Abstract
Cytochrome c peroxidase (CCP) and ascorbate peroxidase (APX) have very similar structures, and yet neither CCP nor APX exhibit each others activities with respect to reducing substrates. APX has a unique substrate binding site near the heme propionates where ascorbate H-bonds with a surface Arg and one heme propionate (Sharp et al. (2003) Nat. Struc. Biol. 10, 303–307). The corresponding region in CCP has a much longer surface loop and the critical Arg residue that is required for ascorbate binding in APX is Asn in CCP. In order to convert CCP into an APX, the ascorbate binding loop and critical arginine were engineered into CCP to give the CCP2APX mutant. The mutant crystal structure shows that the engineered site is nearly identical to that found in APX. While wild type CCP shows no APX activity, CCP2APX catalyzes the peroxidation of ascorbate at a rate of ≈ 12 min−1 indicating that the engineered ascorbate binding loop can bind ascorbate.
Peroxidases are a large ubiquitous family of detoxifying enzymes that catalyze the reduction of H2O2 at the expense of various reducing substrates. Most peroxidases are heme-containing enzymes that use hydrogen peroxide to catalyze a number of oxidative reactions. The general reaction scheme of heme peroxidases is as follows:
The enzyme reacts with H2O2 to form compound I. In compound I, the heme iron is oxidized from Fe3+ to Fe4+ and the porphyrin ring to a π-cationic radical (1). Yeast cytochrome c peroxidase (CCP) is an exception since Trp 191 in the proximal pocket just below the heme and adjacent to the His 175 heme ligand is the site of radical formation (2) and not the porphyrin ring. Compound I is then subsequently reduced back to the resting state in two successive one electron transfer reactions involving two molecules of substrate (S in the above scheme) via another enzyme intermediate called compound II.
In general, peroxidases can oxidize a wide variety of substrates including aromatic amines, indoles, phenols, lignin, halides, and manganese (3). When the first few crystal structures of peroxidases became available it appeared that the most likely region for small aromatic substrates to bind is at the one edge of the heme exposed to solvent which thus allows substrates to directly contact the heme for short and fast electron transfer to the porphyrin radical. Indeed, the crystal structure of horse radish peroxidase complexed with ferulic acid shows binding at the heme edge as expected (4). CCP, however, is poor at oxidizing small aromatic dyes but instead is specialized to oxidize cytochrome c. This is accomplished by providing a surface unique to forming a complex with cytochrome c (5) and by locating the radical in compound I on Trp 191 (2) rather than the porphyrin macrocycle. As more crystal structures became available, however, this simple view that, with the exception of CCP, substrates bind at the heme edge came into question. For example, manganese peroxidase has the substrate, Mn(II), coordinated to one heme propionate (6) thus providing a direct route of electron transfer to heme along the heme propionate group. Chemical modification (7), mutagenesis studies (8), and crystallographic studies (9) showed that ascorbate peroxidase (APX) also uses a similar site near the heme propionates.
A long standing goal in enzyme engineering is to introduce novel activities into enzymes by altering or introducing new substrate binding sites. Given the close similarity between the APX and CCP structures with the exception of ascorbate binding site, it appeared to us that it should be possible to introduce the APX substrate binding site into CCP and convert CCP into an APX. If successful, this would provide an important step in developing strategies of tailoring peroxidases to bind other types of substrates that could prove useful in a variety of practical applications such as bioremediation. Here we report our initial attempts at engineering CCP into an APX.
Materials and Methods
Site-directed Mutagenesis
Oligonucleotide-directed mutagenesis experiments were performed on pT7-7 vector that contained the wild-type CCP. Residues 30–42 (LRED DEYDNYIGY) of WTCCP were replaced with residues 27–32 (IAEKKC) of APX in order to introduce the ascorbate-binding loop. The following 69 base oligonucleotide & its reverse complement was obtained from QIAGEN’s Operon Technologies (Alameda, CA) GTG TAC AAT GCG ATT GCA CTC AAG ATT GCG GAA AAG AAG TGT GGG CCC GTA TTA GTC CGT CTT GCT TGG.
In order to introduce crucial Arg into CCP essential for ascorbate binding and catalysis the following oligonucleotide and its reverse complementary sequence was synthesized and obtained from QIAGEN’s Operon Technologies (Alameda, CA).
ACC CAC TTG AAG CGC TCT GGA TAC GAA
Site-directed mutagenesis was carried out by overlap extension method by PCR (10) using Thermal Ace Polymerase (Invitrogen) with outside primers “T7 ” TAATACGACTCACTATAGG and “Lac-80” CAGTCACGACGTTGTAAAAC, which were also purchased from QIAGEN’s Operon Technologies and the requisite internal primers. These mutations were introduced iteratively in stages to follow properties of the protein as a consequence of mutations. First, the ascorbate-binding loop was introduced into CCP followed by the N184R point mutation. The resultant mutant was called CCP2APX. Although the mutant has fewer amino acids than wild type CCP, the wild type amino acid numbering will be used. In order to enable this mutant to form a porphyrin π-cation radical during catalysis Trp 191 was converted to Phe using CCP2APX as template to give CCP2APX/F191. The mutant clones were analyzed and confirmed by restriction analysis and automated DNA sequencing at the sequencing facility of Biotech Diagnostic, Suite 372 23974, Aliso Creek Road, Laguna Niguel CA 92677-3908.
Protein Expression and Purification
Expression plasmids were transformed into Escherichia coli BL21(DE3) STAR competent cells and the transformants were grown in 7 L of terrific broth with vigorous shaking at 37°C until the cultures reached an A600 of 0.8. At this point, protein production was induced from the T7 promoter by adding 750 µM of IPTG. Cells were allowed to express protein overnight at a reduced temperature of 25°C and reduced shaking to 100 rpm. Cells were harvested, lysed and chromatographed on 2 L Sephadex G-75 column followed by heme incorporation according to published protocols (11, 12). After heme incorporation and dialysis proteins were further purified using DEAE-Sephacel anion exchange chromatography. Fractions were collected, pooled, and concentrated by ultrafiltration using YM30 membrane and the protein was stored in 50 mM potassium phosphate, pH 6.0 after extensive dialysis against the same buffer. Protein concentrations were determined spectrophotometrically using an extinction coefficient ε408 = 96 mM−1 cm−1.
Steady-State Activity Assays
Spectrophotometric assays were carried out at room temperature using a Cary 3E UV-visible spectrophotometer. Steady-state activity of wild-type and mutant proteins was measured using the native substrate of CCP, yeast cyt. c (ferrocytochrome c), as well as L-ascorbic acid. The steady state oxidation of cytochrome c was measured at room temperature using a Δε550 = 19.6 mM−1 cm−1 using 25 µM sodium dithionite reduced cyt. c, 100 mM Tris-phosphate pH 6.0, and 100 µM H2O2. Peroxidation of L-ascorbic acid was measured by following the decrease in optical density at 290 nm using a ε290 = 2.8 mM−1 cm−1 in 50 mM potassium phosphate pH 7.5, 250 µM H2O2, and varying concentrations of ascorbate. Hydrogen peroxide concentrations were standardized with KMnO4 using the method of Fowler and Bright (13).
