Abstract
It has been shown that activating killer Ig-like receptor (aKIR) genes are important for control of CMV reactivation after hematopoietic cell transplantation (HCT). To date, using the broad classification of KIR haplotypes A and B, the precise role of individual KIR genes in control of infection cannot be discerned. To address this, a consecutive case series of 211 non T-cell depleted HCT patients all at risk for CMV, were monitored bi-weekly for CMV DNA in plasma by Q-PCR and at intervals for CMV-specific T cell immunity. Comparing patients with CMV reactivation (n=152) to those with no reactivation (n=59), the presence of specific aKIR haplotypes in the donor, but not in the recipient, were associated with protection from CMV reactivation and control of peak plasma CMV DNA (p< 0.001). A donor aKIR profile, predictive for low risk of CMV reactivation, contained either aKIR2DS2 and aKIR2DS4 or had ≥ 5 aKIR genes. Neither donor nor recipient iKIR played a role in a protective effect. CD4+- and CD8+-specific CMV immunity did not explain reduced CMV infection. The initial control of CMV infection after HCT is managed by aKIR functions, and donor aKIR haplotypes deserve further evaluation in donor selection for optimized HCT outcome.
Introduction
Natural killer (NK) cells are considered a bridge between the innate and adaptive immune systems because of their ability to provide rapid cytotoxic function similar to that of innate immunocytes (e.g. macrophages and granulocytes) and because this cytotoxic function is closely aligned with that of T lymphocytes (e.g. use of interferon-γ, granzymes and perforins) [1]. It has been suggested that the NK cell provides a cytokine milieu that informs and supports the maturation of the adaptive immune system [2, 3]. NK cells destroy cells infected with viruses and other intracellular pathogens, transformed cells, and allogeneic cells, and this involves surface receptors, termed activating and inhibitory killer immunoglobulin-like receptors (KIR) [for review see [4-6].
The iKIR genes encode receptors which recognize defined specificities for HLA class I molecules and block further interaction of NK cell with “self” targets [1, 4, 7]. The incompatibility of the iKIR haplotype with its HLA class I ligand has been the subject of intense interest in influencing the outcome of allogeneic hematopoietic cell transplantation (HCT) with conflicting results [for review see [8]]. The KIR haplotype of donor and recipient can have beneficial or detrimental effects on rate of graft vs. host disease (GVHD), relapse, and survival after HCT [9-13]. In addition, iKIRs have been shown to be involved in improved time to resolution of hepatitis C virus infection [14, 15] and reduced susceptibility to HIV infection [16, 17].
In contrast, aKIR receptor—ligand interactions are poorly understood but appear to have an important effect on control of virus infection. It has been suggested that aKIR haplotypes are inversely associated with CMV reactivation in HCT recipients [18, 19]. In HCT with matched-sibling donors, the presence of more than one aKIR gene (haplotype B) was associated with reduced CMV reactivation [18]. Identity between donor and recipient aKIR genotypes has been reported to be associated with a lower incidence of CMV reactivation and lower transplant-related mortality [19]. The question of which donor aKIR genes are important in protection from CMV infection has not yet been answered.
The KIR haplotype A contains only one aKIR gene (aKIR2DS4), while the haplotype B is composed of both aKIR and iKIR genes [20]. This haplotype classification is limited in that most previous KIR genotyping methods could not distinguish aKIR2DS4 from its deletion mutant, a non-expressing membrane-bound receptor variant. More importantly, the use of the haplotype A and B system does not permit assessment of the role of particular aKIR receptors in control of CMV infection.
In this report, to assess the importance of KIR genotypes in influencing CMV reactivation, the KIR genotype was evaluated in a large series of HCT recipients and donor pairs, all of whom were at risk for CMV infection. A detailed KIR genotype containing both inhibitory and activating KIR was established, and patients were prospectively followed and analyzed for CMV infection and effects on CMV-specific adaptive immunity during 1 year post-transplant.
Patients and Methods
Study patients
A case series of 211 consecutive allogeneic HCT recipients, transplanted from 2001 to2006, were followed for CMV reactivation one year post-transplant. Based on study eligibility, all HCT recipients were at risk for CMV reactivation as determined by CMV antibody seropositivity in either the donor or recipient or both. All subjects, donors and recipients, signed an informed consent for research approved by the City of Hope (COH) Institutional Review Board. The donor/recipient KIR profile was assessed prospectively and haplotype A and B determined accordingly [20]. Patients were managed by the clinical staff without knowledge of the KIR profiles, and transplantation procedures were performed according to Foundation for the Accreditation of Cellular Therapy-approved guidelines. Table 1 describes the general demographics of the study participants in terms of median age, transplant type, disease diagnosis, myeloablative vs. non-myeloablative conditioning, pre-transplant CMV serology of donor and recipient, incidence of acute and chronic GVHD, and corticosteroid therapy for GVHD.
Table 1. Study cohort.
