Abstract
DNA fragmentation is a critical component of apoptosis but it has not been characterized in non-apoptotic forms of cell death, such as necrosis and autophagic cell death. In mammalian apoptosis, caspase activated DNase (CAD) cleaves DNA into nucleosomal fragments in dying cells, and subsequently DNaseII, an acid nuclease, completes the DNA degradation but acts non-cell-autonomously within lysosomes of engulfing cells. Here we examine the requirement for DNases during two examples of programmed cell death (PCD) that occur in the Drosophila melanogaster ovary, starvation-induced death of mid-stage egg chambers and developmental nurse cell death in late oogenesis. Surprisingly, we found that DNaseII was required cell-autonomously in nurse cells during developmental PCD, indicating that it acts within dying cells. Dying nurse cells contain autophagosomes, indicating that autophagy may contribute to these forms of PCD. Furthermore, we provide evidence that developmental nurse cell PCD in late oogenesis shows hallmarks of necrosis. These findings indicate that DNaseII can act cell-autonomously to degrade DNA during non-apoptotic cell death.
Keywords: autophagic cell death, necrosis, Drosophila, ovary, lysosome
Introduction
Programmed cell death (PCD) is an essential process in animal development and an important component of many diseases.1,2 The three major types of PCD are apoptosis, necrosis and autophagic cell death.3 Cells undergoing apoptosis display chromatin condensation, nucleosomal DNA fragmentation and blebbing of the cell membrane, and are eventually engulfed by macrophages or other cells. Necrosis is characterized by the swelling and lysis of cellular organelles and ultimately the cell itself, whereas autophagic cell death is associated with self-digestion via lysosomes. While the molecular mechanisms of apoptosis are increasingly well-understood, the mechanisms of necrosis and autophagic cell death are less clear.
DNA fragmentation is a well-established characteristic of apoptosis, and causes critical damage from which a cell cannot recover. Although DNA fragmentation occurs during apoptosis in C. elegans, flies, and mammals, the mechanisms differ somewhat.4,5 The two major players in this process are DNase II and caspase-activated DNase (CAD). In mammals and Drosophila, CAD is properly folded and sequestered by its inhibitor, ICAD.4,5 Activated caspases cleave ICAD to release CAD, initiating DNA fragmentation by cleaving DNA between nucleosomes. The next step, mediated by DNase II, breaks down DNA into nucleotides.4,5 DNase II is found within lysosomes and carries out DNA degradation in engulfing cells in mammals.4,5 C. elegans engulfment mutants accumulate cell corpses that retain DNA, indicating that DNA degradation cannot be completed within dying cells.5 NUC-1, the C. elegans DNaseII ortholog, can partially degrade DNA in dying cells when engulfment is blocked,6 however, this may not normally occur. Additional nucleases, most notably mitochondrial endo G, have been shown to participate in DNA fragmentation in mammals and C. elegans.4,5
Drosophila DNaseII was identified based on homology with C. elegans NUC-1.7,8 Hypomorphic mutations or RNAi of Drosophila DNaseII result in impaired innate immunity8,9 and persisting nurse cell (NC) nuclei in the ovary.8,10 Interestingly, nucleosomal DNA fragmentation is increased in DNaseII mutants suggesting that another nuclease is responsible for this step during PCD.8,10 Indeed, dCAD (Drosophila CAD; Flybase: Rep4) was identified as this nuclease when a null allele of dICAD (Flybase: Rep1) was found to prevent nucleosomal DNA fragmentation.8,11 These mutants lack dCAD protein because dICAD is required for dCAD stability.8 Developmental NC death occurred normally in dICAD mutant ovaries, suggesting that nucleosomal fragmentation was not required for the elimination of NC nuclei late in oogenesis. Mukae et al. (2002) proposed a two-step process, similar to the one in mammals, leading to DNA fragmentation in the Drosophila ovary. In their model, dCAD is necessary for the initial nucleosomal DNA fragmentation and acts in the dying cell, whereas DNase II acts in the engulfing cell to complete digestion of DNA.
Interpretation of these findings is complicated by the fact that PCD occurs at multiple stages in the Drosophila ovary (Figure 1a).12 Individual egg chambers, consisting of somatic follicle cells (FCs), germline-derived NCs and an oocyte, mature through fourteen stages of oogenesis.13 With limited nutrients, early germline cysts and mid-stage egg chambers undergo PCD.14 The death of NCs at mid-oogenesis in response to nutrient deprivation is caspase-dependent15-17 but displays characteristics of both apoptosis and autophagic cell death.15,18-20 Later in oogenesis, NCs undergo a different type of PCD as part of normal oocyte development (Figure 1a). This developmental NC PCD occurs largely independently of caspases16,17 and is accompanied by autophagosomes in Drosophila virilis, suggesting that NC PCD could occur by autophagic cell death.18
Figure 1.
DNA fragmentation genes are required for cell death in response to nutrient deprivation. (a) Drawing of an ovariole containing egg chambers at different stages of oogenesis (adapted from 39). FCs are dark gray and NC nuclei in mid-stages are colored black. Nutrient-deprivation induces PCD in early cysts and in mid-oogenesis about st8, whereas developmental NC death occurs between st10 to st14. (b-j) Flies were nutrient-deprived and mid-stage egg chambers were stained with DAPI. (b-e) Progression of cell death in wild-type mid-stage egg chambers. (b) Healthy egg chamber shows dispersed chromatin in NC nuclei. (c) The first signs of degeneration are seen as disorganized clumps of NC chromatin within obvious nuclei. NC nuclei then become highly compact (d) and fragmented (e). (f) At late stages, only small NC remnants are visible (arrow) while FCs are still present and the egg chamber becomes elongated. dICADP homozygous egg chambers show abnormal chromatin compaction (g) and NC nuclei persist until late stages of degeneration (h). (i) DNaseIIlo homozygous egg chambers appear to initiate degeneration normally. (j) In late stages, DNaseIIlo homozygous egg chambers become opaque with dispersed fragments of NC DNA (arrow). All egg chambers are st8-9 and the scale bar is 50μm. All images are projections of 10-16 consecutive images.