Transient-State Kinetic Studies
The rate of compound I formation and decay was determined using an Applied Photophysics SX.18MV-R stopped-flow spectrophotometer at room temperature. Wild-type and mutant proteins at a concentration of 6 µM were mixed with H2O2 at concentrations ranging from 6 to 20 µM. The transient-state reaction was examined using a diode array attached to the SX.18MV-R stopped-flow spectrophotometer and was used to determine the maximal change in absorbance for each of the proteins. Buffers used were 50 mM potassium phosphate, pH 7.0 with 0.1 mM EDTA for all proteins except CCP2APX/F191 for which in addition to the previous buffer, 100 mM sodium citrate buffer, pH 5.0 was also used. The formation of compound I was examined for 20 ms and data were fit to a single-exponential curve using Applied Photophysics software. In APX and the mutant where Trp 191 is converted to Phe, there is an initial rapid decrease in absorbance owing to formation of a porphyrin π-cation radical followed by a slower increase in optical density owing to the spontaneous reduction of the porphyrin radical. The kinetics of compound I decay was estimated from the rate of increase in absorbance after the initial formation of the compound I porphyrin π-cation radical.
Electron Paramagnetic Resonance (EPR) Spectroscopy
Quartz EPR tubes (715-PQ-250m) were purchased from WILMAD. EPR spectra were recorded on a Bruker ESP300 spectrophotometer equipped with an Air Products LTR3 liquid helium cryostat. To observe the radical in compound I, 200 µL of 300 µM wild-type or mutant CCP protein was combined with an equal volume of 360 µM H2O2 in 50 mM potassium phosphate, pH 6.5. The solution was mixed and transferred to a quartz EPR tube and flash-frozen in a nhexanes-LN2(1) slurry, a process which took between 18 and 27 s. The EPR spectra were obtained at 10 K as an average of 10 scans using the following instrument parameters: microwave frequency, 9.387 GHz; modulation amplitude, 0.40 G; modulation frequency, 100 kHz; field sweep rate, 23.84 G/s; 0.638mW microwave power, 1.0 ×104 receiver gain, and 5.120ms time constant.
X-ray Crystallography and Structure Refinement
CCP2APX failed to produce decent quality diffracting crystals, while crystals CCP2APX/F191 and CCP/R184 were of high quality. Only peak fractions obtained during purification were used for crystallization. Crystallization conditions were optimized starting with conditions established for wild-type CCP. Sitting-drop vapor-diffusion with a well solution of 30% 2-methyl-2,4-pentanediol (MPD) in 50 mM Tris-phosphate, pH 6.0 was used initially. The best crystals grew in drops consisting of 400 µM protein, 22% MPD, and 50 mM Trisphosphate, pH 6.0 at 4°C. The following day, one round of touch seeding using wild type CCP crystals was done to initiate crystal growth. Immediately after crystal growth mutant crystals were flash-frozen in liquid nitrogen and stored for data collection later.
High-resolution data collection for CCP2APX/F191 and near atomic resolution data for CCP/R184 was collected at SSRL beamline 9-1. Data were reduced using HKL2000 and SCALEPACK (14). CCP2APX/F191 and CCP/R184 crystals were isomorphous with WTCCP crystals and belong to space group P212121. As a result, the structure of WTCCP was used as the starting model for refinement in CNS version 1.1 (15) after molecular replacement. Rigid body refinement was followed by slow-cool simulated annealing starting at 3000 K as implemented in CNS version 1.1 (15) and the remaining cycles that followed consisted of a few cycles of conjugate gradient minimization and water picking. The program O (16) was used for further adjustment and modeling of protein atoms, ligands, and water molecules. The final refinement was carried out using SHELXL (17) and anisotropic B-factors were used for main chain atoms. Refinement of the 1.06 Å structure of the CCP/R184 mutant followed a similar protocol. Data collection and refinement statistics are summarized in Table 1.
Table 1.
Crystallographic Data Collection and Refinement Statistics
| CCP2APX/F191 | CCP/R184 | |
|---|---|---|
| PDB Code | 3E2N | 3E2O |
| Radiation source | SSRL BL9-1 | SSRL BL9-1 |
| Wavelength (Å) | 1.08 | 1.08 |
| Space group | P212121 | P212121 |
| Cell Parameters (Å) | a = 106.78, b = 74.49, c = 50.95 | a = 107.34, b = 75.92, c = 51.22 |
| Resolution range (Å) | 100-1.30 | 100-1.02 |
| Total observations | 481,666 | 731,218 |
| 1Rsymm | 0.05 (0.55) | 0.08(0.39) |
| Unique reflections | 96,961 | 194,391 |
| Completeness (%) | 96.3 (93.1) | 93.3 (94.0) |
| Resolution range for refinement (Å) |
100-1.3 | 10.0-1.06 |
| Reflections used for refinement |
92,280 | 163,883 |
| Mean I/σ (highest resolution) | 32.2 (2.5) | 35.0 (2.3) |
| Rcryst/Rfree | 17.6/22.0 | 16.3/18.8 |
Rsym=∑h ∑i |<I(H)>−I(h)i|/∑h ∑iI(h)I, where I(h) is the intensity of reflection h, ∑h is the sum over all reflections and ∑i is the sum over all I measurements of reflection h.
Molecular Dynamics
Molecular dynamics simulations were carried out with Amber 9.0. Charges and optimal geometry for ascorbate were obtained using the antechamber routine in Amber and AM1-bcc charges with ascorbate assigned a net charge of −1.0. Heme parameters were provided by Dr. Dan Harris (18). The APX-ascorbate structure used was taken from the known crystal structure (9). Ascorbate was modeled into the crystal structure of engineered version of CCP, CCP2APX/F191, assuming that ascorbate binds the same way as in APX. Since we were focusing only on the substrate binding site, a full periodic boundary simulation was not carried out. Instead a sphere of TIP3 water molecules was placed within a 20 Å sphere of the ascorbate, which is sufficient to properly model the interaction of ascorbate and nearby protein groups with solvent. Prior to MD simulations, structures were energy minimized for 1,000 cycles with all H atoms and water molecules allowed to move. Next, all atoms except the ascorbate were allowed to move for 1,000 cycles followed by a final 2,000 cycles of minimization with all atoms allowed to move. For MD runs, atoms greater than 18 Å from the ascorbate were constrained to the starting position using a force constant of 2 kcal/Å. This was done to avoid "boiling off" of solvent since a periodic boundary was not used. MD Simulations were run for 10 ns with structures saved every 10 ps.