| total patients n=211 |
Group I * n=51 |
Group II * n=115 |
Group III * n= 45 |
P-value | |
|---|---|---|---|---|---|
| Patient Age: median (range) | 43.1(18.8, 68.8) | 44.8 (19.5, 67.6) | 42.2 (18.8, 64.8) | 41.9 (20.7, 68.8) | 0.27a |
| Donor Age: median (range) | 42.7 (15.2, 71.4) | 43.0 (18.4, 64.4) | 42.5 (19.0, 71.4) | 39.5 (15.2, 60.2) | 0.10a |
| Donor Type number (%) | |||||
| Sibling | 141 (67%) | 39 (76%) | 73 (63%) | 29 (64%) | 0.25a |
| URD | 70 (33%) | 12 (24%) | 42 (37%) | 16 (36%) | |
| Hematopoietic Progenitor Cell Source: number (%) | |||||
| Bone Marrow | 32 (15%) | 3 (6%) | 20 (17%) | 9 (20%) | 0.09a |
| Peripheral Blood | 179 (85%) | 48 (94%) | 95 (83%) | 36 (80%) | |
| Diagnosis: number (%) | |||||
| Lymphoid Malignancy | 82 (39%) | 18 (35%) | 45 (39%) | 19 (42%) | 0.22a |
| Myeloid Malignancy | 118 (56%) | 30 (59%) | 67 (58%) | 21 (47%) | |
| Other | 11 (5%) | 3 (6%) | 3 (3%) | 5 (11%) | |
| Conditioning Regimen: number (%) | |||||
| Myeloablative: | 134 (64%) | 30 (59%) | 75 (65%) | 29 (64%) | 0.76a |
| Nonmyeloablative: | 77(36%) | 21 (41%) | 40 (35%) | 16 (36%) | |
| CMV Serology: number | |||||
| unknown | 2 | 0 | 1 | 1 | 0.53a |
| D+/R+ | 133 (64%) | 33 (65%) | 72 (63%) | 28 (64%) | |
| D+/R- | 25 (12%) | 9 (17%) | 11 (10%) | 5 (11%) | |
| D-/R+ | 51 (24%) | 9 (17%) | 31 (27%) | 11 (25%) | |
| Acute GVHD : number (%) | |||||
| unknown | 1 | 0 | 1 | 0 | 0.97b |
| None | 58 (28%) | 14 (27%) | 31 (27%) | 13 (29%) | |
| Grade 1 | 24 (11) | 5 (10%) | 14 (12%) | 5 (11%) | |
| Grade 2 | 83 (39%) | 23 (45%) | 42 (37%) | 18 (40%) | |
| Grade 3 | 37 (18%) | 8 (16%) | 21 (19%) | 8 (18%) | |
| Grade 4 | 8 (4%) | 1 (2%) | 6 (5%) | 1 (2%) | |
| Chronic GVHD: number (%) | |||||
| unknown | 4 | 1 | 2 | 1 | 0.94b |
| None | 48 (23%) | 12 (24%) | 24 (21%) | 12 (27%) | |
| Yes: limited | 27 (13%) | 6 (12%) | 16 (14%) | 5 (11%) | |
| Extensive | 132 (64%) | 32(64%) | 73 (68%) | 27 (61%) | |
| Steroids | |||||
| none | 52 (25%) | 15 (29%) | 25 (21%) | 12 (27%) | 0.4b |
| 0<1mg/kg | 99 (47%) | 19 (37%) | 56 (49%) | 24 (53%) | |
| >1mg/kg | 60 (28%) | 17 (33%) | 34 (30%) | 9 (20%) | |
| Overall survival | 0.72 (0.57,0.82) | 0.77(0.68,0.84) | 0.84(0.70,0.92) | 0.2c | |
Group I: no 2DS2 –no 2DS4; Group II: 2DS2 or 2DS4; Group III: 2DS2 and 2DS4 or ≥ 5aKIR
Fisher's exact test/Kruskal-Wallis test
Pearson Chi-square test
Log-rank test
CMV Surveillance
CMV reactivation was monitored by DNA Q-PCR on plasma collected twice weekly up to 100 days post-HCT. CMV surveillance was continued at 1-2 week intervals in “high-risk” patients based on clinical management guidelines at City of Hope (COH). High-risk patients included those with persistent lymphopenia, grade 2-4 GVHD, or those requiring continued immunosuppression for anti-GVHD therapy. The Q-PCR was performed essentially as previously described using the CMV-gB DNA as amplification product [21] and had a limit of detection of 200 genome copies per milliliter plasma (gc/ml). In addition to Q-PCR testing on plasma, all patients had whole blood cultured for CMV infection on the same blood specimen and using a shell vial culture method previously described [22]. Preemptive ganciclovir therapy was implemented based on either one CMV positive shell vial culture or on two consecutive positive Q-PCR assays according to COH guidelines. The applied definition of CMV disease was that of Ljungman and Paya, [23] and all diagnoses were based on correlation of clinical events and documentation of CMV in either bronchoalveolar lavage (BAL) or in tissue biopsies by histology or by tissue culture. In this study, CMV infection was defined as evidence showing CMV in any of three tests, namely, blood culture shell vial, plasma Q-PCR or CMV disease by histology. Time to CMV reactivation was defined as the day of first CMV positive Q-PCR, and the CMV infection was measured quantitatively by plasma DNA gc/ml.
KIR genotyping
DNA extraction from donor and recipient WBCs was performed using Qiamp DNA Blood Mini Kit (Qiagen, Valencia, CA) as previously described [24]. DNA was quantified using a Genequant II assay (Pharmacia Biotech) and adjusted to15 ng/μl. A multiplex PCR-sequence-specific (PCR-SSP) was performed on DNA samples as previously described [24] using 28 primers in four multiplex PCR reactions for detection of 14 functional KIR genes. The thermal cycler was a GeneAmp 9600, and the DNA polymerase was Amplitaq Gold (Applied Biosystems, Foster City, CA). The assays were run with primer combinations, such that primer combination 1 included iKIR2DL1, iKIR2DL2, iKIR2DL4 and aKIR2DS3, primer combination 2 included iKIR2DL3, aKIR2DS2, iKIR3DL1and aKIR3DS1, primer combination 3 included iKIR2DL5, aKIR2DS1 and iKIR3DL2, and primer combination 4 included aKIR2DS4n, aKIR2DS4d and aKIR2DS5. This method identifies all 14 functional KIR genes including the deletion mutant 2DS4d, but not the pseudogenes. The amplified samples were run on a 3% agarose gel for visualization and analysis. Since KIR2DL4, 3DL2 and 3DL3 were detected in 100% of donors and recipients, these were not included in the analyses.
Intracellular Cytokine Assay
In addition to CMV surveillance, blood samples were analyzed for establishment of CMV- specific adaptive immunity by intracellular cytokine assay (ICC), using interferon-γ (IFN-γ) as the marker protein, at the following times post-HCT: day 40, 90, 120, 150, 180, and 360 plus or minus 2 weeks. ICC was performed on T cells as previously described [21] using Brefeldin A (25 μg/ml, Sigma, St. Louis, MO) to block the release of IFN-γ. After stimulation with either CMV Ag (Advanced Biotech, Paterson, NJ) (for CD4+ lymphocytes) and pp65 or IE1 peptides specific for HLA A*0201 or B*07 or Pepmix™ for non-HLA*A0201/or B*07 (JPT Peptide Technologies, Berlin, Germany) (for CD8+ lymphocytes), the cells were fixed, permeabilized then stained with anti-human CD4, CD8 and IFN-γ antibodies. FACS analyses were performed using a FACSCanto flow cytometer (Beckton-Dickinson Biosciences) and data were analyzed using FCS Express software (version3.0; Denovo Software).
