Abstract
Cancer patients show large individual variation in their response to chemotherapeutic agents. Gemcitabine (dFdC) and AraC, two cytidine analogues, have shown significant activity against a variety of tumors. We previously used expression data from a lymphoblastoid cell line-based model system to identify genes that might be important for the two drug cytotoxicity. In the present study, we used that same model system to perform a genome-wide association (GWA) study to test the hypothesis that common genetic variation might influence both gene expression and response to the two drugs. Specifically, genome-wide single nucleotide polymorphisms (SNPs) and mRNA expression data were obtained using the Illumina 550K® HumanHap550 SNP Chip and Affymetrix U133 Plus 2.0 GeneChip, respectively, for 174 ethnically-defined “Human Variation Panel” lymphoblastoid cell lines. Gemcitabine and AraC cytotoxicity assays were performed to obtain IC50 values for the cell lines. We then performed GWA studies with SNPs, gene expression and IC50 of these two drugs. This approach identified SNPs that were associated with gemcitabine or AraC IC50 values and with the expression regulation for 29 genes or 30 genes, respectively. One SNP in IQGAP2 (rs3797418) was significantly associated with variation in both the expression of multiple genes and gemcitabine and AraC IC50. A second SNP in TGM3 (rs6082527) was also significantly associated with multiple gene expression and gemcitabine IC50. To confirm the association results, we performed siRNA knock down of selected genes with expression that was associated with rs3797418 and rs6082527 in tumor cell and the knock down altered gemcitabine or AraC sensitivity, confirming our association study results. These results suggest that the application of GWA approaches using cell-based model systems, when combined with complementary functional validation, can provide insights into mechanisms responsible for variation in cytidine analogue response.
Introduction
The application of high-throughput genomic techniques makes it possible to identify variation across the genome that may help to predict clinical response to drug treatment [1]–[5]. Wide variations in response to many antineoplastic drugs exist in the clinic. Therefore, it is crucial to identify biomarkers that will help make it possible to maximizing efficacy while minimize drug-related toxicity. One possible explanation for variation in response to chemotherapy is genetic variation that influences gene expression and, ultimately, impacts variation in drug response phenotypes [6]–[8]. Most previous studies have focused on biomarkers in tumor tissue and their relationship with therapeutic response [9]–[15]. While identifying genetic biomarkers in the tumor is important for understanding tumorigenesis and for predicting prognosis and treatment response, individual variation in germline DNA can also play an important role in drug response. Gemcitabine and AraC are two cytidine analogues that are widely used to treat a variety of cancers [16]–[19]. Only 20% of the pancreatic cancer patients treated with gemcitabine have life expectancy over 1 year. Many side effects such as GI and hematological side effects are associated with the use of these drugs. Therefore, it would be important to identify biomarkers that might help to select responsive patients and avoid side effects during treatment with these drugs.