Results
Mutant Design
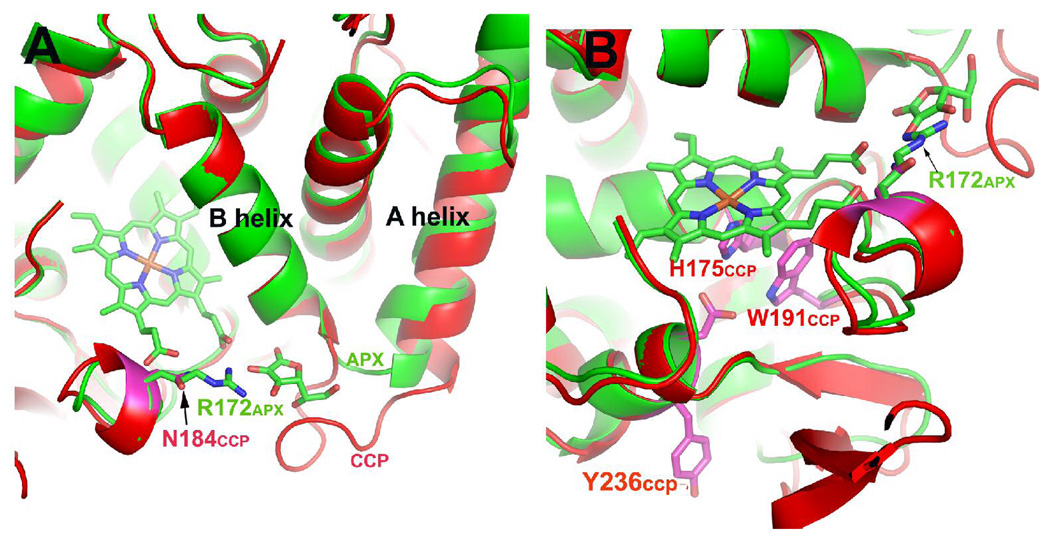
Fig. 1 shows a comparison between CCP (red) and APX-ascorbate complex (green) in the vicinity of the ascorbate binding site identified in the crystal structure of the APX-ascorbate complex (9). A loop on the surface between the A and B helices provides the ascorbate binding pocket. In CCP this loop is 7 amino acids longer than in APX. Based on structural superposition of WTCCP and APX as shown in Fig.1 and after subsequent sequence analysis the ascorbate binding site was introduced into WTCCP. This was achieved by replacing wild type residues 30–42 (LREDDEYDNYIGY) with residues 27–32 (IAEKKC) from APX. In the APX-ascorbate complex, Arg 172 (Fig. 1) H-bonds with the substrate and is known to be critical for activity in APX (8), whereas the corresponding residue in CCP is Asn 184, and thus was replaced with Arg.
Fig. 1.
WTCCP (red) superimposed on APX (green). Panels A and B are two different views.
Crystal Structures and Molecular Dynamics
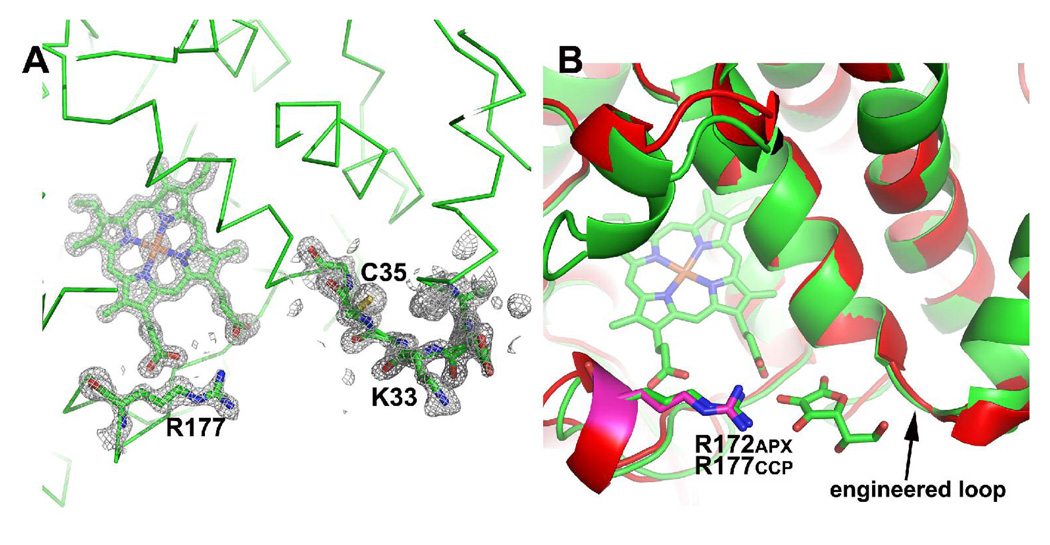
Crystals of CCP2APX/F191 diffracted to 1.3 Å resolution which provided sufficient data to justify the use of anisotropic B-factor refinement for main chain atoms. The fit of the engineered ascorbate binding site to the electron density map including Arg 184 is shown in Fig. 2A. The engineered ascorbate binding loop superimposes well on APX (Fig. 2B).
Fig. 2.
The engineered ascorbate binding loop in CCP2APX. (A) 2Fo−Fc electron density map contoured at 1.0 σ. Note that the engineered loop centered on Lys33 and Arg 172 which is Asn in WTCCP are very well ordered. (B) A superimposition of CCP2APX/F191 (red) on the APX-ascorbate complex (green). The key components of the ascorbate binding site are the same in both enzymes.
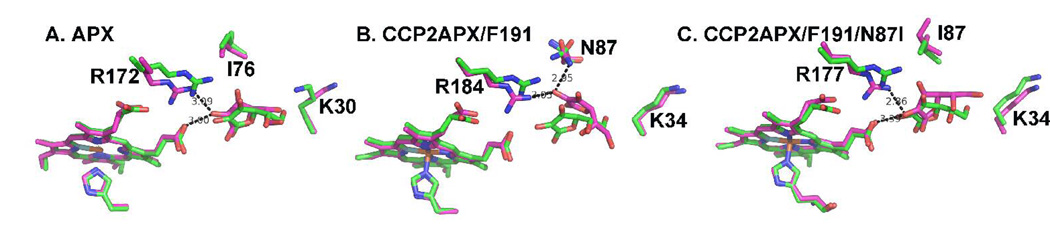
We made several unsuccessful attempts to obtain crystals of the CCP2APX/F191-ascorbate complex. Soaking crystals in ascorbate resulted in crystal cracking which could be due to ascorbate induced structural changes incompatible with crystal lattice. Alternatively, the increase in ionic strength upon addition of sodium ascorbate could be a problem since crystals are grown at low ionic strength. We also attempted to co-crystallize the CCP2APX-ascorbate complex but this, too, failed to generate useful crystals. We therefore used MD simulations to obtain some insight on the structure, stability, and energetics of the engineered ascorbate binding site. The results of MD simulations are summarized in Fig. 3 and Table 2. Structures shown in Fig. 3 were averaged over the last 5 ns of the 10 ns simulation followed by energy minimization. As shown in Fig. 3A and Table 2, the H-bonding interaction between ascorbate, Arg 172, and heme in APX remain intact during the simulation. However, the H-bond between Lys 30 and ascorbate found in the crystal structure of the APX-ascorbate complex (9) breaks early on in the simulation resulting in Lys 30 forming a stable ion pair with Glu 29 indicating that Lys 30 does not play an important role in ascorbate binding. This agrees well with the experimental data since replacing Lys 30 with Ala actually increases kcat (19). In sharp contrast Arg 172 in APX is essential for catalysis (8, 19) and the Arg 172-ascorbate interaction remains intact over the course of the 10 ns simulation (Fig. 3A and Table 2). For CCP2APX/F191 structure the ascorbate rotates up toward Asn 87 (Ile 76 in APX) in order to form a new H-bond with Asn 87 (Fig. 3B). This movement requires breaking of the ascorbate-heme H-bond. In addition the rms fluctuation of ascorbate is higher in CCP2APX/F191 than APX (Table 2). The main difference in the substrate binding pocket and the reason ascorbate moves so much in CCP2APX/F191 is that Asn 80 (Asn 87 in WTCCP numbering) is Ile 76 in APX. In APX Ile 76 lies on top of the ascorbate to assist the ascorbate to remain in position for H-bonding with Arg 172. We therefore carried out an in silico mutagenesis experiment and converted Asn 87 to Ile in CCP2APX/F191 followed by a 10 ns simulation. As expected CCP2APX/F191/N87I more closely resembled APX than CCP2APX/F191. As shown in Fig. 3C and Table 2 ascorbate remains in position for H-bonding with Arg and heme and the rms fluctuation is slightly lower than in CCP2APX/F191 (Table 2). A priori one might conclude that the overall ΔG of binding is about the same for all 3 structures since all 3 have 2 H-bonds between the substrate and neighboring groups. The main difference is the more extensive van der Waals contacts between Ile and ascorbate in APX and CCP2APX/F191/N87I.
