression, we first tested whether the degradation rate of HIF-1α protein was affected by KC7F2 in the presence of cycloheximide (CHX), an inhibitor of protein translation. LN229 cells were pre-incubated under hypoxia for 4 hrs to make HIF-1α levels reach their steady state, then treated with 100 μM of CHX with or without 40 μM of KC7F2. The half-life of pre-stabilized HIF-1α was examined under normoxia (Fig. 4A) and hypoxia (Fig. 4B) by western blot analysis. Accumulated HIF-1α was degraded in less than 10 min under normoxia as expected (45), and the kinetics of protein degradation were not affected by KC7F2. Under hypoxia, HIF-1α showed a ~6 fold longer half-life due to the absence of VHL-mediated proteasomal degradation, but no significant difference in its degradation rate was observed with or without KC7F2. These results indicate that KC7F2 does not affect the HIF-1α protein degradation machineries which operate under normoxic (45) or hypoxic conditions (46).
Fig. 4.
KC7F2 does not affect the protein degradation rate of HIF-1α. LN229 cells were treated with 100 μM of cycloheximide (CHX) to inhibit protein synthesis at time zero. The protein levels of HIF-1α, HIF-1β, α-tubulin and β-actin with or without KC7F2 (40 μM) treatment were analyzed under normoxic (A) or hypoxic conditions (B) over a period of 60–120 min. Note that the degradation rate of HIF-1α is slower under hypoxia than normoxia as expected, but neither is changed by KC7F2.
KC7F2 inhibits HIF-1α protein synthesis but not its mRNA transcription
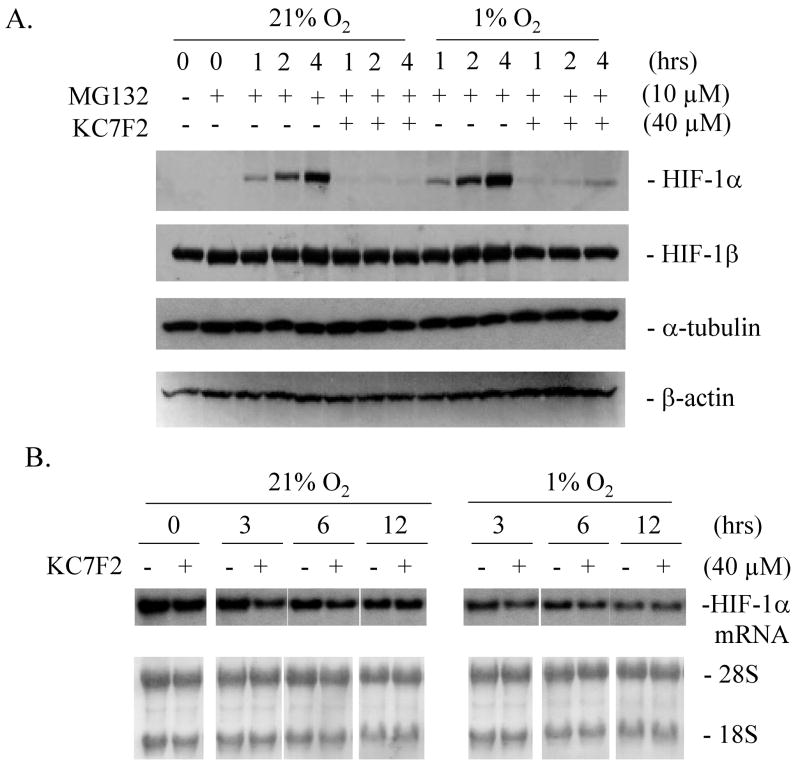
Next, we examined whether KC7F2 could regulate HIF-1α protein synthesis. To address this question, LN229 cells were treated with MG132, a proteasome inhibitor, to interrupt HIF-1α degradation. MG132 was added to cell culture medium at time zero and the accumulation of newly synthesized HIF-1α protein was detected over time (Fig. 5A). Under both normoxic and hypoxic conditions, HIF-1α protein appeared within one hour after MG132 addition, and continued to accumulate up to four hrs. However, in the presence of KC7F2, HIF-1α protein accumulation was strongly suppressed at all time points, while that of control proteins (HIF-1β, α-tubulin and β-actin) were unaffected. Northern blot experiments show that HIF-1α mRNA expression levels were not affected by KC7F2 (Fig. 5B). Overall, these results suggest that KC7F2 can inhibit HIF-1α protein accumulation at the protein translational level, but does not affect its mRNA synthesis.