Statistics
The GraphPad Prism® 4 software (GraphPad Software Inc., La Jolla CA) (www.graphpad.com) was used for the analysis of CMV DNA Q-PCR results. Other analyses were performed using the SAS® statistical package and programming language (SAS Institute Inc., Cary NC). Contingency tables of numbers of patients experiencing CMV reactivation after HCT were analyzed using Pearson's chi-square test, Fisher's exact test, and Mantel-Haenszel's test for ordered categories. The Kruskal-Wallis test was used as a non-parametric analog to oneway analysis of variance. Tests were two-sided and the cutoff for statistical significance was 0.05. Survival outcomes were calculated as time in day's post-HCT until event or last contact. Kaplan-Meier survival estimates were tested using the Mantel-Haenszel (log-rank) test. Logistic regression analyses were performed to identify independent risk factors of CMV reactivation. Cumulative incidence of first time to CMV-PCR positivity and peak-PCR among the 3 KIR groups were evaluated using the last assay date as censored events and relapse or death as competing risks.
Results
KIR genotype profile and CMV reactivation
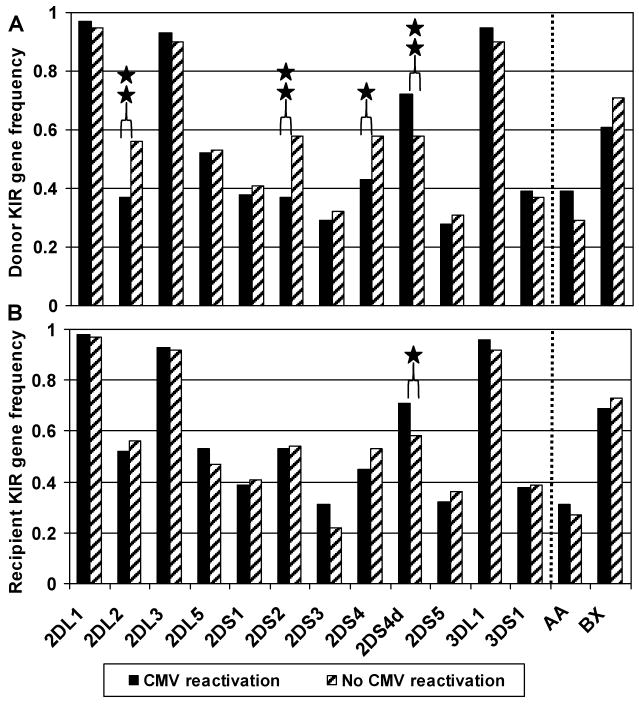
The KIR genotype profile for donors and recipients is shown in Figure 1. The donors (Figure 1A) and the recipients (Figure 1B) were dichotomized, according to the recipients' CMV reactivation status, into either a CMV reactivation group or No CMV reactivation group, and the gene frequency is shown for each group. The aKIR genotypes, aKIR2DS2 and aKIR2DS4, of donors (Figure 1A), but not of recipients (Figure 1B), were more frequent in the CMV-negative group. The donor aKIR2DS2 gene frequency was 37% in the CMV reactivation group vs. 58% in the No CMV group (p < 0.05 by Chi-square test). For the donor aKIR2DS4, the genotype frequency was 43% in the CMV reactivation group vs. 58% in the No CMV group (p = 0.06). Importantly, the aKIR2DS4 deletion mutant (aKIR2DS4d), which does not express membrane-bound aKIR2DS4, was found to be more prevalent in donors in the CMV reactivation group compared to the No CMV group (p≤ 0.05 by Chi-square test). This argues for the importance of donor aKIR2DS2 and aKIR2DS4 in control of CMV reactivation in the recipient. Of note, the iKIR genotype, 2DL2, was also significantly more frequent in the donors in the non-infected group, but 2DL2 iKIR and the aKIR2DS2 aKIR are in strong linkage disequilibrium and are expected to be expressed together. In contrast, the iKIR genes, 2DL1, 2DL3, 2DL5, 3DL1 of both the donors and the recipients distributed similarly between the two groups with and without CMV reactivation (see 2DL1, 2DL2, 2DL3, 2DL5, 3DL1 in Figure 1). Finally, the AA and BX haplotypes did not show any significant difference in proportions between the CMV reactivation group and the No CMV group (39% vs. 29% respectively for donor haplotype AA and 61% vs. 71% respectively for donor haplotype BX). In summary, in answer to the question of which KIR genotypes are most frequently associated with protection from CMV reactivation, donor aKIR2DS4, the absence of the aKIR2DS4 deletion, and the iKIR gene 2DL2 that is linked with aKIR2DS2 were most prevalent.
Figure 1. Donor and recipient KIR gene frequency in HCT recipients with and without CMV reactivation.
The cohort has been dichotomized according to the recipient's CMV reactivation status (■ n=152, having at least one incident of CMV reactivation; and  n=59, having no CMV reactivation). The KIR gene frequency is separately portrayed for donors (Panel A) and recipients (Panel B). aKIR2DS4d represents the deletion variant of aKIR2DS4. iKIR2DL4, iKIR3DL2 and iKIR3DL3 were detected in all members of this cohort, and thus omitted from this figure. AA indicates individuals homozygous for KIR haplotype A, and BX indicates individuals either heterozygous (BA) or homozygous (BB) for KIR haplotype B. * p= 0.06, ** p≤ 0.05 using the Chi-square test.
n=59, having no CMV reactivation). The KIR gene frequency is separately portrayed for donors (Panel A) and recipients (Panel B). aKIR2DS4d represents the deletion variant of aKIR2DS4. iKIR2DL4, iKIR3DL2 and iKIR3DL3 were detected in all members of this cohort, and thus omitted from this figure. AA indicates individuals homozygous for KIR haplotype A, and BX indicates individuals either heterozygous (BA) or homozygous (BB) for KIR haplotype B. * p= 0.06, ** p≤ 0.05 using the Chi-square test.
Donor aKIR profile and CMV reactivation incidence
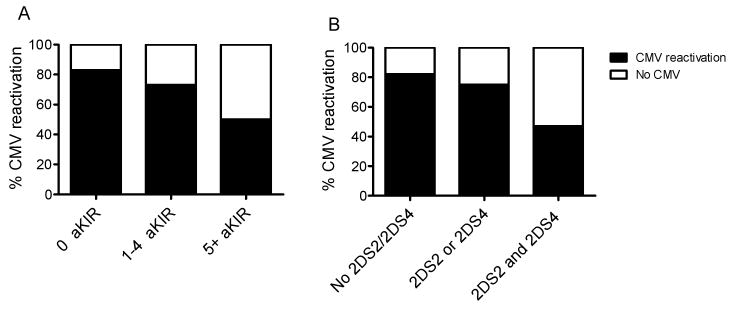
To further examine the contribution of donor aKIR genotype to CMV reactivation in the recipient, CMV reactivation was analyzed based on number of aKIR genes in the donor. As shown in Figure 2A, the prevalence of recipients with CMV reactivation was significantly higher if the number of aKIR genes per donor was less than 5. Only 17% of the recipients whose donors had 0 aKIRs were free of CMV infection post-HCT; 27% of those with 1-4 aKIRs in the donor genotype had no CMV infection, and 52% of those with ≥ 5 aKIRs never developed CMV reactivation (p=0.03 by Chi-square test). These results confirmed that the protection from CMV reactivation was associated with increased numbers of aKIR genes in the donor genotype.