Fig. 3.
Average MD structures (magenta) superimposed on original structures (green) for (A) APX, (B) CCP2APX and (C) CCP2APX with Asn 87 converted to Ile. Note that in APX (panel A) the ascorbate remains stable throughout the MD simulations while in CCP2APX (panel B) the ascorbate moves up toward Asn 87 thus allowing the ascorbate and Asn80 to form an H-bond. The movement of the substrate results in the loss of the heme-ascorbate H-bond. The in silico conversion of Asn 87 to Ile in CCP2APX results in the ascorbate maintaining its interactions with Arg 184 and the heme.
Table 2.
Distances and standard deviations of key H-bonding distances averaged over the 10 ns MD simulation. The fluctuation refers to the average rms deviation of the entire ascorbate molecule.
| Protein | Arg-ascorbate distance (Å) | Heme-ascorbate distance (Å) | Asn80-ascorbate distance (Å) | Fluctuation (Å) |
|---|---|---|---|---|
| APX | 3.19±0.21 | 3.19±0.36 | - | 0.61 |
| CCP2APX/F191 | 3.33±0.35 | 5.64±0.31 | 3.36±0.32 | 1.02 |
| CCP2APX/F191/N87I | 3.0±0.19 | 3.47±0.42 | - | 0.90 |
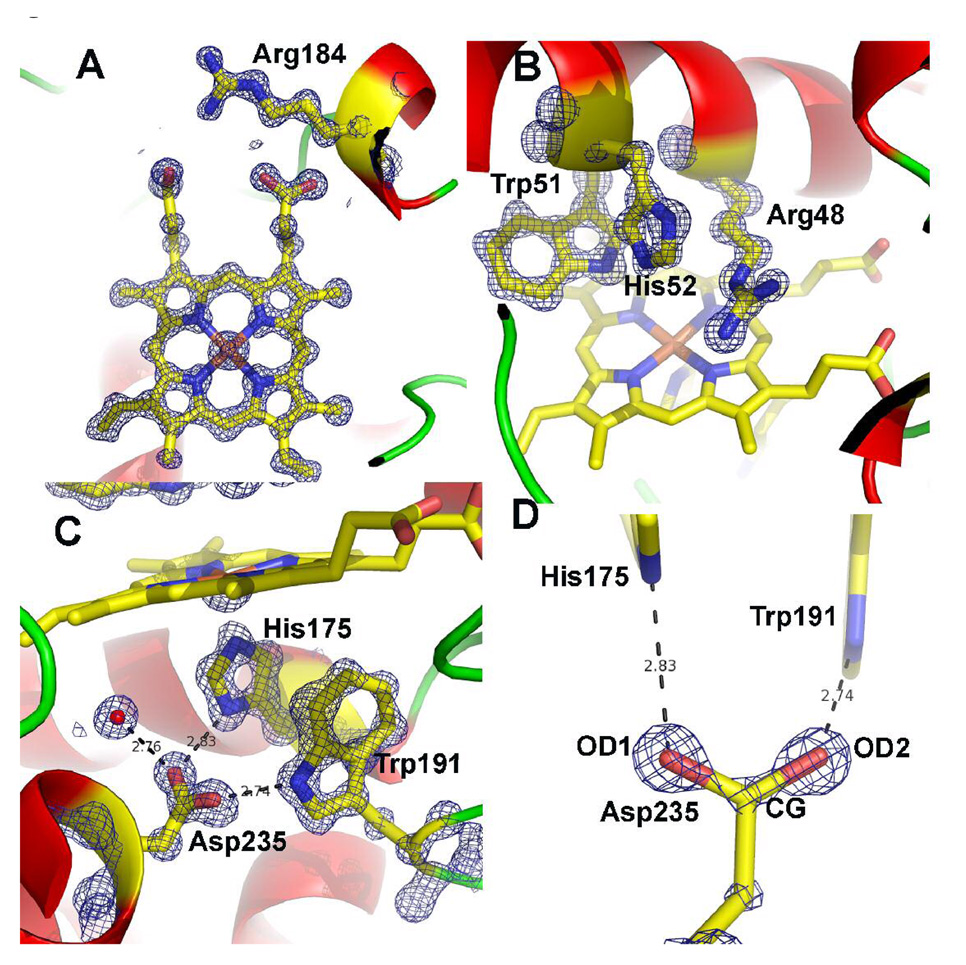
We also solved two other structures: the first with just the ascorbate-binding loop introduced into CCP and the second with just Asn 184 converted to Arg which we call CCP/R184. With respect to engineering CCP into an APX neither structure provides any further insights than CCP2APX/F191 structure. However, crystals of CCP/R184 provided an unexpected benefit of diffracting to atomic resolution which enabled the structure to be refined to a nominal resolution of 1.02 Å (Table 1). Applying a I/σI>2.0 cutoff, the resolution would decrease to 1.06 Å. Although outside the scope of the present paper, it is worth a slight diversion to describe some interesting insights provided by the CCP/R184 structure. Fig. 4 shows the 2Fo−Fc maps in the active site region. The main change induced by the N184R mutation is in Arg 48, a critical active site residue. In WTCCP Arg 48 occupies multiple positions (20) but in CCP/R184 Arg 48 occupies only one position. Both Arg 48 and the mutant Arg 184 side chains interact with the same heme propionate via a network of H-bonded solvent molecules. This additional ordering perhaps accounts for the decrease in flexibility of Arg 48. Such subtle changes apparently have little effect on enzyme activity since the CCP/R184 mutant exhibits about 50% of WTCCP activity.
Fig. 4.