Fig. 5. KC7F2 inhibits HIF-1α protein synthesis but not its mRNA transcription.
A, LN229 cells were treated with 10 μM of MG132 to inhibit protein degradation through the proteasome. The levels of newly synthesized HIF-1α, HIF-1β, α-tubulin and β-actin with or without KC7F2 (40 μM) treatment were determined over a period of 4 hrs. Note that HIF-1α synthesis is dramatically reduced in the presence of KC7F2.
B, The levels of HIF-1α mRNA were not affected by KC7F2 (40 μM) in LN229 cells treated for up to 12 hrs, whether under hypoxic or normoxic conditions as measured by northern blot analysis.
KC7F2 represses the phosphorylation of eukaryotic initiation factor 4E binding protein 1 (4EBP1)
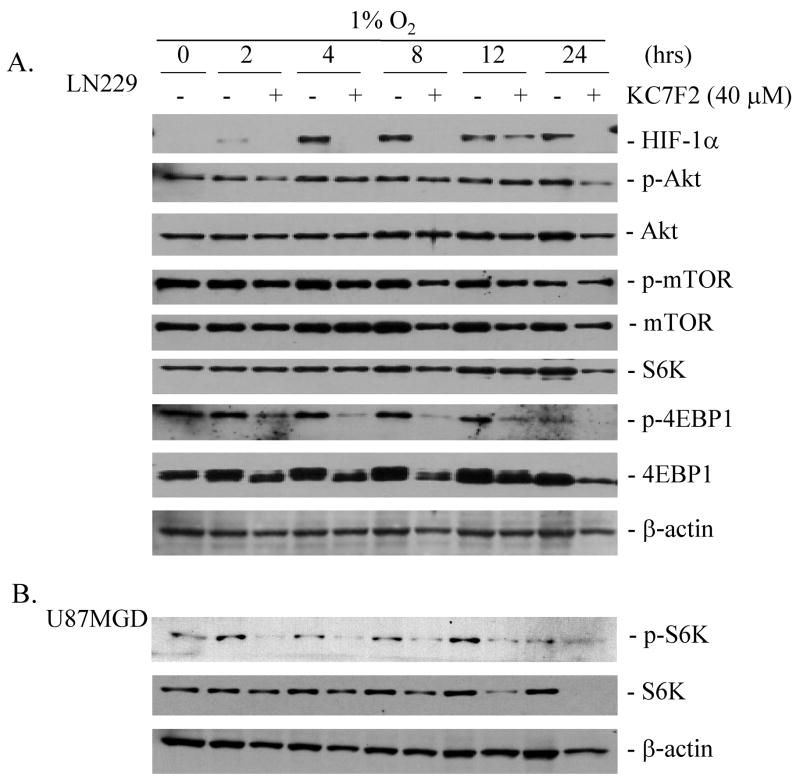
The PI3K-Akt-mTOR pathway plays a key role in the control of HIF-1α translation and synthesis (11). To explore whether the HIF-1α protein inhibition by KC7F2 was linked to the suppression of this pathway, LN229 cells were pre-treated for one hour with 40 μM of KC7F2 followed by hypoxia. Thereafter, the levels of HIF-1α, and the total or phosphorylated forms of Akt, mTOR, S6K, and 4EBP1 were examined at 2 to 24 hrs by western blot (Fig. 6A). HIF-1α protein was clearly detectable after 4 hrs of hypoxia treatment in LN229 cells and, as expected, was suppressed by KC7F2 at each time point. The levels of phosphorylated 4EBP1 were strongly suppressed by KC7F2 as early as 2 hrs and throughout the 24 hrs incubation under hypoxia. The levels of non-phosphorylated 4EBP1 (lower band) were not altered until 12 hrs, then at 24 hrs showed a drop in expression. In contrast, the levels of phosphorylated Akt, total Akt, phosphorylated mTOR, total mTOR, and total S6K were not affected or showed only modest changes up to 12 hrs in response to KC7F2. At 24 hrs, a ~50% drop in Akt and S6K levels was observed. As phosphorylated S6K was not detectable in LN229 as previously observed (31), we analyzed its changes in U87MGD glioma cells upon KC7F2 treatment (Fig. 6B). The level of phosphorylated S6K was affected in a similar fashion to phospho-4EBP1, while total S6K showed a gradual decrease which became more pronounced after 12 hrs. The phosphorylation of 4EBP1 and S6K are important steps for the initiation of protein translation (47); therefore, these findings provide a mechanistic explanation for the inhibition of KC7F2 on HIF-1α protein synthesis.