Figure 2. CMV reactivation and aKIR profiles.
Panel A: Percent CMV reactivation vs. number of donor aKIR genes: 0 donor aKIR (n=30), 1-4 donor aKIR (n=161) and equal to 5 or more aKIR, (n=20). Chi –square T test is significant for independence p=0.034. Panel B: Percent CMV reactivation vs. type of donor aKIR combinations, including No aKIR2DS2 or aKIR2DS4 (n=57), either aKIR2DS2 or aKIR2DS4 (n=118), and both aKIR2DS2 and aKIR2DS4 (n=36). Chi-square T test is significant for independence p=0.001.
Because of the observation for CMV protection in the presence of the aKIR2DS2 and aKIR2DS4 genes in donors, the effect of single appearance or combination of these two genes was also evaluated (see Figure 2B). As shown, only 18% of the recipients having no aKIR2DS2 and aKIR2DS4 in their donor cells remained free of CMV infection; 25% of those with either one of these genes (aKIR2DS2 or aKIR2DS4) did not develop CMV infection; and 53% in those with both aKIR genes remained free of CMV infection. Thus, the presence of aKIR2DS2 and aKIR2DS4 was associated with decreased CMV reactivation (p=0.001 by Chi-square test).
Characterization of CMV infection based on aKIR donor genotype
This formed the basis for a hypothesis that aKIR2DS2 and aKIR2DS4 genes play the major role in protection from CMV reactivation. To test this hypothesis, we investigated CMV infection in recipients whose donors had both of these genes vs. those which did not. Because the data show that the number of donor aKIR (≥ 5) is protective of CMV reactivation as well as the presence in the donor of both 2DS2 and 2DS4 aKIRs, the donor/recipient pairs were grouped according to donor aKIR genotype as follows: Group I had no aKIR2DS2, aKIR2DS4, and <5 aKIR total genes; Group II had either aKIR2DS2 or aKIR2DS4 and <5 aKIR total genes; Group III had one of the possible combinations of aKIR2DS2 plus aKIR2SD4 and/or ≥5 aKIR genes. The CMV reactivation rate for each group is shown in Table 2 and the difference in CMV rates, with Group III having the lowest rate, was statistically significant by Chi-square analysis (p<0.001). In addition, the number of CMV episodes (as measured by Q-PCR) were significantly lower in Group III (p=0.001), with less frequency of both 1 CMV episode (27% in Group III vs. 37% in Group II or 43% in Group I) and of 2 CMV episodes (11% in Group III vs. 23% in Group II or 20% in Group I). So the conclusion can be made that Group III benefited from both a lower CMV reactivation rate and fewer recurrences of CMV infection.
Table 2. Donor KIR profile and CMV reactivation.
| Group I * n=51 |
Group II * n=115 |
Group III * n=45 |
p value | ||
|---|---|---|---|---|---|
| CMV reactivation | |||||
| yes | 44 (86%) | 86 (75%) | 22 (48%) | p<0.001a | |
| no | 7 (14%) | 29 (25%) | 23 (52%) | ||
| CMV episodes | |||||
| none | 13 (25%) | 41 (36%) | 28 (62%) | 0.001b | |
| 1 | 22 (43%) | 43 (37%) | 12 (27%) | ||
| 2 | 10 (20%) | 26 (23%) | 5 (11%) | ||
| 3 | 6 (12%) | 3 (3%) | 0 | ||
| >3 | 0 | 2 (1.6%) | 0 | ||
| CMV Infection: median time to first event in days (range) | |||||
| Q-PCRd | 37 (21-356) | 50 (22-153) | 44 (22-87) | 0.15c | |
| SV assayd | 47 (30-122) | 47 (21-259) | 47 (27-76) | 0.37c | |
| CMV Disease: number (%) | |||||
| CMV pneumonitis | 2 (3.9%) | 3 (2.6%) | 0 | 0.47a | |
| CMV gastroenterocolitis | 4 (7.8%) | 5 (4.3%) | 1(2.2%) | ||
Group I: no 2DS2 –no 2DS4; Group II: 2DS2 or 2DS4; Group III: 2DS2 and 2DS4 or ≥ 5aKIR
Chi-square test
Chi-square test on episodes 0-3
Kruskal-Wallis test
Q-PCR plasma CMV > 200gc/ml; SV: shell vial assay
Of note, as shown in Table 2, the median times to first CMV infection, as detected by Q-PCR or shell vial assay, was similar in all groups I, II and III (p=0.15 and 0.37, respectively, by Kruskal-Wallis test). For groups I, II, and III, the plasma Q-PCR became positive at a median time in days of 37, 50, and 44, respectively, and the shell vial cultures had a median first time positive of 47 days for all groups. Although the incidence of disease was not significantly different across groups due to the small numbers, most disease occurred in Groups I and II, with 5 cases of CMV pneumonitis. There was no CMV pneumonitis and only one case of CMV gastroenteritis in Group III (p=0.47 by Chi-square test). Therefore, it appeared that there was a difference in the percentage of recipients with CMV reactivations for the three groups but, if infection was to occur, the time to first reactivation was not different in Groups I, II, and III (Table 2).