1.06 Å 2Fo−Fc maps for the CCP/R184 mutant contoured at 2.5 σ in panels A, B, and C and 5.0 σ in panel D. A) The region around the mutant side chain Arg 184. B) The distal pocket showing the conserved residues Arg 48 and His 52 that are the catalytic groups responsible for heterolytic cleavage of the peroxide O–O bond and formation of compound I. C) The proximal binding pocket showing the conserved His175 ligand and its H-bonding partner Asp235. Trp 191 is the site of free radical formation in compound I. D). The 2Fo−Fc map contoured at 5.0 σ after 10 rounds of refinement with no angle or distance restraints applied to Asp 235. Note the continuous density along the CG-OD2 bond suggesting a double bond while the weaker connectivity between CG and OD1 indicates a single bond.
One last feature to be highlighted is the level of detail provided for key active site H-bonding residues. Asp 235 (Fig. 4C) is an invariant residue that H-bonds with the His 175 heme ligand. At atomic resolution it is possible to discern the difference between C-O single and double bonds in well ordered carboxylate side chains. To obtain an objective picture on the precise bond distances in Asp 235, 10 cycles of SHELXL refinement were carried out with no distance or angle restraints imposed on Asp 235. The resulting 2Fo−Fc electron density map is shown in Fig. 4D. Note that the electron density contoured at 5σ is continuous along the CG-OD2 bond but broken along the CG-OD1 bond. In addition, the CG-OD2 unrestrained bond distance is 1.228 Å compared to 1.290 Å for the CG-OD1 bond. In order to assess the significance of this difference we carried out a round of full matrix least squares refinement. Standard uncertainties on bond lengths can be calculated by inversion of the normal matrix using SHELXL. All restraints on positional parameters were excluded for the calculation. The matrix included all of the positional parameters and none of the thermal parameters. The resulting bond distances and standard deviations are CG-OD1 1.290±0.017 Å and CG-OD2 1.228±0.014 Å. Thus the difference in bond lengths is more than 3 standard deviations above the error and hence is significant. Both the map and distances indicate that the CG-OD1 bond has less double bond character with more electron density localized on OD1 than OD2. Therefore, OD1 should be a particularly strong H-bond acceptor from His175. Indeed, the H-bond angle between Asp235 and His175 is a near ideal 119° while the H-bond angle between Asp 235 and Trp 191 is 127°. The strong His 175-Asp 235 H-bond found in many heme peroxidase structures is generally consideredto be an important factor in the low heme redox potential of peroxidases compared to other heme proteins with His ligands such as the globins. However, there is nothing unusual about the Asp-His H-bond other than ideal geometry and distance which thus precludes an unprotonated His ligand or low barrier H-bond.
Spectroscopy
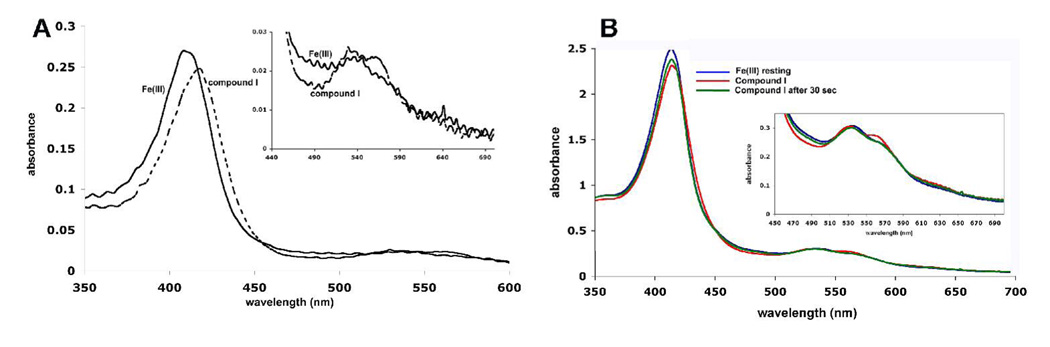
Fig. 5a shows the absorption spectrum of CCP2APX before, immediately after, and one hour after the addition of one equivalent of H2O2. Because the Trp 191 radical in compound I does not contribute significantly to the absorption spectrum, the spectrum of compounds I and II are very similar and thus are characteristic of the Fe4+=O center. As with WTCCP, the compound I Soret band red shifts relative to the Fe3+ resting enzyme with new bands at 530 nm and near 560 nm. The spectrum is quite stable and slowly relaxes back toward the Fe3+ spectrum. However, there is a decrease in absorption of the main Soret band immediately after addition of peroxide and remains below the starting level. This indicates that a small fraction of heme may be destroyed during the decay of compounds I and II. Fig. 5b shows the EPR spectrum of CCP2APX immediately after the addition of peroxide. The compound I signature of WTCCP is typically axially symmetric with a broad envelope due to a Trp 191 π-cation radical, which is in magnetic exchange with S = 1.0 oxyferryl heme iron center with a g = 2.01–2.04. The spectrum for CCP2APX is very similar to WTCCP indicating that engineering the ascorbate-binding loop into CCP does not effect formation of Trp 191 cation radical. Taken together the EPR and UV-vis data indicate that compounds I and II of CCP2APX are long lived with very similar spectral properties of WTCCP although there does appear to be a small amount of heme lost during the decay of compounds I and II.
Fig. 5.
A) UV-vis spectra of CCP2APX Fe3+ resting state (blue), immediately after the addition of 1 equivalent of H2O2 to give compound I (red), and one hour after the addition of H2O2 (green). B) EPR spectrum of CCP2APX immediately after the addition of 1 equivalent of H2O2.
Stopped Flow Studies
As shown in Table 3 the rate of compound I formation in the mutants is very similar to wild type CCP. With CCP2APX/F191 we expected the initial reaction with H2O2 to generate a porphyrin π-cation radical. In order to capture this reaction intermediate we employed diode array stopped flow. As shown in Fig. 6a the initial product formed 6 ms after mixing with H2O2 exhibits decreased absorption in the Soret band and a red shift in the maximum to longer wavelengths. In addition, there are distinct changes in the 500–580 nm region. These changes are very similar to what was observed for the porphyrin π-cation radical in the W191F CCP mutant (21) but with clear differences. With CCP containing the W191F mutation the first spectrum captured immediately after mixing with peroxide exhibits a decrease in the Soret maximum of about 35% (taken from Fig. 4 in ref. (21)) while CCP2APX/F191 exhibits only a 10% decrease. This together with the less well defined α and β bands in the 500–580 nm region indicates that CCP2APX/F191 forms a porphyrin π-cation radical to less of an extent than the single W191F mutant. It could be that one full peroxide oxidizing equivalent resides on the porphyrin immediately after mixing but rapidly migrates elsewhere. APX compound I spontaneously converts to compound II and the rate of compound I decay can be estimated from the compound I to II spectral changes (22). CCP2APX/F191 compound I decays about 160 fold faster than APX compound I (Table 3). Fig. 6b shows the spectrum of CCP2APX/F191 before, immediately after, and about 30 seconds after the addition of one equivalent of H2O2 which should generate the compound II spectrum. However, the spectrum closely resembles the starting Fe3+ enzyme with a diminished Soret maximum although there is the band near 560 nm is indicative of compound II. This indicates that CCP2APX/F191 forms a short-lived porphyrin radical which does not decay to a clearly defined and stable compound II species.