Nucleases that carry out DNA fragmentation during necrotic and autophagic cell death have not been reported. Because of the requirement for lysosomes in autophagy, it is plausible that acid nucleases could degrade DNA within dying cells. Furthermore, as lysosomes have been reported to rupture during necrosis, acid nucleases could play a role in necrosis. Given the requirement for DNase II in NC degradation, and the unusual mechanisms of NC PCD, we sought to determine whether DNase II acted in engulfing cells, or cell-autonomously in dying NCs.
Here we demonstrate that DNase II is required for the cell-autonomous degradation of DNA in dying NCs during developmental PCD. Further, we find that both dCAD and DNase II are required for distinct steps of NC nuclear breakdown during mid-oogenesis. Cell death in mid-oogenesis has characteristics of both apoptosis and autophagy, whereas developmental NC PCD appears to involve a distinct mechanism. We find that NCs become acidified during the final stages of PCD, and lysosomal genes are required for this process, suggesting that developmental NC PCD shows characteristics of programmed necrosis. These findings indicate that DNase II can act cell-autonomously to degrade DNA during non-apoptotic cell death.
Results
The role of DNA fragmentation genes in mid-oogenesis
We wished to examine the roles of two nucleases, DNaseII and dCAD, during developmental cell death in late oogenesis (discussed below) and starvation-induced cell death in mid-oogenesis. To induce mid-oogenesis PCD, we subjected flies to yeast deprivation. We then stained egg chambers with DAPI to visualize nuclear structure.
Wild-type egg chambers display a characteristic progression during mid-oogenesis PCD (Figure 1b-f). The first signs of degeneration are disordered chromatin in NCs (Figure 1c), followed by condensation of NC nuclei (Figure 1d). Subsequently, NC nuclei become fragmented and disappear, while FCs persist (Figure 1e-f). Eventually the FCs also degenerate and disappear (data not shown). In dICADP mutant egg chambers, which fail to produce stable dCAD,8 chromatin did not condense in the usual manner (Figure 1g). As degeneration proceeded, NC nuclei persisted while cytoplasm dispersed, as evidenced by an elongate egg chamber morphology (Figure 1h). Eventually most FCs degenerated, leaving small sacs of NC nuclei (data not shown). DNaseIIlo homozygous mutants, which have reduced DNase II activity,7-9,21 had degenerating mid-stage egg chambers with condensed and fragmented NC nuclei similar to controls (Figure 1i). However, in later stages of degeneration, DAPI staining became diffuse and egg chambers appeared opaque (Figure 1j). This diffuse DAPI staining is indicative of NC DNA fragments dispersed throughout the cytoplasm. These results suggest that the two-step model outlined by Mukae et al. (2002) may apply to mid-oogenesis DNA fragmentation, where dCAD first fragments the DNA and DNase II acts secondarily to further degrade the DNA.
The role of DNA fragmentation genes in late oogenesis
During late oogenesis, NC cytoplasmic contents are transferred to the oocyte during stage (st) 11, such that by st12, only NC nuclei remain in the anterior portion of the egg chamber (Figure 2a).13 During st13, NC nuclei become condensed and eventually disappear (Figure 2b).12 By st14, NC nuclei have completely disappeared in 93-95% of wild-type egg chambers (Figure 2c, Table 1).16,22
Figure 2.
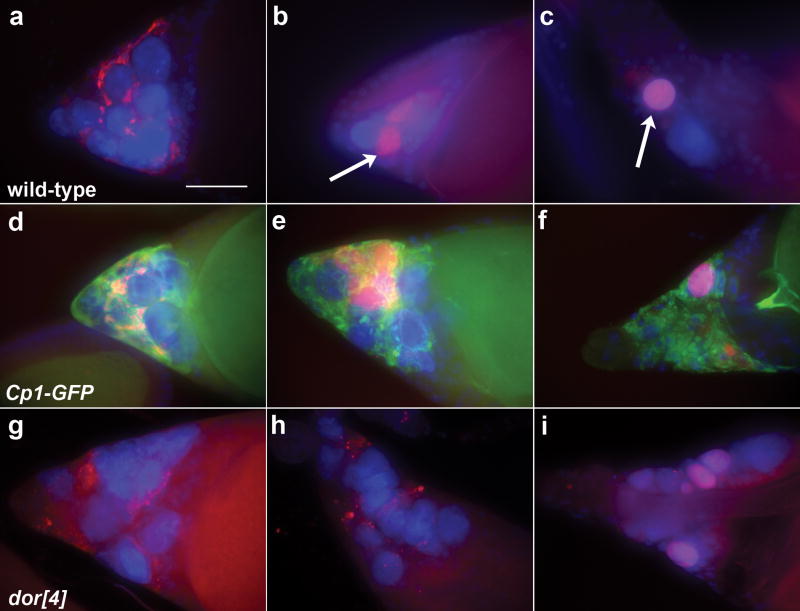
DNase II is required for developmental nurse cell programmed cell death. (a-f and k-l) Egg chambers were stained with DAPI and stages were determined by growth of dorsal appendages. (a-c) Progression of developmental NC death in wild-type egg chambers. (a) At st12, the NCs have transferred their cytoplasm to the oocyte and NC nuclei are still large. (b) By st13, some NC nuclei have disappeared and the remaining NC nuclei are condensed. (c) By st14 NC nuclei cannot be detected. (d) A dICADP homozygous st14 egg chamber with a persisting NC nucleus. (e) A DNaseIIlo st14 egg chamber contains NC nuclei and smeared DNA. (f) dICADP; DNaseIIlo double mutant egg chambers resemble DNaseIIlo mutants. (g-h) Egg chambers labeled with TUNEL (green) and DAPI (blue). (g) TUNEL labels some NC nuclei in a wild-type st13 egg chamber (arrows). (h) Reduced TUNEL labeling is seen in dICADP mutant egg chambers (arrow indicates one weakly labeled nucleus). (i) TUNEL labeling is seen around the periphery of NC nuclei in DNaseIIlo homozygous egg chambers (arrow). Long tubular structures in (g) and (h) are dorsal appendages displaying background fluorescence. Egg chambers from DNaseIIlo/Df(3R)sr16 (j) and DNaseIIlo GLC (k) flies show persisting NC nuclei at st14. (l) A degenerating mid-stage egg chamber from a DNaseIIlo GLC shows a less cloudy appearance than that seen in DNaseIIlo homozygotes (Figure 1i). Egg chambers in (a-l) are projections of 10-16 consecutive images. Scale bars are 50μm; images (c-f and j-k) correspond to the scale bar in b and images (g-i and l) correspond to the scale bar in (a).