Fig. 6. The inhibition of KC7F2 on HIF-1α protein synthesis involved inhibition of the phosphorylation of 4EBP1 and S6K.
A, LN229 cells were pre-treated for one hour with 40 μM of KC7F2 followed by 2–24 hrs of hypoxia. The levels of HIF-1α, and the total and phosphorylated forms of Akt, mTOR, S6K, and 4E binding protein 1 (4EBP1), were examined over time by western blot. The levels of p-Akt, Akt p-mTOR, mTOR, S6K were unchanged or showed only modest change up to 12 hrs in response to KC7F2. In contrast, the phosphorylated 4EBP1 was strongly suppressed by KC7F2 as early as 2 hrs and throughout the 24 hrs incubation under hypoxia. LN229 cells do not express detectable p-S6K as we previously reported (31).
B, to examine the effect of KC7F2 on a cell line expressing constitutively p-S6K we treated U87MGD glioma cells for 2–24 hrs with KC7F2 (40 μM) and examined total and phospho-forms of S6K by western blot. Note that phospho-S6K was decreased as early as 2 hrs post-treatment.
Discussion
Through the screening of a natural product-like chemical compound library, we have found a family of compounds with a cystamine structure that inhibits the HIF-1 transcriptional pathway. Compounds containing cystamine have been previously found to be metabolically stable (48). Herein, we described the KC7F2 compound, showed its preferential cytotoxicity to cancer cells and examined its mechanism of action. We found that KC7F2 cytotoxicity was aggravated under hypoxia and since HIF-1α is the major regulator for hypoxic cell growth and survival, the suppression of HIF-1α by KC7F2 may contribute to its enhanced cytotoxicity under hypoxia. However, it should be noted that the cytotoxicity/antitumor effects of KC7F2 may not only be caused by its suppression on HIF-1α. Mechanistically, KC7F2 dramatically suppressed the protein accumulation of HIF-1α in cancer cell lines derived from several different organ types (brain, breast and prostate). KC7F2 did not accelerate HIF-1α degradation, but rather inhibited its protein synthesis at the translation level. Further work will be needed to precisely determine the mechanism of KC7F2 cytotoxicity, its relationship to the HIF family of transcription factors and whether it can produce other antitumor effects similar to those identified for Psammaplin A, given their similarity in chemical structure. Psammaplin A is a natural product isolated from a two-sponge association (49) which exhibits antitumor effects through the inhibition of topoisomerase II (50), mammalian aminopeptidase N (51) and histone deacetylase (52).