In a present study, we took advantage of ethnically diverse Coriell “Human Variation Panel” lymphoblastoid cell lines to test the hypothesis that genetic variation across the genome in germline DNA might impact gemcitabine and AraC cytotoxicity. These cell lines were obtained from three different ethnic groups: Caucasian-Americans, African-Americans and Han Chinese-Americans. Although they are Epstein-Barr virus (EBV) transformed cell lines with expression profiles distinctive from those in tumor cells, these cell lines provide an opportunity to query the impact of common genomic variation on drug response [5], [20]–[25]. Specifically, we have now used these cell lines to perform GWA experiments designed to identify genetic variation that might be associated with variation in gene expression and then, subsequently, with variation in gemcitabine and AraC IC50 values. We identified 19 and 30 SNPs associated with gemcitabine and AraC IC50 that were also associated with the expression of 29 and 30 genes, respectively. At the same time, this variation in gene expression was associated with IC50 values for the two drug. We then focused on 2 SNPs with unknown function in the intron and 5′-flank region (FR) of a gene encoding the IQ motif containing GTPase activating protein 2 (IQGAP2),(rs3797418) and a gene encoding transglutaminase 3 (TGM3), (rs6082527), respectively. IQGAP2 (rs3797418) was significantly associated with IC50 for both drugs and TGM3 (rs6082527) was significantly associated with the expression of multiple genes and with gemcitabine IC50 values. These two SNPs are not in linkage disequilibrium (LD) with each other. Because gemcitabine is used to treat a variety of solid tumors including pancreatic cancer, in order to validate this association obtained using lymphoblastoid cell lines, we selected two pancreatic tumor cell lines to perform siRNA knock down of three selected genes (MGMT, VAV3 and GPM6A) with expression levels that were associated with either or both of the rs3797418 and rs6082527 SNPs. Those results indicated that down-regulation of VAV3 and GPM6A altered gemcitabine or AraC response, as predicted by the association studies. In addition, we also took the advantage of the fact that we had information with regard to SNPs for all of the genes involved in the cytidine analogue activation and deactivation pathway, and performed SNP expression as well as drug cytotoxicity analyses with those genes. These results suggest that a genome-wide approach can help to identify genetic variation that might be important for variation in drug response through the trans-regulation of the expression of multiple genes.
Results
Genome-Wide SNP vs. Gemcitabine IC50 Association Study
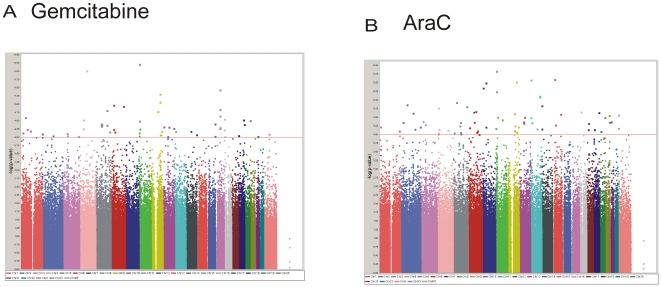
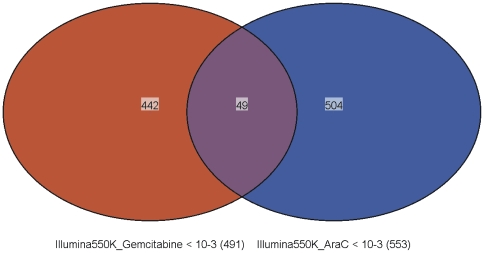
A total of 561,464 SNPs were genotyped on the Illumina 550K SNP array and were subjected to quality control prior to performing the association study. As a result, we first removed the 8,222 SNPs that had call rate <95% as well as the 33,564 SNPs with Minor Allele Frequencies (MAFs) <5%. Finally, we removed 4,639 SNPs that deviated from Hardy-Weinberg Equilibrium (HWE) using a stringent threshold of p<0.001. Therefore, a total of 515,039 SNPs were used in a genome-wide SNP analyses for 171 cell lines. Figure 1 depicts graphically and Table S1 and S2 list the 492 SNPs associated with gemcitabine IC50 and the 553 SNPs associated with AraC IC50 with p-values <10−3. 58 and 68 SNPs among these 492 and 553 SNPs had p-values <10−4, respectively. These 58 and 68 SNPs were in or near 47 or 59 unique genes on the basis of the annotation provided by Illumina, genome build 36.1. 14 of those 58 SNPs were in 3′-flanking regions, 19 in 5′-flanking regions, 23 in intragenic regions of the closest gene, while only two were in the coding regions for the PIGB and HLA-DRA genes. The top three SNPs from the GWAS, rs4272382 (p = 6.14×10−7), rs3775182 (9.67×10−7) and rs2290344 (3.65×10−6), were in genes encoding claudin 23, MAPK10 and PIGB. For the 68 top SNPs associated with AraC IC50 in 59 unique genes, 28 SNPs were in 3′-flanking regions, 17 in 5′-flanking regions and 23 in introns. None were in coding regions. The top three SNPs for AraC were rs2604376 (p = 1.84×10−6), rs9512755 (p = 2.54×10−6) and rs2595500 (p = 2.63×10−6) in the TUSC3, LNX2 and ZNF215 genes. When we compared gemcitabine and AraC, among the top SNPs that were associated with drug IC50 values (p<0.001), only 49 were common to both drugs (Table S3 and Figure 2 ).