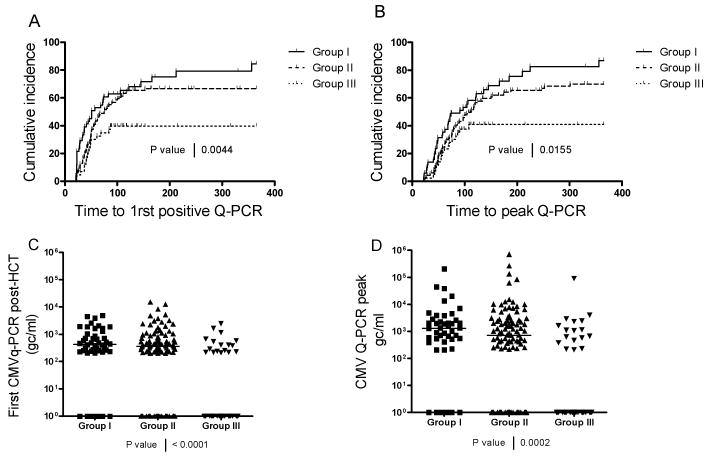
To analyze the kinetics of appearance of CMV reactivation in these three groups after HCT, a cumulative incidence curve was established plotting the first time to reach CMV Q-PCR positivity for each subject (Figure 3A). The subjects with no CMV infection were censored at the last day of follow-up, with relapse and death treated as competing risks. As shown, the occurrence of CMV infection was significantly less in Group III (p<0.001 by log-rank test). In addition, the occurrence of first infection went beyond day 100 post-HCT in Groups I and II (150-200 days post-HCT) whereas CMV reactivation no longer occurred by day 100 in Group III. In Group III, CMV reactivation appeared to be controlled, as detected by Q-PCR, in 53 % of the recipients as compared to only 14% and 25% of Groups I and II, respectively.
Figure 3. CMV reactivation profile for each group I, II and III.
Panel A: Time to first CMV positivity by Q-PCR is shown for subjects in each group (Log-Rank test p=0.004). Panel B: Time to peak Q-PCR is shown for subjects in each group (Log-Rank test p=0.015). Panel C: Amounts of CMV genome copies/ml are shown at time of first detected CMV reactivation (Kruskal-Wallis test p=0.0001). Panel D: Peak amounts of CMV genome copies/ml are shown (Kruskal-Wallis test p<0.0002).
To check whether the severity of CMV viremia was different in each group, the proportion reaching peak CMV DNA plasma level was analyzed using a similarly censored cumulative incidence plot which showed a pattern similar to time to first Q-PCR positivity (see Figure 3B, p<0.001 by log-rank test). To further characterize the nature of the infection in these 3 groups, the plasma CMV load was expressed as genome copies/ml plasma (gc/ml) for first CMV detection (Figure 3C) or for peak CMV load (Figure 3D) using a scatter plot that included the 0 gc/ml values for the patients who did not reactivate CMV. Here again, the median CMV loads at first detection and at peak level were lowest in group III (Figure 3C, p < 0.001, and Figure 3D, p<0.001 by Kruskal-Wallis tests). In summary, recipients of donors having aKIR haplotypes containing aKIR2DS2 and aKIR2DS4 or ≥5 aKIR genes had proportionately less CMV reactivation and earlier control of infection compared to those with only a single aKIR, aKIR2DS2 or aKIR2DS4, or less than 5 aKIR genes.
Comparison of clinical characteristics among the aKIR Groups
A comparison of the clinical characteristics among the aKIR groups was performed to ensure that no baseline differences existed among these factors (see Table 1). The comparison was performed using contingency tables, and the characteristics studied were: donor and recipient age, type of HCT, source of blood progenitor cells, underlying diagnosis, conditioning regimen, pre-HCT CMV antibody in donor and recipient, incidence of acute and chronic GVHD, and use of corticosteroid therapy. As shown in Table 1, the results of this evaluation show that there were no significant differences across the clinical characteristics among the aKIR groups. In addition, there were no differences in the groups with regard to overall survival as reported at three years post transplant.
Logistic regression analysis of factors affecting CMV reactivation
The univariable logistic regression analysis, as shown in Table 3, examined factors previously shown to impact CMV reactivation. Among the variables tested, the most important was the KIR Risk Groups, Group III being highly significant (p=0.0003) and other variables such as acute GVHD (p=0.006). In addition, the material transplanted, bone marrow vs. PBSCs (p=0.03), and donor KIR2DS2 (p=0.01) and donor KIR2DS4 (p=0.06, borderline) were also important independent variables in this model. All other variables such as age of donors or recipients, related vs. unrelated donor type, conditioning intensity, CMV serology of at-risk donor-recipient pairs, or prednisone dose were not associated with CMV reactivation. The study excluded donor-recipient pairs that were CMV seronegative pre-transplant, and therefore the baseline chosen for the univariate comparison of CMV serology was D+/R-. This may explain the lack of significant difference based on serostatus of donor and recipient. In addition, the prednisone dose was based on the highest dose given to a recipient but time to highest dose was not included in the model (overall p= 0.26).
Table 3. Univariable logistic regression affecting the percentage of CMV reactivation.
| Variables | N | Odds Ratio (95% CI) | P-value |
|---|---|---|---|
| Recipient Age at Transplant (continuous) | 210 | 1.00 (0.98, 1.03) | 0.85 |
| Donor Age at Transplant (continuous) | 210 | 1.01 (0.98, 1.04) | 0.58 |
| Patient Sex | |||
| Female | 99 | (baseline) | 0.68 |
| Male | 111 | 0.88 (0.48, 1.61) | |
| Donor Sex | |||
| Female | 83 | (baseline) | 0.33 |
| Male | 127 | 1.35 (0.73, 2.50) | |
| Donor Type | |||
| Sibling Donor | 140 | (baseline) | 0.83 |
| Matched Unrelated Donor | 70 | 0.93 (0.49, 1.76) | |
| Transplant source | |||
| Bone Marrow | 32 | (baseline) | 0.03 |
| Peripheral Blood Stem Cells (PBSCs) | 178 | 2.37 (1.09, 5.15) | |
| Conditioning intensity | |||
| Fully Ablative | 133 | (baseline) | 0.69 |
| Non-myeloablative Reduced Intensity | 77 | 1.14 (0.60, 2.15) | |
| Diagnosis | 0.98* | ||
| Lymphoid Malignancy | 81 | (baseline) | |
| Myeloid Malignancy | 118 | 1.07 (0.57, 2.00) | 0.84 |
| Other | 11 | 1.06 (0.26, 4.34) | 0.94 |
| CMV Serology | 0.59* | ||
| D+/R- | 25 | (baseline) | |
| D+/R+ | 132 | 1.56 (0.63, 3.85) | 0.34 |
| D-/R+ | 51 | 1.64 (0.59, 4.61) | 0.34 |
| Acute GVHD II+ (grade II-IV) | |||
| No | 77 | (baseline) | 0.006 |
| Yes | 122 | 2.47 (1.30, 4.69) | |
| Chronic GVHD | |||
| No | 41 | (baseline) | 0.42 |
| Yes | 165 | 1.36 (0.64, 2.86) | |
| Prednisone Dose | 0.26 | ||
| No Prednisone (0) | 41 | (baseline) | |
| Low Dose Prednisone (< 1) | 104 | 1.49 (0.69, 3.21) | 0.31 |
| High Dose Prednisone (>= 1) | 64 | 2.06 (0.86, 4.91) | 0.10 |
| Donor aKIR | 0.006* | ||
| No KIR2DS2 and no KIR 2DS4 | 57 | (baseline) | |
| KIR2DS4 and no KIR2DS2 | 64 | 0.70 (0.28, 1.70) | 0.43 |
| KIR2DS2 and no KIR 2DS4 | 53 | 0.59 (0.24, 1.48) | 0.26 |
| KIR2DS2 and KIR2DS4 | 35 | 0.20 (0.08,0.52) | 0.0009 |
| KIR Risk Groups | 0.0006* | ||
| I (No 2DS2 or 2DS4) | 51 | (baseline) | |
| II (2DS2 or 2DS4 or < 5 aKIR) | 115 | 0.47 (0.19, 1.16) | 0.10 |
| III (2DS2 and 2DS4 and /or ≥ 5aKIR) | 44 | 0.16 (0.06, 0.43) | 0.0003 |
P-value of the model.