Investigation of the molecular mechanism through which KC7F2 inhibits HIF-1α protein synthesis led us to the mTOR pathway. In mammalian cells, the PI3K-Akt-mTOR pathway plays key roles in cell metabolism, nnutrition regulation, protein synthesis, and tumorigenic processes (53). mTOR plays its functions through two different complexes, mTORC1 and mTORC2 (54, 55). mTORC1 regulates protein synthesis through its downstream targets, 4EBP1 and S6K. 4EBP1 is a negative factor for protein translation initiation as it binds tightly and blocks the function of eukaryotic translation initiation factor 4E (eIF-4E). The phosphorylation of 4EBP1 by the TORC1 complex inactivates it, freeing eIF-4E to perform its normal function in translation initiation (47). mTORC1 can also phosphorylate S6K, which stimulates the S6 ribosomal protein and other components of the translational machinery, including eIF4b and eEF2 (56). Compared to mTORC1, mTORC2 is mainly involved in cytoskeleton regulation (55) and Akt activation (57). To detect possible targets of KC7F2 in the mTOR pathway, we have analyzed the changes in total and phosphorylated forms of Akt, mTOR, S6K, and 4EBP1 upon treatment. Results showed that KC7F2 significantly reduces the phosphorylation of 4EBP1 and S6K, while it has no obvious effects on the other signaling molecules. These findings suggest that KC7F2 specifically targets the mTOR complex 1 pathway, which leads to the inhibition of protein synthesis. Maintenance of 4EBP1 in a non-phosphorylated form is expected to stabilize its binding to eIF-4E, thus making the latter unavailable for the initiation of protein translation. Inhibition of the phosphorylation of S6K also makes it inactive in protein synthesis. These effects may account for the down-regulation of HIF-1α protein synthesis by KC7F2. It is interesting to note that another inhibitor of HIF-α protein synthesis, the thioredoxin inhibitor PX-1, also has the disulfide core unit (58). Whether PX-1 and KC7F2 have similar molecular targets need to be further investigated.
Although KC7F2 inhibits the phosphorylation of 4EBP1 and S6K, what is currently considered as a general step in protein translation and dramatically down-regulated the protein levels of HIF-1α, it had only modest effects on the steady state levels of other proteins, including Akt, mTOR, total 4EBP1, total S6K, HIF-1α, β-actin, and α-tubulin. These results are similar in part to those of our recently identified anti-HIF-1α compound, 103D5R, which also inhibited the phosphorylation of Akt, Erk1/2, and stress-activated protein kinase/c-jun-NH2-kinase (31). Both 103D5R and KC7F2 specifically inhibit protein synthesis of HIF-1α, yet the levels of other control proteins were only minimally affected. These findings raise the question as to whether the translational process of HIF-1α is more susceptible for drug inhibition. As the main regulator of rapid adaptation to hypoxia, HIF-1α has a rapid turnover rate. It is possible that the translation of HIF-1α mRNA involves a somewhat distinct mechanism which could be more susceptible to the action of certain inhibitors, while the protein translational machinery for other major proteins is largely unaffected. In support of this possibility, it was found that HIF-1α mRNA contains an internal ribosome entry site that allows its efficient translation under both normoxic and hypoxic conditions (59). Under hypoxia, HIF-1α mRNA translation continues despite the global inhibition of the protein translation process (60). Therefore, the regulation of HIF-1α translation may be more complex than we expected. Further investigations are needed to address how KC7F2, 103D5 and other compounds (31, 61, 62) preferentially impair the translation of HIF-1α mRNA.
Activation of the HIF pathway is observed in many solid tumors through hypoxia, as well as the result of growth-promoting stimuli and aberrant oncogenic signaling (63, 64). Activation of HIF transcription can lead to the expression of over 100 HIF target genes that are implicated in adaptive mechanisms such as erythropoiesis, angiogenesis, invasion, metabolic adaptation, glucose transport and acidification (11). The spectrum of target genes activated varies upon tissue type and cancer examined, and it is currently unknown what defines the selectivity of target genes activated and which encode the critical players that are important in tumor development and progression. We showed that KC7F2 was able to abolish the stabilization of HIF-1α protein in different cancer cell lines, and examined more specifically in gliomas which downstream genes were affected. These studies established that Carbonic Anhydrase IX, MMP2, Enolase 1 and Endothelin 1 were strongly down-regulated. These proteins contribute to acidification of the tumor environment, tumor invasion, glycolysis and angiogenesis (13–17; 65, 66), suggesting that KC7F2 has potential to impact tumor growth. It will be promising to evaluate the antitumor efficacy of KC7F2 in preclinical models once its toxicity and pharmacological profile have been established.
In summary, we have identified a novel small molecule KC7F2 targeting HIF pathway inhibition. Our results showed that KC7F2 exhibited enhanced cytotoxicity in various cancer cells under hypoxia and inhibited HIF transcriptional activity through the down-regulation of the protein levels of the HIF-1α subunit, the result of reduced translation of its mRNA. Further investigation found that KC7F2 dramatically repressed the phosphorylation of 4EBP-1 and S6K, which may explain its inhibition of HIF-1α protein synthesis. Precisely how KC7F2 affects 4EBP-1 and S6K and why this inhibitory effect shows specificity remains to be further elucidated. Efforts in better understanding the mechanism of action of KC7F2 will undoubtedly help define its potential as a novel therapeutic for cancer.