Table 3.
Rates of formation and decay of compound I as determined by rapid kinetic measurements. Experimental conditions are provided in Materials and Methods.
| Enzyme | Compound I Formation [M−1 s−1] | Spontaneous Decay of Compound I [s−1] |
|---|---|---|
| APX | 3.8 × 107 | 0.23 |
| wild type CCP | 1.25 × 108 | --- |
| CCP/W191F | 3.5 × 106 | 57 |
| CCP2APX/F191 | 5.4 × 106 | 37 |
| CCP2APX | 9.1 × 107 | --- |
Fig. 6.
A) Diode-array spectrum of CCP2APX/F191 before (labeled as Fe3+ and 6 ms after mixing (labeled as compound I) with H2O2. B) UV/Vis spectrum of CCP2APX/F191 before (blue), immediately after the addition of 1 equivalent of H2O2 (red), and about 30 seconds after the addition of H2O2 (green).
Steady State Activity
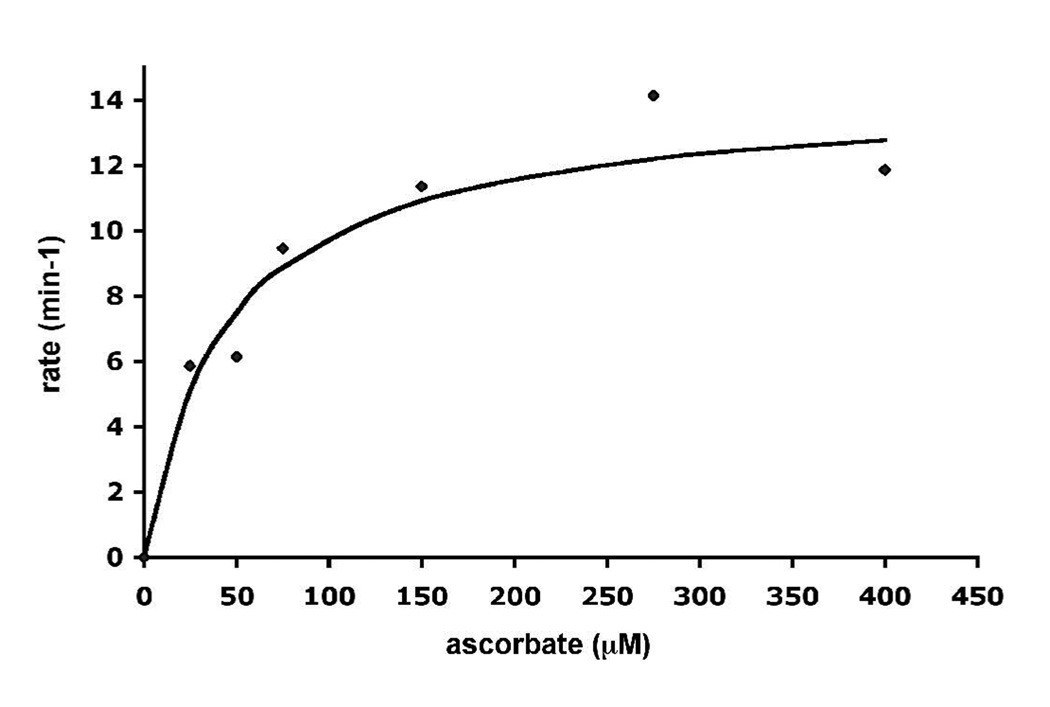
CCP2APX exhibits about ~ 2% WTCCP activity which is not too surprising since the loop engineered out in the mutant provides direct contact with cyt. c in the CCP-cyt. c complex (5). Under the assay conditions employed in this study WTCCP exhibits no ascorbate peroxidase activity. However, as shown in Fig. 7, CCP2APX exhibits reasonably good ascorbate peroxidase activity. From where the curve in Fig. 7 plateaus, we can estimate a kcat ≈ 12 min−1. CCP2APX/F191 exhibited activity as well but only about half the level of CCP2APX.
Fig. 7.
Ascorbate peroxidase activity of CCP2APX. 1.0 µM enzyme was added to a cuvette containing 250 µM H2O2 and varying concentrations of ascorbate. The rate was determined by the decrease in absorbance at 290 nm using a ε290 = 2.8 mM−1 cm−1.
The MD work suggested that the Ile in APX which replaces Asn 87 in CCP helps to hold the ascorbate in place for proper H-bonding with the heme propionate (Fig. 3). We therefore replaced Asn 87 with Ile in CCP2APX/F191 and found no improvement in enzyme activity.
Discussion
Our results show that it is relatively straight forward to engineer the ascorbate binding site found in APX into CCP. While it is true that CCP and APX are structurally very similar, it may seem somewhat surprising that the removal of a 7 residue surface loop in CCP required to form the ascorbate site that has very little effect on structure, stability, or the ability to form well ordered crystals. However, loop insertions or deletions between stable elements of secondary structure is a common strategy employed by nature to alter function. Loop swapping also has been successfully employed in the redesign of other metalloproteins (23). The fact that engineered ascorbate-binding site in CCP2APX/F191 is the same as in APX and the mutants exhibit ascorbate peroxidase activity strongly suggests that ascorbate is binding to the engineered site and is responsible for the observed ascorbate peroxidase activity in the mutants.
The rate of ascorbate peroxidation for CCP2APX is 12 min−1, while WTCCP exhibits no activity under the same experimental conditions clearly demonstrating that we were successful in engineering a new activity. Even so this value is well below the kcat of 40–100 sec−1 observed for plant APX. One possible reason is that the engineered site may bind ascorbate but not very well. The MD simulations showed that Ile 76 in APX helps to hold the substrate in place such that it H-bonds with both Arg 172 and one heme propionate. This provides a direct connection between the substrate and heme. The MD work also indicates that in CCP2APX/F191 the substrate fluctuates more and rotates up toward Asn 87 and no longer maintains a stable short contact with the heme propionate. Even so the substrate maintains a close interaction with the engineered Arg residue (Table 2) and it is doubtful that a slight reorientation of the substrate in CCP2APX compared to APX is responsible for the lower activity in CCP2APX. Moreover, replacing Asn 87 with Ile resulted in no further improvement in activity.
A second possibility is that in CCP2APX the radical site in compound I is stably located on Trp 191 while in APX the radical is located on porphyrin macrocycle. Thus the electron transfer distance is much longer in CCP2APX, about 9 Å, and does not follow a direct route to the radical site as in APX. However, a rough estimate of the expected electron transfer rate at a distance of 9.0Å can be made using the web-based electron transfer calculator (http://www.uphs.upenn.edu/biocbiop/local_pages/dutton_lab/golden.html). To make such an estimate the overall thermodynamic driving force of the reaction and reorganization energy must be known. The typical values used for reorganization energy are 07.-1.0V. The redox potential of the ascorbate/ascorbate radical couple is +282mV (24) while the Trp 191/Trp 191 radical couple is in the range of 1V (25). Although there are uncertainties in the actual redox potential of Trp 191 in the protein the overall driving force for the oxidation of ascorbate is large and in the range of 400–700mV. Even assuming that the driving force is 0V and large errors in reorganization energy, the rate of electron transfer over a distance of 9.0Å is orders of magnitude more than the observed turnover so it is doubtful that the distance between the ascorbate and Trp 191 radical is limiting. Even so anticipating this problem is the reason we also prepared the CCP2APX/F191 mutant since this mutant should form a porphyrin radical. However, based on the magnitude of spectral changes obtained in our diode array experiments, the extent of porphyrin radical formation is much less in CCP2APX/F191 compared to plant APX and the decay of porphyrin radical is about 160 times faster in CCP2APX/F191 than in APX. Thus CCP2APX/F191 forms neither a stable porphyrin radical, an amino acid radical, nor a stable compound II which can account for its lower ascorbate peroxidase activity in comparison to CCP2APX mutant.