Table 1.
Quantification of developmental nurse cell death
| Stage 14 egg chambers | ||||
|---|---|---|---|---|
| Genotype | % normal | % persisting nuclei | % dumplessa | nb |
| w1118 | 93 | 7 | 0 | 780 |
| DNaseIIlo/DNaseIIlo | 51 | 49 | 0 | 142 |
| DNaseIIlo/Df(3R)sr16 | 25 | 75 | 0 | 119 |
| DNaseIIlo GLCs | 25 | 75 | 0 | 51 |
| dICADP | 72 | 28 | 0 | 141 |
| dICADP; DNaseIIlo | 29 | 68 | 3 | 241 |
| Atg1Δ3D GLCs | 62 | 36 | 2 | 472 |
| dor4/dor4 | 8 | 92 | 0 | 218 |
| spinP1/spinP1 | 16 | 84 | 0 | 90 |
| spinP1/SM1 | 56 | 44 | 0 | 89 |
| spinP1 GLCs | 33 | 67 | 0 | 9 |
| spinEP822/spink09905 | 10 | 65 | 24 | 86 |
| spinEP822/spin10403 | 0 | 48 | 52 | 23 |
| spinEP822 GLCs | 0 | 73 | 27 | 60 |
| spink09905 GLCs | 9 | 77 | 14 | 119 |
| cathD1/cathD1 | 65 | 33 | 2 | 123 |
| cathD6/cathD6 | 37 | 45 | 19 | 112 |
| cathD21/cathD21 | 38 | 52 | 10 | 84 |
| cathD24/cathD24 | 41 | 56 | 3 | 105 |
adumpless refers to egg chambers where NC cytoplasm failed to be transferred to the oocyte.
bn= number of egg chambers
To characterize DNA fragmentation genes in late oogenesis, we examined st14 egg chambers stained with DAPI. Mukae et al. (2002) reported that dICADP mutant egg chambers did not display any apparent phenotype. Somewhat consistent with this, we found a mild phenotype in st14 egg chambers (28% persisting nuclei compared to 7% in controls, Table 1, Figure 2d). Interestingly, this percentage is comparable to that seen when caspase inhibitors are over-expressed, or in mutants of the caspase activator ark.16,22 These findings are consistent with the model that caspases are partially required for NC PCD, and that the effects of caspases on nuclei are mediated by dCAD.
In agreement with Stone et al. (1983) and Mukae et al. (2002), we observed a more dramatic phenotype in st14 DNaseIIlo egg chambers. 49% of st14 mutant egg chambers contained persisting NC nuclei (Figure 2e; Table 1). This phenotype was enhanced in DNaseIIlo/deficiency flies (Figure 2j, Table 1) consistent with DNaseIIlo being a hypomorphic allele. The persisting DNA in DNaseIIlo mutants was often smeared, suggesting that nuclei were partially degraded, unlike the discrete nuclei observed in wild-type or dICADP mutants (Figure 2b, d, e). Double dICADP; DNaseIIlo mutants showed the smeared DNA phenotype similar to DNaseIIlo single mutants (Figure 2f; Table 1). The partial nuclear breakdown in these double mutants could be due to residual activity in the DNaseIIlo hypomorph, or another nuclease like endo G.
Mukae et al. (2002) found that dICADP mutants had a block in nucleosomal fragmentation whereas DNaseIIlo mutants had enhanced nucleosomal fragmentation. To determine precisely which ovarian cell types and stages had alterations in nucleosomal fragmentation, we used the TUNEL technique on egg chambers. Positive TUNEL staining is often equated with CAD activity in apoptosis, as it labels the 3′-hydroxyl ends of DNA generated by nucleosomal cleavage of chromatin. TUNEL-positive labeling was observed in wild-type st13 NC nuclei (Figure 2g).12 Some labeling was also observed in dICADP st13 egg chambers, but it was often less intense (Figure 2h), and TUNEL was undetectable in a higher percentage of st13/14 egg chambers compared to controls (89% TUNEL+ in w1118 versus 27% in dICADP, n= 38 and 66 egg chambers respectively). In DNaseIIlo mutants, TUNEL was detected in most egg chambers, however labeled DNA was often at the nuclear periphery (Figure 2i), suggesting that DNase II is required for other nucleases to access the inner regions of the compact NC nuclei. Additionally, DNaseIIlo mutants showed enhanced TUNEL labeling of some FCs and muscle sheath compared to wild-type (data not shown). These findings are consistent with both dCAD and DNase II participating in developmental NC death in late oogenesis.