The multivariable logistic regression analysis of factors affecting CMV reactivation, as a model of goodness-of-fit (p=0.001) included the donor KIR2DS2 genotype, donor KIR 2DS4 genotype, the combination donor KIR2DS2 and KIR2DS4 genotypes, acute GVHD, and the transplant source (Table 4). The individual donor aKIRs were included in this model, and for this reason the KIR groups I, II, and III, which duplicate the KIR variables, could not be added into the analysis. However, this model establishes the importance of donor aKIR2DS2 and aKIR2DS4 genotype as factors in decreasing the risk of CMV reactivation in recipients.
Table 4. Multivariable Logistic Regression affecting the percentage of CMV reactivation.
| Variables | N | Odds Ratio (95% CI) | P-value |
|---|---|---|---|
| Model Goodness-of-fit | 198 | 0.001* | |
| Donor KIR-2DS2 and KIR 2DS4 | |||
| No KIR2DS2 and no KIR2DS4 | 53 | (baseline) | |
| KIR2DS4 and no KIR2DS2 | 59 | 0.87 (0.32, 2.32) | 0.78 |
| KIR2DS2 and no KIR2DS4 | 52 | 0.59 (0.22, 1.55) | 0.28 |
| KIR2DS2 and KIR2DS4 | 34 | 0.21 (0.08, 0.58) | 0.003 |
| AGVHD II+ (grade II-IV) | |||
| No | 76 | (baseline) | 0.006 |
| Yes | 122 | 2.64 (1.33, 5.23) | |
| Transplant source | |||
| Bone Marrow | 28 | (baseline) | 0.06 |
| Peripheral Blood Stem Cells | 170 | 2.36 (0.97, 5.76) | |
Wald test
Adaptive immune response within donor KIR Groups
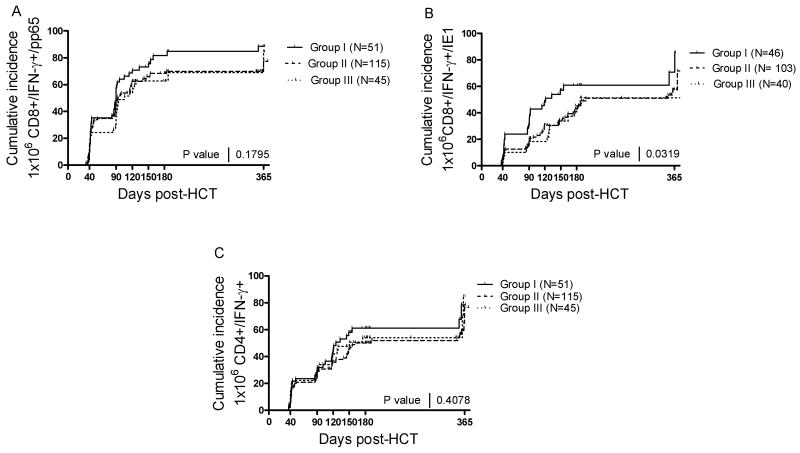
The next question was whether the influence of the donor aKIR genotype on CMV infection was associated with an enhanced development of CMV-specific T cell immunity post-HCT. A cumulative incidence plot was established for recipients monitored prospectively for adaptive immune response by intra-cellular cytokine assay (ICC) at day 40, 90, 120, 150, 180 and 365 days post-HCT. The immune responses were compared by arbitrarily defining the acquisition of immunity at 1×106 CD4+/IFN-γ+ or CD8+/IFN-γ+ cells/L when stimulated in vitro with CMV antigen (CD4+ cell assay) or a mixture of overlapping CMV-pp65 peptides or CMV-IE1 peptides (CD8+ cell assay). The HLA A*0201 and B*07 were stimulated with pp65 peptides specific to their alleles. As shown in Figure 4, Group I consistently developed immunity prior to groups II and III, possibly due to the increased infection rate in this group, but this was significantly different only for CD8 response to CMV-IE1 (Figure 4B, p=0.03). CD8+ cell reactivity to CMV pp65 (Figure 4A) and CD4+ cell immunity to CMV antigens (Figure 4C) was not significantly impacted by the donor aKIR haplotype. Interestingly, the cumulative incidence profile for immune response recovery in all 3 CMV assays were very similar for Group II and Group III (see Figure 4A-C), suggesting that the presence of donor aKIR2DS2 and aKIR2DS4 KIR genotype, or ≥ 5 donor aKIRs, affected the adaptive response in the same fashion.
Figure 4. Adaptive immune response in Group I, II, and III.
The adaptive immune response was analyzed at days 40, 90, 120, 150, 180, and 360 post-HCT for groups I, II, and III. Panel A: Time to first 1×106 CD8+/IFN-γ+ cells/L as determined by in vitro assay after stimulation with a CMV-pp65 peptide mixture or 1-2 peptides specific for HLA A*0201 and or B*07. Panel B: Time to first 1×106 CD8+/IFN-γ+ cells/L as determined in vitro after stimulation with a CMV-IE1 peptide mixture or 3 peptides specific for HLA A*0201 and or *B08. Panel C: Time to first 1×106 CD4+/IFN-γ+ cells/L as determined by in vitro assay after stimulation with a CMV antigen cell lysate.