Figure 1. Genome-wide SNP association with drug IC50.
(A). Genome-wide SNP-gemcitabine IC50 association data. (B). Genome-wide SNP-AraC IC50 association data. Genome-wide SNPs were obtained for all 171 cell lines using Illumina 550K Beadchips. Genome-wide SNP association analysis was then performed with gemcitabine IC50 as the drug-response phenotype (n = 171). The y-axis is the −log10 (p-value) for the SNP. SNPs are plotted on the x-axis on the basis of their chromosomal locations. The “cut-off” p-value of 0.001 is highlighted with a red line.
Figure 2. Top candidate SNPs that overlapped between gemcitabine and AraC.
49 SNPs were common (p<0.001) among 442 unique SNPs associated with gemcitabine and 504 associated with AraC.
SNP and Gene Expression Association
Among the above associated 492 associated with gemcitabine IC50, we observed that 100 unique SNPs, located within or near 78 genes on the basis of the Illumina annotation were associated with variation in the expression of 296 probesets (220 unique genes) with a cutoff of p-values <10−6. Among the 553 SNPs associated with AraC IC50, 140 unique SNPs within or near 123 genes were associated with variation in expression of 843 (563 unique genes) expression probesets (Table S4 and S5). Only 12 out of the 100 SNPs or 32 out of the 140 SNPs were in cis-regulatory regions for gemcitabine and AraC, respectively, defined as 5 Mb on either side of the gene, while the rest of the SNPs were localized in trans-regulatory regions, consistent with previous findings [5], [26]–[28].
Gene Expression and IC50 Association
Among the 296 expression probesets associated with the 100 unique SNPs for gemcitabine, we determined that 35 expression probesets were also associated with gemcitabine IC50 values (p<0.001). These 35 probesets represented 29 genes. By performing the same analysis for AraC, we identified 60 expression probesets that represented 54 genes which were associated with AraC IC50 values (p<0.001). Figure 3 showed our integrated analysis strategy, a strategy that included SNP, gemcitabine and gene expression data. This analysis resulted in the identification of 19 SNPs close to or within 16 genes that were associated with gemcitabine IC50 values through an influence on the expression of 29 genes represented by 35 expression probesets ( Table 1 ). The same type of analysis resulted in the identification of 30 SNPs close to or within 30 genes that were associated with AraC IC50 values through an influence on the expression of 54 genes represented by 60 expression probesets ( Table 2 ). Among the top candidate SNPs, rs3797418 (A/C), a SNP located in an intron of IQGAP2, was significantly associated with the expression of multiple genes and IC50 values for both drug. Therefore, this SNP and gene were selected for further functional verification. In addition, a second SNP, rs6082527 (A/G), located in the 5′-FR of TGM3, which was only associated with gemcitabine IC50 values, was also selected for further functional study because of its significant association with the expression of multiple genes, and these genes were also significantly associated with gemcitabine cytotoxicity, with p values ranging from 10−4 to 10−8 ( Tables 1 and 2 ). Although the SNP in TGM3 was not significantly associated with AraC IC50 values, given the similarity between these two drugs, we also performed functional studies for TGM3 and its downstream gene GPM6A with AraC. Specifically, rs3797418 was associated with the expression of 10 individual genes for gemcitabine and 7 for AraC. rs6082527 was associated with 6 probesets that targeted 5 individual genes.