Tissue specificity of DNase II
Based on the mammalian DNA fragmentation model, it was hypothesized that DNase II is required in engulfing cells in Drosophila oogenesis.8 If this were the case, DNase II would be active in FCs which engulf NC remnants.12 To determine whether DNase II is required in the NCs or FCs, we generated DNaseIIlo germline clones (GLCs), mosaics where the germline is completely homozygous mutant and other tissues are predominantly heterozygous.23 Egg chambers dying in mid-oogenesis displayed a milder phenotype in GLCs than homozygotes (Figure 2l), indicating that engulfing follicle cells are likely to contribute to nurse cell DNA fragmentation in mid-oogenesis. However, DNaseIIlo GLCs had 75% of st14 egg chambers with persisting NC nuclei (Figure 2k, Table 1). The same phenotypes in homozygotes and GLCs indicate that DNase II is required cell-autonomously in the germline for DNA fragmentation during developmental PCD.
The role of lysosomal genes in developmental PCD in the ovary
Because DNase II is an acid DNase active within lysosomes, and DNase II acted cell-autonomously within NCs, we reasoned that lysosomes would play an important role during PCD in oogenesis. Furthermore, because lysosomes are involved in both necrosis and autophagic cell death,3 we reasoned that either autophagy or necrosis would be occurring during ovarian PCD. Consistent with this hypothesis, autophagosomes have been detected during ovarian PCD in Drosophila virilis,18 and autophagy occurs in conjunction with caspase activation during starvation-induced PCD in mid-oogenesis.19,20 However, unlike mid-oogenesis PCD, developmental NC death is largely caspase-independent,16,17 indicating that non-apoptotic mechanisms of cell death may be critically important. Therefore, we sought to determine whether lysosome function is required for PCD in late oogenesis.
To investigate the role of lysosomes and autophagy during late oogenesis, we obtained mutants associated with these processes. deep orange (dor) and spinster (spin) are required generally for lysosome fusion,24-27 whereas Autophagy-specific gene 1 (Atg1) encodes a kinase that acts upstream in the autophagy pathway.28CathepsinD (cathD) encodes a lysosomal cathepsin shown to act during necrotic cell death in other systems.29spin homozygous females had previously been shown to have defects in NC death,27 but the effects of the other mutants had not been determined.
We found that all mutants examined showed a significant disruption in developmental NC death, displaying persisting NC nuclei at st14 (Figure 3; Table 1). Additionally, we found that Atg1 and spinster act cell-autonomously within NCs as the effects were seen in GLCs (Table 1). However, the phenotype of Atg1 mutants was significantly milder than that of dor and spin mutants, indicating that the function for lysosomes in NC PCD might extend beyond autophagy. dor4 and spin mutants frequently displayed a persistence of all fifteen NC nuclei whereas Atg1 GLCs typically showed only a few persisting nuclei (Figure 3a-c). Similarly, cathD mutants showed a partial block to NC PCD (Figure 3d), suggesting that other proteases can degrade NC remnants. Many of the mutants also showed abnormal mid-oogenesis PCD (data not shown), consistent with the reported role for lysosomes and autophagy in mid-oogenesis cell death.
Figure 3.
Lysosome mutants exhibit a disruption in developmental programmed cell death. Persisting NC nuclei are observed in DAPI-stained st14 egg chambers from (a) spinP1 homozygotes, (b) dor4 homozygotes, (c) Atg1Δ3D germline clones, and (d) cathD24 homozygotes. Scale bar is 50μm. Egg chambers are projections of 11-16 consecutive images.
Acidification of nurse cell remnants in late oogenesis
To determine whether autophagy was occurring in late oogenesis, we utilized GFP markers.30 In Drosophila, GFP fusions to Atg5, human LC3 and the Drosophila LC3 ortholog Atg8a have been developed, and become punctate during starvation-induced autophagy in the larval fat body,28,31,32 and during PCD in mid-oogenesis.19,20 In contrast, during late oogenesis, we were unable to detect punctate labeling of either Atg5-GFP or LC3-GFP (data not shown). However, LC3-GFP flies showed significant defects in late oogenesis so we examined GFP-Atg8a flies32 which showed normal oogenesis. GFP-Atg8a was uniform in early stages, but puncta became apparent by st12 (Figure 4a). By st13, GFP puncta were largely absent (Figure 4b), suggesting that autophagy occurs transiently in NC PCD. We additionally examined st12 and st13 egg chambers for evidence of autophagy using transmission electron microscopy (EM) and found that the NCs in st12 egg chambers did contain autophagosomes, as well as lysosomes (Figure 4c-f). However, at later stages, very few organelles were seen.
Figure 4.