Discussion
NK cells play a vital role in the innate immune response to viral infections. The mechanism(s) that control this important protective response are only beginning to be understood. Innate immunity against viral infections is mediated through NK cells, whose function is dependent in part upon KIR genes from the Lymphocyte Receptor Complex on chromosome 19. When NK cells have been implicated in HCT outcomes, e.g. survival, relapse, and GVHD, it has been the iKIR donor and recipient ligand—ligand compatibility that appeared most important [8, 10]. However, the effect of KIR haplotypes on outcome remains controversial with beneficial and detrimental effects observed depending on whether it was a related sibling transplant [13] or unrelated transplant [8, 25] whether it was T-cell depleted [10] or T-cell replete transplant [12], and other aspects such as myeloid diagnosis, use of anti-T cell globulin, and age of patient [8]. It is likely that the variability of the KIR repertoire, the significant gene polymorphisms, and the variable NK responses also contribute to these different outcomes [26, 27].
With the advent of methods for fine genetic characterization of donor and recipient [24], it is possible to determine the contributions of the various aKIR genes. Our data indicate that the aKIR genotype of the donor is important in limiting CMV reactivation and that iKIR, in either the donor or recipient, are not associated with reduction in CMV reactivation. This allowed the testing of a hypothesis that specific donor aKIR genotypes had prominence in control of CMV reactivation. Our data indicate that both aKIR2DS2 plus aKIR2DS4, in the donor genotype, are associated with reduced CMV activation in HCT recipients. In addition, among other aKIR genes, the same association was found regardless of which aKIR genes were present in the donor genotype, as long as there were at least 5 or more aKIR alleles present. A protective donor genotype was defined as that in which there are both aKIR2DS2 and aKIR2DS4 or ≥5 aKIR genes. The recipients of HCT from donors with the beneficial aKIR haplotype had significantly less CMV reactivation, had no reactivation after day 100, and had significantly lower median peak CMV load. More importantly, although not significantly different because of small numbers, CMV disease was minimal in this group and no CMV pneumonia occurred.
A complexity of interpretation is introduced when the broad haplotyping system is used that divides all patients in haplotypes A and B. Individuals genotyped positive for the so-called “framework genes”, plus only an aKIR2DS4 locus, are KIR haplotype A homozygotes. Those with additional aKIR loci are either AB or BB haplotypes and are grouped together as KIR haplotype B. Therefore, KIR haplotype B carriers potentially express multiple aKIRs. Yet, the classification of haplotype A and B does not define the full complement of aKIR and iKIR genes. Therefore, the contribution of individual genes to antiviral effects has not been precisely evaluated. Although this system has be useful in defining risk for GVHD and for survival in matched related HCT [13], it does not allow for evaluation of specific aKIR gene relationships.
The aKIR genes do not appear to play a role in altering GVHD or relapse after HCT, but they have been associated with decreased reactivation of CMV [18, 19]. In the study of Cook et al., it was reported that in T-cell replete HCT in matched sibling donor transplants, the presence of donor KIR haplotype B was associated with a 65% reduction in CMV reactivation [18]. Our results confirmed a small protective effect of haplotype B, but showed a major protective effect of donor aKIR genotype aKIR2DS2 and aKIR2DS4.
Chen et al. were the first to characterize the actual number of donor and recipient KIR genes relative to CMV infection, but they did not distinguish between the aKIR2DS4 and its inactive deletion aKIR2DS4d [19]. In their paper, HCT recipients of a T-cell replete transplant from HLA identical sibling donors having more aKIR genes were shown to have lower transplant related mortality, better survival, and a reduced incidence of CMV reactivation. Our data includes T-replete transplant recipients with unrelated donors and demonstrates that the protective effect of increased donor aKIR genotypes on CMV infection extends to this population.
It is important to note, however, that, although the CMV reactivation was significantly less, approximately half of the recipients with donors having the favorable donor aKIR genotype still reactivated CMV. CMV reactivation occurred in 48% of those in the protected group and, when this occurred, there was no difference in time to first infection or in median CMV plasma DNA levels. However, significantly fewer episodes of CMV infection occurred in this group. It is possible that there is variable expression of aKIR receptors, as has been shown with iKIR [27], and this could explain the break-through of CMV reactivation despite the donor aKIR genotype. KIR expression in mice has been shown to vary with promoter polymorphisms and epigenetic silencing events [28], and it is possible that similar variations in expression of aKIR will be linked to the control of CMV reactivation.
NK cells are not the only cells expressing KIR receptors, which are also expressed on CD8+ T-cells (NKT), and there is evidence that NK cells interact in support of adaptive immunity [2, 6]. Thus, the question was raised as to whether the aKIR contribution is mediated by an enhanced adaptive immune response in our patients. In the murine CMV (MCMV) model, the continuum of interaction between innate processes, involving dendritic cells and NKT cells, produces an initial INF-α response which appears to activate NK cells and leads to control of MCMV [29]. In this model, it has been proposed that the potential immunosuppressive effect of INF-α is relieved by the NK-induced reduction of MCMV infection, and this allows for expansion of MCMV-specific CD4 and CD8 lymphocytes [30]. Of note, in the mouse model, NK cells can also limit the expansion of the CD4/CD8 responses by unknown mechanisms [31]. In our study, there was no enhancement of the CD4+ or CD8+ lymphocyte response to CMV that would explain the reduced infection in the group with donor enhanced aKIR haplotype. Neither the CD4 responses to CMV antigens nor the CD8 responses to CMV pp65 peptides were different among the groups with varying donor aKIR genotypes. As part of a separate study, IFNγ+ CD8+ cells from this same patient cohort were further evaluated for the expression of simultaneous multiple immunologic functions using a CMV- pp65 peptide library for induction of MIP-1β, TNF-α, and CD107 degranulation. Similarly, there was no effect of aKIR donor genotype [W. Zhou, personal communication]. We did observe an association of the aKIR donor haplotype with significant reduction in CD8+ lymphocyte responses to CMV-IE1 (see Figures 3A and 3B). It is possible that this is due to the reduction in CMV infection by aKIR effects, but the actual mechanism is unknown. Of note, the fact that reduced CMV infection occurred in the absence of enhanced CD8-specific immunity, further supports the overall importance of NK cells, acting by means of the aKIR receptor—ligand, in initial control of CMV.
These data support the conclusion that both aKIR2DS2 plus aKIR2DS4, or a total of at least 5 aKIR in the donor KIR haplotype, are associated with reduced CMV infection in T-replete allogeneic HCT transplantation. Even in those with reactivation of CMV, the favorable donor haplotype appeared to exert a better control of the infection. These data better define the aKIR contribution to control of CMV in this population and suggest that donor aKIR genotype be included in algorithms for optimizing the outcome of HCT in related and unrelated T-replete allogeneic donor transplantation. Larger studies will be necessary to determine whether selective consideration of donor aKIR repertoire might be of benefit to the success of allogeneic transplants.