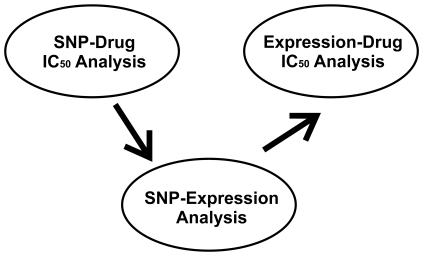
Figure 3. Strategy applied in these studies.
A genome-wide SNP association study was performed with a drug-related phenotype, gemcitabine or AraC cytotoxicity (IC50). SNPs associated with cytotoxicity (p<10−3) were used to perform an association study with 54,000 expression probesets to identify SNPs that were associated with both gene expression (p<10−6) and with gemcitabine or AraC IC50. Finally, an association study was performed with gene expression and gemcitabine or AraC IC50 to identify SNPs that might be associated with drug IC50 values through an influence on gene expression.
Table 1. Top candidate SNPs and probe sets that were associated with gemcitabine IC50.
| SNPs | Probe sets | SNP vs Expression | Expression vs IC50 | SNP vs IC50 | |||||||||
| rsID | Chr | Closest gene | SNP Location | MAF | Probeset | Chr | Gene Symbol | r-value | p-value | r-value | p-value | r-value | p-value |
| rs1423300 | 5 | ADAMTS12 | intron | 0.526 | 209303_at | 5 | NDUFS4 | −0.40 | 2.00E-07 | 0.32 | 2.27E-05 | −0.28 | 3.57E-04 |
| rs2118524 | 12 | BTG1 | flanking_5UTR | 0.088 | 219104_at | 11 | RNF141 | 0.39 | 5.74E-07 | −0.33 | 1.07E-05 | −0.26 | 9.95E-04 |
| rs2118524 | 12 | BTG1 | flanking_5UTR | 0.088 | 231186_at | 14 | FLJ43390///LOC100128793 | 0.39 | 5.95E-07 | −0.29 | 1.09E-04 | −0.26 | 9.95E-04 |
| rs1809180 | 15 | CHSY1 | flanking_3UTR | 0.173 | 201462_at | 7 | SCRN1 | −0.41 | 8.28E-08 | 0.34 | 6.47E-06 | −0.27 | 9.12E-04 |
| rs453169 | 16 | FLJ32252 | flanking_3UTR | 0.056 | 231851_at | 1 | RAVER2 | 0.45 | 3.23E-09 | −0.30 | 6.63E-05 | −0.27 | 7.79E-04 |
| rs11215406 | 11 | IGSF4 | intron | 0.14 | 1556095_at | 15 | UNC13C | 0.39 | 3.22E-07 | −0.32 | 1.57E-05 | −0.27 | 6.70E-04 |
| rs11215406 | 11 | IGSF4 | intron | 0.14 | 1569969_a_at | 15 | UNC13C | 0.38 | 9.91E-07 | −0.30 | 5.46E-05 | −0.27 | 6.70E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 203726_s_at | 18 | LAMA3 | 0.41 | 9.07E-08 | −0.44 | 1.23E-09 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 204105_s_at | 7 | NRCAM | 0.43 | 1.65E-08 | −0.28 | 1.75E-04 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 204880_at | 10 | MGMT | −0.41 | 1.22E-07 | 0.40 | 5.49E-08 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 205925_s_at | 1 | RAB3B | 0.39 | 3.94E-07 | −0.29 | 1.35E-04 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 205931_s_at | 7 | CREB5 | 0.38 | 9.37E-07 | −0.26 | 5.71E-04 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 210058_at | 6 | MAPK13 | 0.39 | 3.20E-07 | −0.27 | 3.09E-04 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 210059_s_at | 6 | MAPK13 | 0.46 | 1.29E-09 | −0.31 | 4.70E-05 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 213056_at | 3 | FRMD4B | 0.39 | 5.54E-07 | −0.30 | 6.37E-05 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 216959_x_at | 7 | NRCAM | 0.43 | 1.54E-08 | −0.29 | 9.36E-05 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 218651_s_at | 15 | LARP6 | 0.40 | 2.39E-07 | −0.31 | 3.71E-05 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 218806_s_at | 1 | VAV3 | 0.41 | 8.14E-08 | −0.37 | 5.79E-07 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 218807_at | 1 | VAV3 | 0.49 | 9.16E-11 | −0.39 | 1.49E-07 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 227123_at | 1 | RAB3B | 0.47 | 4.56E-10 | −0.26 | 4.85E-04 | −0.27 | 8.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.102 | 232280_at | 14 | SLC25A29 | 0.39 | 3.24E-07 | −0.35 | 2.99E-06 | −0.27 | 8.11E-04 |
| rs2290344 | 15 | PIGB | coding | 0.254 | 242760_x_at | 15 | PIGB | −0.41 | 1.43E-07 | −0.29 | 8.98E-05 | 0.36 | 3.65E-06 |
| rs8024695 | 15 | PIGB | intron | 0.287 | 242760_x_at | 15 | PIGB | −0.38 | 7.05E-07 | −0.29 | 8.98E-05 | 0.34 | 1.43E-05 |
| rs12188464 | 5 | PIK3R1 | flanking_3UTR | 0.459 | 201334_s_at | 11 | ARHGEF12 | 0.38 | 9.03E-07 | −0.34 | 5.35E-06 | −0.32 | 4.57E-05 |
| rs7713001 | 5 | PIK3R1 | flanking_3UTR | 0.459 | 201334_s_at | 11 | ARHGEF12 | 0.38 | 9.03E-07 | −0.34 | 5.35E-06 | −0.32 | 4.57E-05 |
| rs6625401 | 23 | PJA1 | flanking_3UTR | 0.351 | 214908_s_at | 7 | TRRAP | 0.39 | 4.31E-07 | −0.27 | 3.28E-04 | −0.31 | 8.03E-05 |
| rs2217881 | 1 | PLD5 | intron | 0.08 | 231186_at | 14 | FLJ43390///LOC100128793 | 0.44 | 1.46E-08 | −0.29 | 1.09E-04 | −0.29 | 3.35E-04 |
| rs7313937 | 12 | PTPRR | intron | 0.088 | 211572_s_at | 20 | SLC23A2 | 0.41 | 8.23E-08 | −0.33 | 7.50E-06 | −0.27 | 7.17E-04 |
| rs4896870 | 6 | RAB32 | flanking_5UTR | 0.117 | 223194_s_at | 6 | SLC22A23 | 0.45 | 2.92E-09 | −0.29 | 1.24E-04 | −0.27 | 6.66E-04 |
| rs4896870 | 6 | RAB32 | flanking_5UTR | 0.117 | 242629_at | 1 | --- | 0.40 | 2.83E-07 | −0.28 | 2.21E-04 | −0.27 | 6.66E-04 |
| rs4896870 | 6 | RAB32 | flanking_5UTR | 0.117 | 47560_at | 19 | LPHN1 | 0.38 | 9.73E-07 | −0.28 | 1.73E-04 | −0.27 | 6.66E-04 |
| rs907609 | 11 | SYT8 | coding | 0.082 | 205925_s_at | 1 | RAB3B | 0.38 | 6.49E-07 | −0.29 | 1.35E-04 | −0.28 | 4.12E-04 |
| rs6082527 | 20 | TGM3 | flanking_5UTR | 0.058 | 204880_at | 10 | MGMT | −0.41 | 8.06E-08 | 0.40 | 5.49E-08 | −0.28 | 4.86E-04 |