Transient autophagy occurs in dying nurse cells. (a-b) Egg chambers from ovaries of UASp-GFP-Atg8a/NGT; nanos-GAL4/+ flies stained with DAPI. GFP-Atg8a signal can be seen as small puncta in st12 (a), but not st13 (b) egg chambers. Yolk granules within the oocyte also fluoresce. Both images are projections of 12 slices. (c-d) Wild-type st12 and st13 egg chambers (1 μm sections) stained with 1% toluidine blue. N = nucleus; N1-3 are NC nuclei analyzed below. oo = oocyte, DA = dorsal appendage. (e-g) Electron micrographs of organelles in the cytoplasm surrounding NC nucleus 1 (N1) from Figure 4C. FC = Follicle cells, L = lysosome, * = autophagosome. PM = plasma membrane. (h-j) Electron micrographs reveal the progression of nuclear breakdown in NCs. (h) N1 from Figure 4c. Nucleus with compacted chromatin and many surrounding organelles. (i) N2 from Figure 4c. NC with fragmented chromatin and vacuole-like structures. (j) N3 from Figure 4d. Terminal NC with little chromatin remaining. Scale bars in (a-d) are 50 μm. (e-g) at 10,000×, scale bars are 500 nm; (h-j) at 2000×, scale bar is 2 μm.
Given that autophagy was observed only transiently in late oogenesis, yet there was a strong requirement for the lysosome fusion gene dor, we examined lysosomes directly using LysoTracker (LT). LT was at low levels in early oogenesis, except in degenerating mid-stages or in the germarium, as previously reported.19 During early st12, punctate LT surrounded NC nuclei (Figure 5a). Later in st12, punctate staining was lost, and large domains of LT were observed (Figure 5b). In st13 egg chambers, large domains of LT staining continued to be observed (Figure 5c) and staining became undetectable by st14 (data not shown). The LT-positive domains usually overlapped with NC nuclei (Figure 5b-c).
Figure 5.
Lysosome clustering and acidification of nurse cell remnants. All egg chambers stained with DAPI (blue) and LysoTracker (LT, red). (a-c) Progression of LT labeling in wild-type egg chambers. (a) In early st12, increased LT labeling is observed in clusters around NC nuclei. (b) In late st12, acidification of NC nuclei is indicated by large LT-positive domains (arrow). (c) Large LT-positive domains are also observed in st13 egg chambers (arrow). (d-f) Egg chambers from a GFP gene trap in Cp1 (cathepsinL). (d) In early st12, Cp1-GFP clusters around NC nuclei and partially overlaps with LT. (e) In late st12, most Cp1-GFP remains punctate and LysoTracker is detected in large domains. (f) In st13, no significant overlap is detected between CP1-GFP and LT. (g-i) LT labeling in dor4 homozygous mutants reveals a delay in acidification of NC remnants. LT is punctate in st12 (g) and st13 (h) dor4 homozygous egg chambers. (i) A dor4 homozygous st14 egg chamber shows acidification of some persisting NC nuclei. Egg chambers in a-i are projections of 11-21 consecutive images. Scale bar is 50μm.
LysoTracker does not detect lysosomes per se, but is an acidotropic probe.30 Thus the LT staining we observed could represent very large lysosomes, or an acidification of NC remnants. To determine which of these possibilities was more likely, we examined CP1-GFP flies, which carry a GFP fusion to the lysosomal cathepsinL (Flybase: CP1, Cysteine Proteinase-1). As seen in Figure 5d, CP1-GFP also clustered around NC nuclei during st12, partially overlapping with punctate LT. However, during later st12 and st13, CP1-GFP remained largely punctate and failed to overlap the large domains seen with LT (Figure 5e-f). Thus, we believe that the large domains of LT staining represent an acidification of NC remnants, perhaps caused by a rupture of lysosomes, as occurs during necrosis.
We examined egg chambers closely for evidence of necrosis using EM and we found a progression of nuclear degradation associated with loss of organelles. Degenerating NC nuclei initially showed compacted chromatin (Figure 4h), consistent with fluorescent imaging. Numerous organelles were observed surrounding these nuclei, including autophagosomes and lysosomes, as well as larger vacuolar structures, similar to Drosophila virilis.18 Some lysosomes showed discontinuous membranes, suggesting lysosomal rupture (Figure 4g). NC nuclei progressed to a phase in which the chromatin was highly dispersed with few surrounding organelles (Figure 4i). The organelles that remained were primarily empty vacuole-like structures. At terminal stages, a devoid pocket remained with small bits of chromatin (Figure 4j) and little else other than multilayered membranous organelles (data not shown). This progression suggests that early phases of NC nuclear breakdown involve lysosome clustering, while later nuclear degradation occurs after lysosomes have ruptured, similar to what has been reported for necrotic cell death.29
If acidification of NC remnants is critical for proper NC PCD, we would expect this process to be disrupted in the lysosome fusion mutants. Indeed, dor4 homozygotes showed a delay in NC acidification compared to controls (Figure 5g-I; Figure 6). While 36% of control st12 egg chambers showed large domains of LT, only 3% of dor4 homozygous st12 egg chambers showed this staining pattern. Similarly, among st13 and st14 egg chambers with persisting NC nuclei, 4% of control egg chambers showed punctate labeling, whereas 24% of dor4 mutant egg chambers did (Figure 5h; Figure 6b). Atg1 and spin mutants showed similar but milder effects in st13 and 14 (Figure 6b). Taken together, these results suggest that lysosome fusion genes are required for the timely acidification of NC remnants in late oogenesis. This acidification would allow for DNase II activity and subsequent degradation of NC nuclei.
Figure 6.