Acknowledgments
This work was supported by US Public Health Service grant no. RO1 AI58148 to JAZ; P01-CA30206 to S.J.F. and D.J.D.; R01-CA77544 to D.J.D.; and M01-RR00043-38 to the General Clinical Research Center at City of Hope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Gerosa F, Gobbi A, Zorzi P, et al. The reciprocal interaction of NK cells with plasmacytoid or myeloid dendritic cells profoundly affects innate resistance functions. J Immunol. 2005;174:727–734. doi: 10.4049/jimmunol.174.2.727. [DOI] [PubMed] [Google Scholar]
- 3.Moretta A, Marcenaro E, Parolini S, Ferlazzo G, Moretta L. NK cells at the interface between innate and adaptive immunity. Cell Death Differ. 2008;15:226–233. doi: 10.1038/sj.cdd.4402170. [DOI] [PubMed] [Google Scholar]
- 4.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 5.Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol. 2006;298:225–257. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- 6.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 8.Witt CS, Christiansen FT. The relevance of natural killer cell human leucocyte antigen epitopes and killer cell immunoglobulin-like receptors in bone marrow transplantation. Vox Sang. 2006;90:10–20. doi: 10.1111/j.1423-0410.2005.00712.x. [DOI] [PubMed] [Google Scholar]
- 9.Ruggeri L, Capanni M, Casucci M, et al. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94:333–339. [PubMed] [Google Scholar]
- 10.Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295:2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 11.Giebel S, Locatelli F, Lamparelli T, et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood. 2003;102:814–819. doi: 10.1182/blood-2003-01-0091. [DOI] [PubMed] [Google Scholar]
- 12.Sun JY, Dagis A, Gaidulis L, et al. Detrimental effect of natural killer cell alloreactivity in T-replete hematopoietic cell transplantation (HCT) for leukemia patients. Biol Blood Marrow Transplant. 2007;13:197–205. doi: 10.1016/j.bbmt.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 13.McQueen KL, Dorighi KM, Guethlein LA, Wong R, Sanjanwala B, Parham P. Donor-recipient combinations of group A and B KIR haplotypes and HLA class I ligand affect the outcome of HLA-matched, sibling donor hematopoietic cell transplantation. Hum Immunol. 2007;68:309–323. doi: 10.1016/j.humimm.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khakoo SI, Thio CL, Martin MP, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 15.Romero V, Azocar J, Zuniga J, et al. Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol. 2008;45:2429–2436. doi: 10.1016/j.molimm.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jennes W, Verheyden S, Demanet C, et al. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J Immunol. 2006;177:6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]
- 17.Alter G, Martin MP, Teigen N, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook M, Briggs D, Craddock C, et al. Donor KIR genotype has a major influence on the rate of cytomegalovirus reactivation following T-cell replete stem cell transplantation. Blood. 2006;107:1230–1232. doi: 10.1182/blood-2005-03-1039. [DOI] [PubMed] [Google Scholar]
- 19.Chen C, Busson M, Rocha V, et al. Activating KIR genes are associated with CMV reactivation and survival after non-T-cell depleted HLA-identical sibling bone marrow transplantation for malignant disorders. Bone Marrow Transplant. 2006;38:437–444. doi: 10.1038/sj.bmt.1705468. [DOI] [PubMed] [Google Scholar]
- 20.Uhrberg M, Valiante NM, Shum BP, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 21.Gallez-Hawkins G, Thao L, Lacey SF, et al. Cytomegalovirus immune reconstitution occurs in recipients of allogeneic hematopoietic cell transplants irrespective of detectable cytomegalovirus infection. Biol Blood Marrow Transplant. 2005;11:890–902. doi: 10.1016/j.bbmt.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Gleaves CA, Smith TF, Shuster EA, Pearson GR. Comparison of standard tube and shell vial cell culture techniques for the detection of cytomegalovirus in clinical specimens. J Clin Microbiol. 1985;21:217–221. doi: 10.1128/jcm.21.2.217-221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ljungman P, Griffiths P, Paya C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin Infect Dis. 2002;34:1094–1097. doi: 10.1086/339329. [DOI] [PubMed] [Google Scholar]
- 24.Sun JY, Gaidulis L, Miller MM, et al. Development of a multiplex PCR-SSP method for Killer-cell immunoglobulin-like receptor genotyping. Tissue Antigens. 2004;64:462–468. doi: 10.1111/j.1399-0039.2004.00303.x. [DOI] [PubMed] [Google Scholar]
- 25.Sun JY, Gaidulis L, Dagis A, et al. Killer Ig-like receptor (KIR) compatibility plays a role in the prevalence of acute GVHD in unrelated hematopoietic cell transplants for AML. Bone Marrow Transplant. 2005;36:525–530. doi: 10.1038/sj.bmt.1705089. [DOI] [PubMed] [Google Scholar]
- 26.Hsu KC, Liu XR, Selvakumar A, Mickelson E, O'Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169:5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, Heller G, Chewning J, Kim S, Yokoyama WM, Hsu KC. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J Immunol. 2007;179:5977–5989. doi: 10.4049/jimmunol.179.9.5977. [DOI] [PubMed] [Google Scholar]
- 28.Uhrberg M. Shaping the human NK cell repertoire: an epigenetic glance at KIR gene regulation. Mol Immunol. 2005;42:471–475. doi: 10.1016/j.molimm.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 29.Hokeness KL, Kuziel WA, Biron CA, Salazar-Mather TP. Monocyte chemoattractant protein-1 and CCR2 interactions are required for IFN-alpha/beta-induced inflammatory responses and antiviral defense in liver. J Immunol. 2005;174:1549–1556. doi: 10.4049/jimmunol.174.3.1549. [DOI] [PubMed] [Google Scholar]
- 30.Robbins SH, Bessou G, Cornillon A, et al. Natural killer cells promote early CD8 T cell responses against cytomegalovirus. PLoS Pathog. 2007;3:e123. doi: 10.1371/journal.ppat.0030123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su HC, Nguyen KB, Salazar-Mather TP, Ruzek MC, Dalod MY, Biron CA. NK cell functions restrain T cell responses during viral infections. Eur J Immunol. 2001;31:3048–3055. doi: 10.1002/1521-4141(2001010)31:10<3048::aid-immu3048>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]