A third possibility relates to the reactivity of compound II. The reduction of compound II is often the rate limiting step in peroxidases and thus is the obvious place to focus for improving activity. Here the efforts of Yeung et al. are highly relevant (26). In this work the manganese peroxidase Mn(II) site was introduced into CCP. The initial design effort resulted in a mutant CCP that oxidizes Mn(II) at 15 min−1 compared to 14,500 min−1 for authentic manganese peroxidase. In subsequent studies the activity was improved to about 240 min−1 by converting both Trp 191 and the distal pocket Trp 51 to Phe (27, 28). These mutations have nothing to do with improving Mn(II) binding but do affect the reactivity of compounds I and II. Pfister et al. (29) argued that the improved activity in the Trp 191Phe/Trp51Phe double mutant is due to an increased reactivity of compound II which is consistent with other studies (30). Trp 51 directly contacts the Fe-linked oxygen atom in compound I (20) so it is not surprising that the conversion of Trp 51 to Phe dramatically alters reactivity. Similar arguments could hold for our present work. The spectral data suggest that Fe4+=O center in CCP2APX is stable and thus may lack the reactivity required for fast ascorbate oxidation. Indeed, CCP compounds I and II are unusually stable among heme peroxidases. This could be biologically advantageous since more reactive compounds I and II would be subject to non-specific reduction by small molecule reductants. Thus CCP had to evolve a somewhat more elaborate electron transfer process to ensure only cyt. c is oxidized. One novel feature that helps to control selectivity in CCP is both electrons from cyt. c required to reduce compound I back to the resting Fe(III) enzyme must pass through Trp 191 (31). This means that after reduction of the Trp 191 radical in compound I, there is an internal electron transfer from Trp 191 to Fe4+=O to give Trp 191.+/Fe3+-OH and it is this species that accepts the second electron from cyt. c. The cyt. c heme directly contacts Ala 193 (5) in CCP and thus there is a direct polypeptide electron transfer path to Trp 191. The binding of cyt. c also could effect the energetics of Trp 191-to-Fe4+ internal electron transfer process and thus the energetics of compound II reduction. High resolution structures (20) show that the Ala193 section of polypeptide is disordered in resting Fe3+ CCP but becomes well ordered in compound I such that the Trp 191 cationic radical is more effectively stabilized by the surrounding protein. The crystal structure of the covalent CCP-cyt. c complex (32) shows that this region is well ordered in CCP indicating that the binding of cyt. c can affect the local Trp 191 environment and could promote the formation of compound II. None of these effects associated with cyt. c binding is possible in the CCP2APX mutant alone. As a result, the reactivity of compound II is too low to support rapid oxidation of ascorbate. This appears to be the most reasonable explanation and is consistent with previous studies (27).
A low compound II reactivity at first may seem puzzling since Fe4+=O is a powerful oxidant and there is no thermodynamic barrier to electron transfer from ascorbate. However, the reduction of Fe4+=O to Fe3+-OH is a proton coupled electron transfer process which can present a substantial kinetic barrier to electron transfer. Here again the internal electron transfer from Trp 191 to Fe4+=O is the key to understanding compound II reactivity since once Trp 191.+/Fe3+-OH is formed there is very little thermodynamic or kinetic barrier to reducing the Trp cation radical. The rate limiting and least understood step is the coupling of electron transfer from Trp 191 with protonation of Fe4+=O oxygen atom and how this process may be affected by cyt. c binding. The success in substantially increasing the Mn(II) peroxidation activity of engineered versions of CCP with Trp 51 converted to Phe could have directly affected the kinetic barrier to protonation of Fe4+=O resulting in increased Mn(II) peroxidation rates. There thus appears to be a path to further improving CCP2APX activity by making additional distal pocket mutants that increase the reactivity of compound II. In conclusion, it appears that engineering novel peroxidase activities is a two part problem. First, introducing the proper substrate binding site and second, altering the activity of enzyme oxidant, usually compound II, whose reduction is rate limiting. The first goal is relatively straight forward since loop swapping is sufficient to provide a novel binding pocket without altering the core structure. The second goal is more difficult since this is less of a structural problem and more of a chemical reactivity issue where the effects of mutagenesis are not so simple to interpret. Nevertheless, engineered peroxidases with altered activity may provide a window into understanding the more challenging problem of proton coupled electron transfer.
Acknowledgments
This work was supported by National Institutes of Health Grant GM42614.
Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Abbreviations
- CCP
cytochrome c peroxidase
- APX
ascorbate peroxidase
- WTCCP
wild-type CCP
- MD
molecular dynamics
- CCP2APX
CCP mutant engineered to bind ascorbate
- CCP2APX/F191
CCP2APX with Trp 191 converted to Phe
- EPR
electron paramagnetic resonance
- IPTG
isopropyl-β-D-thiogalactopyranoside
- MPD
2-methyl-2,4-pentanediol
- EDTA
ethylenediaminetetraacetic acid
- PDB
coordinates and structural factors have been deposited in the Protein Data Bank under accession numbers 3E2N and 3E2O.