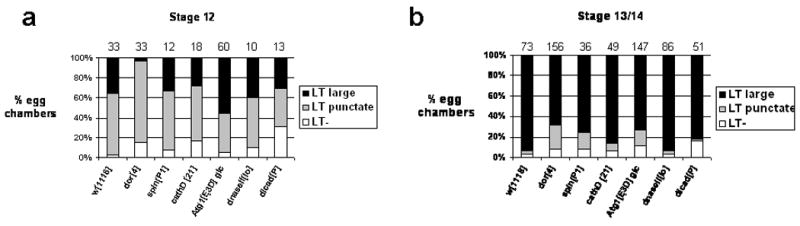
Quantification of LysoTracker staining. Egg chambers from the genotypes listed on the bottom of each graph were staged and scored for LysoTracker (LT) staining in large domains, punctate spots, or absent/diffuse (LT-). Egg chambers displaying >3 puncta were scored as punctate. Egg chambers displaying both puncta and large domains were scored as having large domains. Separate graphs are shown for st12 (a) and st13/14 (b). Numbers of egg chambers scored are indicated above each column. w1118 was used as a wild-type control.
Discussion
DNA fragmentation is a critical point of no return for dying cells. Mice that are unable to degrade DNA to completion die during embryogenesis, or if cytokine signaling is disrupted will survive but develop arthritis.4 Flies with disrupted DNA fragmentation show persisting DNA and defects in innate immunity.8,9 These effects have been largely attributed to phagocytic cells that fail to degrade the DNA of apoptotic cells. However, the importance of DNA fragmentation in non-apoptotic cell deaths, which may not be phagocytosed, is unknown.
In mammals, a two-step model for DNA fragmentation in apoptosis has been proposed, where caspases activate CAD to degrade DNA in dying cells, and DNase II finishes the degradation within lysosomes of engulfing cells.4 We find a similar two-step process during mid-oogenesis PCD. dICADP mutants, deficient in dCAD, show initial defects in nuclear compaction and retain NC nuclei in late egg chambers. DNaseIIlo mutants show a strikingly different phenotype where there is normal degeneration at early stages, but a block in complete DNA degradation, causing a uniform distribution of DNA throughout the egg chamber. A milder phenotype is observed in DNaseIIlo GLCs, suggesting that DNase II can act in the engulfing follicle cells.
Unlike cell death in mid-oogenesis, developmental NC death in late oogenesis is only partially affected when caspases are inhibited.16 This suggests that caspases play a minor role, or redundant or compensatory pathways are used. Consistent with this minor requirement for caspases, dICADP mutants have a weak phenotype in late oogenesis. However, DNaseIIlo mutants show a significant disruption in nurse cell death in late oogenesis. Further, DNaseIIlo GLCs show the same phenotype as DNaseIIlo homozygotes, indicating that DNase II acts in the dying NCs themselves. This cell-autonomous requirement for DNase II suggests a prominent role for lysosomes in dying nurse cells. Autophagosomes have been reported in late oogenesis in Drosophila virilis,18 suggesting that autophagy could be the major cell death mechanism in late oogenesis. However, Atg1 GLCs show only a partial disruption of NC PCD and autophagic markers and EM indicate that autophagy is occurring only transiently.
The third major form of cell death is necrosis. Since the coining of the term apoptosis, necrosis has been used to define accidental, traumatic cell death, rather than an intrinsic cell suicide program. This view has been brought into question recently by studies of necrosis in several organisms.29 We propose that developmental NC death in late oogenesis displays characteristics of programmed necrosis. Several hallmarks of necrosis, such as calcium release, ATP depletion and lysosomal clustering,29 occur in dying NCs. During st11, free calcium is released from nuclear stores,33 which could activate calcium dependent proteases like calpains. During NC dumping in st11, mitochondria are transferred to the oocyte,13 which could cause a massive depletion in ATP. Here we have shown that lysosomes cluster around NC nuclei during st12, and acidification of NC remnants is observed beginning in late st12. Furthermore, several lysosomal mutants have defective NC PCD. The requirement for these lysosomal proteins could implicate either an autophagic or necrotic type of PCD. However, the large-scale acidification suggests that lysosome rupture, a hallmark of necrosis, is a terminal event in NC PCD. Interestingly, a DNase II paralog in mammals, DNase IIβ or DLAD, acts cell-autonomously during differentiation of the eye lens, where nuclei and other organelles are degraded.34,35 As nuclei degenerate during lens differentiation, lysosomes cluster around nuclei,35 similar to what we have observed.
Cytosol acidification has been reported in some examples of cell death, and LysoTracker has been reported to label apoptotic cells in chick and mouse.36 The decreases in cellular pH of apoptotic cells are thought to be mediated by changes in mitochondria and disruption of the Na+/H+ exchanger. Although we cannot rule out a role for these factors in developmental NC PCD, the loss of individual LysoTracker-positive puncta in late oogenesis suggests that remaining lysosomes cannot maintain a lower pH than surrounding NC remnants. Further, the requirement for dor in the acidification process suggests a prominent role for lysosomes. Taken together, our findings indicate that developmental NC death involves early apoptotic and autophagic events, followed by necrotic events. Necrotic events, such as lysosome rupture, lead to intracellular acidification which enables lysosomal enzymes to complete the degradation of NC remnants.
Materials and methods
Drosophila strains and genetic manipulations
DNaseIIlo21 and dICADP mutants8 were obtained from Shigekazu Nagata. spinP1 mutants27 were obtained from Daisuke Yamamoto. Atg1Δ3D mutants28 were received from Tom Neufeld and cathD1 mutants37 were obtained from Mel Feany. The other cathD mutants were generated by imprecise excision of EP element 2151 from the cathDEP2151 insertion line. Sequence analysis of cathD6, cathD21, and cathD24 revealed insertion mutations in the 5′ untranslated region of each of these alleles. Western blots confirmed cathD21 and cathD24 to be null alleles, while cathD6 is an extreme hypomorph (data not shown). The UASp-Atg8a-GFP transgenic line, obtained from Harald Stenmark,32 was crossed to NGT;nosGAL416 to generate germline-specific expression. The CP1-GFP line (CC01377)38 was obtained from Allan Spradling. All other strains were obtained from the Bloomington Drosophila Stock Center. spin alleles EP822, k09905 and 0140326,27 and the dor4 allele25 have been previously described.