Supplementary Material
Acknowledgments
We thank Dr Vladimir E. Belozerov for advice with experiments. This research effort was supported in part by grants to E.G.V.M. from the National Institute of Health (DCB APRC supplement to CA86335, CA116804), the American Brain Tumor Association, the Brain Tumor Foundation for Children, the Charlotte Geyer Foundation, EmTechBio, the Southeastern Brain Tumor Foundation, and the Emory University Research Council. Financial support to K.C.N. was from the NIH (USA) as well as a Governor’s Fellowship from the Kellogg School of Science and Technology, The Scripps Research Institute (to C.F.G.).
Footnotes
Note: T Narita and S Yin designed and performed experiments. CS Moreno analyzed microarray data, M Yepes provided the primary mouse neuron cultures and KC Nicolaou and CF Gelin synthesized the chemical compounds. EG Van Meir conceived the project, discussed the experimental design, analyzed and interpreted the results and wrote the article with T Narita and S Yin.
Statement of Translation Relevance
Solid tumors are hypoxic and this feature renders them more resistant to radiation and chemotherapy. Hypoxia activates the HIF transcriptional pathway which drives the synthesis of key factors that promote cancer cell survival and tumor growth. Therefore, targeting the HIF pathway is a promising anti-cancer strategy. In this study, we have identified a novel small molecule KC7F2 as a potent HIF-1α translation inhibitor. This lead compound shows promise for further development into an antitumor agent for clinical cancer therapy.
References
- 1.Semenza GL. Hypoxia and cancer. Cancer Metastasis Rev. 2007;26:223–4. doi: 10.1007/s10555-007-9058-y. [DOI] [PubMed] [Google Scholar]
- 2.Gray LH. Radiobiologic basis of oxygen as a modifying factor in radiation therapy. Am J Roentgenol Radium Ther Nucl Med. 1961;85:803–15. [PubMed] [Google Scholar]
- 3.Amellem O, Pettersen EO. Cell inactivation and cell cycle inhibition as induced by extreme hypoxia: the possible role of cell cycle arrest as a protection against hypoxia-induced lethal damage. Cell Proliferation. 1991;24:127–41. doi: 10.1111/j.1365-2184.1991.tb01144.x. [DOI] [PubMed] [Google Scholar]
- 4.Comerford KM, Wallace TJ, Karhausen J, Louis NA, Montalto MC, Colgan SP. Hypoxia-inducible factor-1-dependent regulation of the multidrug resistance (MDR1) gene. Cancer Res. 2002;62:3387–94. [PubMed] [Google Scholar]
- 5.Kim H, Peng G, Hicks JM, et al. Engineering human tumor-specific cytotoxic T cells to function in a hypoxic environment. Mol Ther. 2008;16:599–606. doi: 10.1038/sj.mt.6300391. [DOI] [PubMed] [Google Scholar]
- 6.Griguer CE, Oliva CR, Gobin E, et al. CD133 is a marker of bioenergetic stress in human glioma. PLoS ONE. 2008;3:e3655. doi: 10.1371/journal.pone.0003655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenza GL. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol Med. 2002;8:S62–7. doi: 10.1016/s1471-4914(02)02317-1. [DOI] [PubMed] [Google Scholar]
- 8.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Graeber TG, Osmanian C, Jacks T, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 10.Shahrzad S, Quayle L, Stone C, et al. Ischemia-induced K-ras mutations in human colorectal cancer cells: role of microenvironmental regulation of MSH2 expression. Cancer Res. 2005;65:8134–41. doi: 10.1158/0008-5472.CAN-05-0713. [DOI] [PubMed] [Google Scholar]
- 11.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–32. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 12.Weidemann A, Johnson RS. Biology of HIF-1alpha. Cell Death Differ. 2008;15:621–7. doi: 10.1038/cdd.2008.12. [DOI] [PubMed] [Google Scholar]