References
- 1.Dolphin D, Forman A, Borg DC, Fajer J, Felton RH. Compounds I of catalase and horseradish peroxidase: π-cation radicals. Proc. Natl. Acad. Sci. USA. 1991;68:614–618. doi: 10.1073/pnas.68.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sivaraja M, Goodin DB, Smith M, Hoffman BM. Identification by ENDOR of Trp 191 as the free-radical site in cytochrome c peroxidase compound ES. Science. 1989;245:738–740. doi: 10.1126/science.2549632. [DOI] [PubMed] [Google Scholar]
- 3.Dunford HB. In: Horseradish peroxidase: structure and kinetic properties. Everse J, Everse KE, Grisham MB, editors. Boca Raton: CRC Press; 1991. pp. 1–24. [Google Scholar]
- 4.Henriksen A, Smith AT, Gajhede MY. The structures of the horseradish peroxidase C-ferulic acid complex and the ternary complex with cyanide suggest how peroxidases oxidize small phenolic substrates. J. Biol. Chem. 1999;274:35005–35011. doi: 10.1074/jbc.274.49.35005. [DOI] [PubMed] [Google Scholar]
- 5.Pelletier H, Kraut J. Crystal structure of a complex between electron transfer partners, cytochrome c peroxidase and cytochrome c. Science. 1992;258:1748–1755. doi: 10.1126/science.1334573. [DOI] [PubMed] [Google Scholar]
- 6.Sundaramoorthy M, Kishi K, Gold MH, Poulos TL. The crystal structure of manganese peroxidase from Phanerochaete chrysosporium at 2.06 Å resolution. J. Biol. Chem. 1994;269:32759–32767. [PubMed] [Google Scholar]
- 7.Mandelman D, Jamal J, Poulos TL. Identification of two electron transfer sites in ascorbate peroxidase. Biochemistry. 1998;37:17610–17617. doi: 10.1021/bi981958y. [DOI] [PubMed] [Google Scholar]
- 8.Bursey EH, Poulos TLY. Two substrate binding sites in ascorbate peroxidase: the role of arginine 172. Biochemistry. 2000;39:7374–7379. doi: 10.1021/bi000446s. [DOI] [PubMed] [Google Scholar]
- 9.Sharp KH, Mewies M, Moody PC, Raven EL. Crystal structure of the ascorbate peroxidase-ascorbate complex. Nat. Struc. Biol. 2003;10:303–307. doi: 10.1038/nsb913. [DOI] [PubMed] [Google Scholar]
- 10.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 11.Fishel LA, Villafranca JE, Mauro JM, Kraut J. Yeast cytochrome c peroxidase: mutagenesis and expression in Escherichia coli show tryptophan-51 is not the radical site in compound I. Biochemistry. 1987;27:351–360. doi: 10.1021/bi00376a004. [DOI] [PubMed] [Google Scholar]
- 12.Choudhury K, Sundaramoorthy M, Hickman A, Yonetani T, Woehl E, Dunn MF, Poulos TL. Role of the proximal ligand in peroxidase catalysis. Crystallographic, kinetic, and spectral studies of cytochrome c peroxidase proximal ligand mutants. J. Biol. Chem. 1994;269:20239–20249. [PubMed] [Google Scholar]
- 13.Fowler RM, Bright HA. J. Res. Natl. Bur. Standards. 1935;15:493–575. [Google Scholar]
- 14.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 16.Jones TA, Kjeldgaard M. Uppsala: Uppsala University Press; 1994. [Google Scholar]
- 17.Sheldrick GM, Schneider TR. SHELXL: high resolution refinement. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 18.Harris DL, Park JY, Gruenke L, Waskell L. Theoretical study of the ligand-CYP2B4 complexes: effect of structure on binding free energies and heme spin state. Proteins. 2004;15:895–914. doi: 10.1002/prot.20062. [DOI] [PubMed] [Google Scholar]
- 19.Macdonald IK, Badyal SK, Ghamsari L, Moody PCE, Raven EL. Interaction of ascorbate peroxidase with substrates: A mechanistic and structural analysis. Biochemistry. 2006;45:7808–7817. doi: 10.1021/bi0606849. [DOI] [PubMed] [Google Scholar]
- 20.Bonagura CA, Bhaskar B, Shimizu H, Li H, Sundaramoorthy M, McRee D, Goodin DB, Poulos TL. High resolution crystal structures and spectroscopy of native and Compound I cytochrome c peroxidase. Biochemistry. 2003;42:5600–5608. doi: 10.1021/bi034058c. [DOI] [PubMed] [Google Scholar]
- 21.Erman JE, Vitello LB, Mauro JM, Kraut J. Detection of an oxyferryl porphyrin π-cation-radical intermediate in the reaction between hydrogen peroxide and a mutant yeast cytochrome c peroxidase. Evidence for tryptophan-191 involvement in the radical site of compound I. Biochemistry. 1989;28:7992–7995. doi: 10.1021/bi00446a004. [DOI] [PubMed] [Google Scholar]
- 22.Patterson WR, Poulos TL, Goodin DB. Identification of a porphyrin π cation radical in ascorbate peroxidase compound I. Biochemistry. 1995;34:4342–4345. doi: 10.1021/bi00013a024. [DOI] [PubMed] [Google Scholar]
- 23.Lu Y. Biosynthetic inorganic chemistry. Angew. Chim. Intl. Ed. 2006;45:5588–5601. doi: 10.1002/anie.200600168. [DOI] [PubMed] [Google Scholar]
- 24.Buettner GR. The pecking order of free radicals and antioxidants: lipid peroxidation, α-tocopherol, and ascorbate. Arch. Biochem. Biophys. 1993;300:535–543. doi: 10.1006/abbi.1993.1074. [DOI] [PubMed] [Google Scholar]
- 25.DeFelippis MR, Murthy CP, Faraggi M, Klapper MH. Pulse radiolytic measurement of redox potentials: the tyrosine and tryptophan radicals. Biochemistry. 1989;28:4847–4853. doi: 10.1021/bi00437a049. [DOI] [PubMed] [Google Scholar]
- 26.Yeung BK, Wang X, Sigman JA, Petillo PA, Lu Y. Construction and characterization of a manganese-binding site in cytochrome c peroxidase: towards a novel manganese peroxidase. Chem. and Biol. 1997;4:215–221. doi: 10.1016/s1074-5521(97)90291-x. [DOI] [PubMed] [Google Scholar]
- 27.Gengenbach AJ, Syn S, Wang X, Lu Y. Redesign of cytochrome c peroxidase into a manganese peroxidase: Role of tryptophans in peroxidase activity. Biochemistry. 1999;38:11425–11432. doi: 10.1021/bi990666+. [DOI] [PubMed] [Google Scholar]
- 28.Pfister TD, Mirarefi AY, Gengenbach AJ, Zhao X, Danstrom C, Conatser N, Gao Y-G, Robinson H, Zukoski CF, Wang AH-J, Lu Y. Kinetic and crystallographic studies of a redesigned manganese-binding site in cytochrome c peroxidase. J. Inorg. Biol. Chem. 2007;12:126–137. doi: 10.1007/s00775-006-0171-0. [DOI] [PubMed] [Google Scholar]
- 29.Pfister TD, Gengenbach AJ, Syn S, Lu Y. The role of redox-active amino acids on compound I stability, substrate oxidation, and protein cross-linking in yeast cytochrome C peroxidase. Biochemistry. 2001;40:14942–14951. doi: 10.1021/bi011400h. [DOI] [PubMed] [Google Scholar]
- 30.Miller VP, DePillis GD, Ferrer JC, Mauk AG, Ortiz de Montellano PR. Monooxygenase activity of cytochrome c peroxidase. J. Biol. Chem. 1994;267:8936–8942. [PubMed] [Google Scholar]
- 31.Miller MA. A complete mechanism for steady-state oxidation of yeast cytochrome c by yeast cytochrome c peroxidase. Biochemistry. 1996;35:15791–15799. doi: 10.1021/bi961488c. [DOI] [PubMed] [Google Scholar]
- 32.Guo M, Bhaskar B, Li H, Barrows T, Poulos TLY. Crystal structure and characterization of a cytochrome c peroxidase-cytochrome c site specific cross-link. Proc. Natl. Acad. Sci. USA. 2004;101:5940–5945. doi: 10.1073/pnas.0306708101. [DOI] [PMC free article] [PubMed] [Google Scholar]