All crosses were carried out at 25°C on standard cornmeal-molasses medium. GLCs were generated using the FLP/FRT/ovoD method as previously described.23 Expression of FLP was induced by heatshocking larvae on days 4 and 5 after egg laying for one hour in a 37°C water bath.
Ovary dissections and staining
Flies were conditioned on standard fly food supplemented with wet yeast paste. For nutrient deprivation experiments, flies were transferred to apple juice agar media lacking yeast for 1-2 days before dissection. Alternatively, flies were fed on the same yeast paste for several days. For DAPI and GFP visualization, ovaries were fixed in PIPES buffer as described15 and mounted in Vectashield with DAPI (Vector Labs). For LT staining, ovaries were dissected in Ringer's, transferred to 20μM LysoTracker Red (Invitrogen) in PBS for 3 min, washed several times in PBS over 30 min, and fixed in Buffer B as described.22 TUNEL (Terminal deoxynucleotidyl Transferase dUTP Nick End Labeling) was carried out with the ApopTag Fluorescein Direct In Situ Apoptosis Detection Kit (Millipore, Billerica, MA) as described.14 Staging of st12-14 egg chambers was determined by the size and shape of dorsal appendages. Images were taken on an Olympus BX51 microscope with DSU spinning disc attachment using a 40× water immersion lens. Images were taken at 2μm intervals using Slidebook (3I) software and combined into a single projection image.
Electron microscopy
Flies were conditioned on wet yeast paste for 2-3 days prior to dissection. To maximize the number of st10-13 egg chambers, flies were switched to fresh yeast paste every 4 hours for the 24 hours prior to dissection. Ovaries were dissected in Ringer's solution and roughly separated before fixation. Ovaries were fixed in 2% glutaraldehye in 0.1M sodium cacodylate buffer for 2 hours on ice, followed by 3 ten min washes in 0.1 M cacodylate buffer. Ovaries were then post-fixed in 2% osmium tetroxide for 2 hours at room temperature or overnight at 4° C, washed, then dehydrated through a series of ethanol and acetone washes prior to infiltration with resin. Individual egg chambers were staged and embedded in Epon812 resin and polymerized at 60° C. The resulting blocks were sectioned 1 μm thick, and stained with 1% toluidine blue for accurate staging. Ultrathin 70 nm sections were collected on copper grids followed by a counter-stain with uranyl acetate and lead citrate. Samples were imaged using a JEOL JEM2010 transmission electron microscope operated at an acceleration voltage of 80 kV.
Acknowledgments
We thank Shigekazu Nagata, Daisuke Yamamoto, Tom Neufeld, Mel Feany, Tor Erik Rusten, Harald Stenmark, the FlyTrap Collection and the Bloomington Drosophila Stock Center for fly strains. We thank Don Gantz at Boston University Medical Center for technical assistance with the electron microscopy. We are grateful to Horacio Frydman for providing the image in Figure 1A and helpful comments on the manuscript. We additionally thank Sharon Gorski and members of the laboratory for helpful discussions. We apologize to authors whose work we unable to cite due to space limitations. This work was supported by NIH grants to KM (R01 GM60574) and PD (EY016747). ET was supported in part by NICHD training grant 2T 32 HD007387.
References
- 1.Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 2.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagata S. DNA degradation in development and programmed cell death. Annu Rev Immunol. 2005;23:853–875. doi: 10.1146/annurev.immunol.23.021704.115811. [DOI] [PubMed] [Google Scholar]
- 5.Parrish JZ, Xue D. Cuts can kill: the roles of apoptotic nucleases in cell death and animal development. Chromosoma. 2006;115:89–97. doi: 10.1007/s00412-005-0038-0. [DOI] [PubMed] [Google Scholar]
- 6.Wu YC, Stanfield GM, Horvitz HR. NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev. 2000;14:536–548. [PMC free article] [PubMed] [Google Scholar]
- 7.Evans CJ, Merriam JR, Aguilera RJ. Drosophila acid DNase is a homolog of mammalian DNase II. Gene. 2002;295:61–70. doi: 10.1016/s0378-1119(02)00819-3. [DOI] [PubMed] [Google Scholar]
- 8.Mukae N, Yokoyama H, Yokokura T, Sakoyama Y, Nagata S. Activation of the innate immunity in Drosophila by endogenous chromosomal DNA that escaped apoptotic degradation. Genes Dev. 2002;16:2662–2671. doi: 10.1101/gad.1022802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seong CS, Varela-Ramirez A, Aguilera RJ. DNase II deficiency impairs innate immune function in Drosophila. Cell Immunol. 2006;240:5–13. doi: 10.1016/j.cellimm.2006.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone JC, Dower NA, Hauseman J, Cseko YM, Sederoff R. The characterization of a mutant affecting DNA metabolism in the development of D. melanogaster. Can J Genet Cytol. 1983;25:129–138. doi: 10.1139/g83-025. [DOI] [PubMed] [Google Scholar]
- 11.Mukae N, Yokoyama H, Yokokura T, Sakoyama Y, Sakahira H, Nagata S. Identification and developmental expression of inhibitor of caspase- activated DNase (ICAD) in Drosophila melanogaster. J Biol Chem. 2000;275:21402–21408. doi: 10.1074/jbc.M909611199. [DOI] [PubMed] [Google Scholar]