- 13.Pastorekova S, Zatovicova M, Pastorek J. Cancer-associated carbonic anhydrases and their inhibition. Curr Pharm Des. 2008;14:685–98. doi: 10.2174/138161208783877893. [DOI] [PubMed] [Google Scholar]
- 14.Kenny HA, Kaur S, Coussens LM, Lengyel E. The initial steps of ovarian cancer cell metastasis are mediated by MMP-2 cleavage of vitronectin and fibronectin. J Clin Invest. 2008;118:1367–79. doi: 10.1172/JCI33775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–20. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Brennan PA, Zaki GA. Angiogenesis in cancer: the role of endothelin-1. Ann R Coll Surg Engl. 2000;82:363–4. [PMC free article] [PubMed] [Google Scholar]
- 17.Guo P, Imanishi Y, Cackowski FC, et al. Up-regulation of angiopoietin-2, matrix metalloprotease-2, membrane type 1 metalloprotease, and laminin 5 gamma 2 correlates with the invasiveness of human glioma. Am J Pathol. 2005;166:877–90. doi: 10.1016/s0002-9440(10)62308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov S, Liao SY, Ivanova A, et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol. 2001;158:905–19. doi: 10.1016/S0002-9440(10)64038-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zelzer E, Levy Y, Kahana C, Shilo BZ, Rubinstein M, Cohen B. Insulin induces transcription of target genes through the hypoxia-inducible factor HIF-1alpha/ARNT. Embo J. 1998;17:5085–94. doi: 10.1093/emboj/17.17.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feldser D, Agani F, Iyer NV, Pak B, Ferreira G, Semenza GL. Reciprocal positive regulation of hypoxia-inducible factor 1alpha and insulin-like growth factor 2. Cancer Res. 1999;59:3915–8. [PubMed] [Google Scholar]
- 21.Richard DE, Berra E, Pouyssegur J. Nonhypoxic pathway mediates the induction of hypoxia-inducible factor 1alpha in vascular smooth muscle cells. J Biol Chem. 2000;275:26765–71. doi: 10.1074/jbc.M003325200. [DOI] [PubMed] [Google Scholar]
- 22.Fukuda R, Hirota K, Fan F, Jung YD, Ellis LM, Semenza GL. Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J Biol Chem. 2002;277:38205–11. doi: 10.1074/jbc.M203781200. [DOI] [PubMed] [Google Scholar]
- 23.Treins C, Giorgetti-Peraldi S, Murdaca J, Semenza GL, Van Obberghen E. Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J Biol Chem. 2002;277:27975–81. doi: 10.1074/jbc.M204152200. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt MH, Furnari FB, Cavenee WK, Bogler O. Epidermal growth factor receptor signaling intensity determines intracellular protein interactions, ubiquitination, and internalization. Proc Natl Acad Sci U S A. 2003;100:6505–10. doi: 10.1073/pnas.1031790100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001;12:363–9. [PubMed] [Google Scholar]
- 26.Maynard MA, Ohh M. The role of hypoxia-inducible factors in cancer. Cell Mol Life Sci. 2007;64:2170–80. doi: 10.1007/s00018-007-7082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J Biol Chem. 1999;274:32631–7. doi: 10.1074/jbc.274.46.32631. [DOI] [PubMed] [Google Scholar]
- 28.Sang N, Stiehl DP, Bohensky J, Leshchinsky I, Srinivas V, Caro J. MAPK signaling up-regulates the activity of hypoxia-inducible factors by its effects on p300. J Biol Chem. 2003;278:14013–9. doi: 10.1074/jbc.M209702200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong H, De Marzo AM, Laughner E, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5. [PubMed] [Google Scholar]
- 30.Talks KL, Turley H, Gatter KC, et al. The expression and distribution of the hypoxia-inducible factors HIF-1alpha and HIF-2alpha in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157:411–21. doi: 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tan C, de Noronha RG, Roecker AJ, et al. Identification of a novel small-molecule inhibitor of the hypoxia-inducible factor 1 pathway. Cancer Res. 2005;65:605–12. [PubMed] [Google Scholar]