- 12.McCall K. Eggs over easy: cell death in the Drosophila ovary. Dev Biol. 2004;274:3–14. doi: 10.1016/j.ydbio.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 13.King RC. Ovarian Development in Drosophila melanogaster. New York: Academic Press; 1970. [Google Scholar]
- 14.Drummond-Barbosa D, Spradling AC. Stem cells and their progeny respond to nutritional changes during Drosophila oogenesis. Dev Biol. 2001;231:265–278. doi: 10.1006/dbio.2000.0135. [DOI] [PubMed] [Google Scholar]
- 15.Peterson JS, Barkett M, McCall K. Stage-specific regulation of caspase activity in Drosophila oogenesis. Dev Biol. 2003;260:113–123. doi: 10.1016/s0012-1606(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 16.Baum JS, Arama E, Steller H, McCall K. The Drosophila caspases Strica and Dronc function redundantly in programmed cell death during oogenesis. Cell Death Differ. 2007;14:1508–1517. doi: 10.1038/sj.cdd.4402155. [DOI] [PubMed] [Google Scholar]
- 17.Mazzalupo S, Cooley L. Illuminating the role of caspases during Drosophila oogenesis. Cell Death Differ. 2006;13:1950–1959. doi: 10.1038/sj.cdd.4401892. [DOI] [PubMed] [Google Scholar]
- 18.Velentzas AD, Nezis IP, Stravopodis DJ, Papassideri IS, Margaritis LH. Apoptosis and autophagy function cooperatively for the efficacious execution of programmed nurse cell death during Drosophila virilis oogenesis. Autophagy. 2007;3:130–132. doi: 10.4161/auto.3582. [DOI] [PubMed] [Google Scholar]
- 19.Hou YC, Chittaranjan S, Barbosa SG, McCall K, Gorski SM. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis. J Cell Biol. 2008;182:1127–1139. doi: 10.1083/jcb.200712091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nezis IP, Lamark T, Velentzas AD, Rusten TE, Bjorkoy G, Johansen T, et al. Cell death during Drosophila melanogaster early oogenesis is mediated through autophagy. Autophagy. 2009;5 doi: 10.4161/auto.5.3.7454. [DOI] [PubMed] [Google Scholar]
- 21.Grell EH. Genetics of some deoxyribonucleases of Drosophila melanogaster. Genetics. 1976;83:s28–s29. [Google Scholar]
- 22.Peterson JS, Bass BP, Jue D, Rodriguez A, Abrams JM, McCall K. Noncanonical cell death pathways act during Drosophila oogenesis. Genesis. 2007;45:396–404. doi: 10.1002/dvg.20306. [DOI] [PubMed] [Google Scholar]
- 23.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindmo K, Simonsen A, Brech A, Finley K, Rusten TE, Stenmark H. A dual function for Deep orange in programmed autophagy in the Drosophila melanogaster fat body. Exp Cell Res. 2006;312:2018–2027. doi: 10.1016/j.yexcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Sevrioukov EA, He JP, Moghrabi N, Sunio A, Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- 26.Sweeney ST, Davis GW. Unrestricted synaptic growth in spinster-a late endosomal protein implicated in TGF-beta-mediated synaptic growth regulation. Neuron. 2002;36:403–416. doi: 10.1016/s0896-6273(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 27.Nakano Y, Fujitani K, Kurihara J, Ragan J, Usui-Aoki K, Shimoda L, et al. Mutations in the novel membrane protein spinster interfere with programmed cell death and cause neural degeneration in Drosophila melanogaster. Mol Cell Biol. 2001;21:3775–3788. doi: 10.1128/MCB.21.11.3775-3788.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Golstein P, Kroemer G. Cell death by necrosis: towards a molecular definition. Trends Biochem Sci. 2007;32:37–43. doi: 10.1016/j.tibs.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 33.Matova N, Mahanjan-Miklos S, Mooseker MS, Cooley L. Drosophila Quail, a villin-related protein, bundles actin filaments in apoptotic nurse cells. Development. 1999;126:5645–5657. doi: 10.1242/dev.126.24.5645. [DOI] [PubMed] [Google Scholar]
- 34.Nishimoto S, Kawane K, Watanabe-Fukunaga R, Fukuyama H, Ohsawa Y, Uchiyama Y, et al. Nuclear cataract caused by a lack of DNA degradation in the mouse eye lens. Nature. 2003;424:1071–1074. doi: 10.1038/nature01895. [DOI] [PubMed] [Google Scholar]
- 35.Nakahara M, Nagasaka A, Koike M, Uchida K, Kawane K, Uchiyama Y, et al. Degradation of nuclear DNA by DNase II-like acid DNase in cortical fiber cells of mouse eye lens. FEBS J. 2007;274:3055–3064. doi: 10.1111/j.1742-4658.2007.05836.x. [DOI] [PubMed] [Google Scholar]
- 36.Counis MF, Torriglia A. Acid DNases and their interest among apoptotic endonucleases. Biochimie. 2006;88:1851–1858. doi: 10.1016/j.biochi.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Myllykangas L, Tyynela J, Page-McCaw A, Rubin GM, Haltia MJ, Feany MB. Cathepsin D-deficient Drosophila recapitulate the key features of neuronal ceroid lipofuscinoses. Neurobiol Dis. 2005;19:194–199. doi: 10.1016/j.nbd.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 38.Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, et al. The Carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frydman HM, Spradling AC. The receptor-like tyrosine phosphatase lar is required for epithelial planar polarity and for axis determination within Drosophila ovarian follicles. Development. 2001;128:3209–3220. doi: 10.1242/dev.128.16.3209. [DOI] [PubMed] [Google Scholar]