- 32.Rapisarda A, Uranchimeg B, Scudiero DA, et al. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 2002;62:4316–24. [PubMed] [Google Scholar]
- 33.Mabjeesh NJ, Post DE, Willard MT, et al. Geldanamycin induces degradation of hypoxia-inducible factor 1alpha protein via the proteosome pathway in prostate cancer cells. Cancer Res. 2002;62:2478–82. [PubMed] [Google Scholar]
- 34.Kung AL, Zabludoff SD, France DS, et al. Small molecule blockade of transcriptional coactivation of the hypoxia-inducible factor pathway. Cancer Cell. 2004;6:33–43. doi: 10.1016/j.ccr.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Melillo G. Inhibiting hypoxia-inducible factor 1 for cancer therapy. Mol Cancer Res. 2006;4:601–5. doi: 10.1158/1541-7786.MCR-06-0235. [DOI] [PubMed] [Google Scholar]
- 36.Giaccia A, Siim BG, Johnson RS. HIF-1 as a target for drug development. Nat Rev Drug Discov. 2003;2:803–11. doi: 10.1038/nrd1199. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Qian DZ, Tan YS, et al. Inaugural Article: Digoxin and other cardiac glycosides inhibit HIF-1alpha synthesis and block tumor growth. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0809763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belozerov VE, Van Meir EG. Inhibitors of hypoxia-inducible factor-1 signaling. Curr Opin Investig Drugs. 2006;7:1067–76. [PubMed] [Google Scholar]
- 39.Belozerov VE, Van Meir EG. Hypoxia inducible factor-1: a novel target for cancer therapy. Anticancer Drugs. 2005;16:901–9. doi: 10.1097/01.cad.0000180116.85912.69. [DOI] [PubMed] [Google Scholar]
- 40.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–26. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 41.Verheul HM, Salumbides B, Van Erp K, et al. Combination Strategy Targeting the Hypoxia Inducible Factor-1alpha with Mammalian Target of Rapamycin and Histone Deacetylase Inhibitors. Clin Cancer Res. 2008;14:3589–97. doi: 10.1158/1078-0432.CCR-07-4306. [DOI] [PubMed] [Google Scholar]
- 42.Faivre S, Kroemer G, Raymond E. Current development of mTOR inhibitors as anticancer agents. Nat Rev Drug Discov. 2006;5:671–88. doi: 10.1038/nrd2062. [DOI] [PubMed] [Google Scholar]
- 43.Post DE, Van Meir EG. Generation of bidirectional hypoxia/HIF-responsive expression vectors to target gene expression to hypoxic cells. Gene Ther. 2001;8:1801–7. doi: 10.1038/sj.gt.3301605. [DOI] [PubMed] [Google Scholar]
- 44.Nicolaou KC, Hughes R, Pfefferkorn JA, Barluenga S, Roecker AJ. Combinatorial synthesis through disulfide exchange: discovery of potent psammaplin A type antibacterial agents active against methicillin-resistant Staphylococcus aureus (MRSA) Chemistry. 2001;7:4280–95. doi: 10.1002/1521-3765(20011001)7:19<4280::aid-chem4280>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 45.Jaakkola P, Mole DR, Tian YM, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 46.Liu YV, Baek JH, Zhang H, Diez R, Cole RN, Semenza GL. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol Cell. 2007;25:207–17. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pause A, Belsham GJ, Gingras AC, et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature. 1994;371:762–7. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 48.Eker P, Pihl A. Studies on the growth-inhibiting and radioprotective effect of cystamine, cysteamine, and AET on mammalian cells in tissue culture. Radiat Res. 1964:165–79. [PubMed] [Google Scholar]
- 49.Jung JH, Sim CJ, Lee CO. Cytotoxic compounds from a two-sponge association. J Nat Prod. 1995;58:1722–6. doi: 10.1021/np50125a012. [DOI] [PubMed] [Google Scholar]
- 50.Kim D, Lee IS, Jung JH, Lee CO, Choi SU. Psammaplin A, a natural phenolic compound, has inhibitory effect on human topoisomerase II and is cytotoxic to cancer cells. Anticancer Res. 1999;19:4085–90. [PubMed] [Google Scholar]
- 51.Shim JS, Lee HS, Shin J, Kwon HJ. Psammaplin A, a marine natural product, inhibits aminopeptidase N and suppresses angiogenesis in vitro. Cancer Lett. 2004;203:163–9. doi: 10.1016/j.canlet.2003.08.036. [DOI] [PubMed] [Google Scholar]
- 52.Ahn MY, Jung JH, Na YJ, Kim HS. A natural histone deacetylase inhibitor, Psammaplin A, induces cell cycle arrest and apoptosis in human endometrial cancer cells. Gynecol Oncol. 2008;108:27–33. doi: 10.1016/j.ygyno.2007.08.098. [DOI] [PubMed] [Google Scholar]
- 53.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Kim DH, Sarbassov DD, Ali SM, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 55.Sarbassov DD, Ali SM, Kim DH, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 56.Peterson RT, Schreiber SL. Translation control: connecting mitogens and the ribosome. Curr Biol. 1998;8:R248–50. doi: 10.1016/s0960-9822(98)70152-6. [DOI] [PubMed] [Google Scholar]
- 57.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 58.Welsh SJ, Williams RR, Birmingham A, Newman DJ, Kirkpatrick DL, Powis G. The thioredoxin redox inhibitors 1-methylpropyl 2-imidazolyl disulfide and pleurotin inhibit hypoxia-induced factor 1alpha and vascular endothelial growth factor formation. Mol Cancer Ther. 2003;2:235–43. [PubMed] [Google Scholar]
- 59.Lang KJ, Kappel A, Goodall GJ. Hypoxia-inducible factor-1alpha mRNA contains an internal ribosome entry site that allows efficient translation during normoxia and hypoxia. Mol Biol Cell. 2002;13:1792–801. doi: 10.1091/mbc.02-02-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koh MY, Spivak-Kroizman TR, Powis G. HIF-1 regulation: not so easy come, easy go. Trends Biochem Sci. 2008 doi: 10.1016/j.tibs.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 61.Cao Z, Fang J, Xia C, Shi X, Jiang BH. trans-3,4,5′-Trihydroxystibene inhibits hypoxia-inducible factor 1alpha and vascular endothelial growth factor expression in human ovarian cancer cells. Clin Cancer Res. 2004;10:5253–63. doi: 10.1158/1078-0432.CCR-03-0588. [DOI] [PubMed] [Google Scholar]
- 62.Jin X, Jin HR, Lee D, Lee JH, Kim SK, Lee JJ. A quassinoid 6alpha-tigloyloxychaparrinone inhibits hypoxia-inducible factor-1 pathway by inhibition of eukaryotic translation initiation factor 4E phosphorylation. Eur J Pharmacol. 2008;592:41–7. doi: 10.1016/j.ejphar.2008.06.104. [DOI] [PubMed] [Google Scholar]
- 63.Post DE, Devi NS, Li Z, et al. Cancer therapy with a replicating oncolytic adenovirus targeting the hypoxic microenvironment of tumors. Clin Cancer Res. 2004;10:8603–8612. doi: 10.1158/1078-0432.CCR-04-1432. [DOI] [PubMed] [Google Scholar]
- 64.Rong Y, Brat DJ. Vaso-occlusive mechanisms that initiate hypoxia and necrosis in glioblastoma: the role of thrombosis and tissue factor. In: Van Meir EG, editor. CNS cancer: Models, Markers, Prognostic Factors, Targets and Therapeutic Approaches. New York: Humana Press; 2009. pp. 507–528. [Google Scholar]
- 65.Rong Y, Post DE, Pieper RO, Durden DL, Van Meir EG, Brat DJ. PTEN and hypoxia regulate tissue factor expression and plasma coagulation by glioblastoma. Cancer Res. 2005;65:1406–1413. doi: 10.1158/0008-5472.CAN-04-3376. [DOI] [PubMed] [Google Scholar]
- 66.Post DE, Sandberg EM, Devi SN, et al. Targeted cancer-gene therapy using a HIF-dependent oncolytic adenovirus armed with interleukin-4. Cancer Res. 2007;67:6872–81. doi: 10.1158/0008-5472.CAN-06-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.