| rs6082527 | 20 | TGM3 | flanking_5UTR | 0.058 | 209469_at | 4 | GPM6A | 0.42 | 4.67E-08 | −0.35 | 2.08E-06 | −0.28 | 4.86E-04 |
| rs6082527 | 20 | TGM3 | flanking_5UTR | 0.058 | 209470_s_at | 4 | GPM6A | 0.40 | 1.99E-07 | −0.34 | 6.37E-06 | −0.28 | 4.86E-04 |
| rs6082527 | 20 | TGM3 | flanking_5UTR | 0.058 | 224325_at | 10 | FZD8 | 0.41 | 1.01E-07 | −0.30 | 5.69E-05 | −0.28 | 4.86E-04 |
| rs6082527 | 20 | TGM3 | flanking_5UTR | 0.058 | 224499_s_at | 12 | AICDA | −0.40 | 2.07E-07 | 0.31 | 3.71E-05 | −0.28 | 4.86E-04 |
| rs6082527 | 20 | TGM3 | flanking_5UTR | 0.058 | 225784_s_at | 23 | KIAA1166 | −0.43 | 2.33E-08 | 0.28 | 2.41E-04 | −0.28 | 4.86E-04 |
| rs1153942 | 1 | TP53BP2 | intron | 0.118 | 218801_at | 13 | UGCGL2 | −0.45 | 3.26E-09 | −0.25 | 8.80E-04 | 0.27 | 9.38E-04 |
| rs612578 | 19 | ZNF626 | intron | 0.24 | 202092_s_at | 16 | ARL2BP | 0.39 | 3.50E-07 | −0.38 | 2.62E-07 | −0.29 | 3.33E-04 |
| rs11666947 | 19 | ZNF626 | flanking_3UTR | 0.06 | 231186_at | 14 | FLJ43390///LOC100128793 | 0.41 | 1.91E-07 | −0.29 | 1.09E-04 | −0.28 | 6.40E-04 |
35 probe sets (29 unique genes) associated with 19 SNPs (16 unique SNP-nearby genes) were significantly correlated with gemcitabine IC50. Genes that were associated with the two SNPs in IQGAP2 and TGM3 are bolded. p-value for each association are indicated. R values represent the correlation coefficient for each association.
Table 2. Top candidate SNPs and probe sets that were associated with AraC IC50.
| SNPs | Probesets | SNP vs Expression(54K) | Expression (54K) vs GI50 | SNP vs GI50 | |||||||||
| rsID | Chr | Closest Gene | SNP Location | MAF | Probeset | Chr | Gene Symbol | r-value | p-value | r-value | p-value | r-value | p-value |
| rs11215406 | 11 | IGSF4 | intron | 0.139 | 1556095_at | 15 | UNC13C | 0.394 | 3.22E-07 | −0.351 | 7.64E-06 | −0.354 | 1.60E-06 |
| rs11215406 | 11 | IGSF4 | intron | 0.139 | 1569969_a_at | 15 | UNC13C | 0.379 | 9.91E-07 | −0.351 | 7.64E-06 | −0.332 | 7.56E-06 |
| rs10208516 | 2 | CTNNA2 | flanking_3UTR | 0.052 | 214018_at | 12 | GRIP1 | 0.410 | 9.73E-08 | 0.342 | 1.37E-05 | 0.270 | 3.23E-04 |
| rs10208516 | 2 | CTNNA2 | flanking_3UTR | 0.052 | 1565812_at | 5 | TRIM36 | 0.414 | 7.34E-08 | 0.342 | 1.37E-05 | 0.268 | 3.53E-04 |
| rs10208516 | 2 | CTNNA2 | flanking_3UTR | 0.052 | 205088_at | 23 | MAMLD1 | 0.409 | 1.02E-07 | 0.342 | 1.37E-05 | 0.263 | 4.64E-04 |
| rs10208516 | 2 | CTNNA2 | flanking_3UTR | 0.052 | 203443_at | 11 | EML3 | 0.422 | 3.57E-08 | 0.342 | 1.37E-05 | 0.255 | 6.84E-04 |
| rs10208516 | 2 | CTNNA2 | flanking_3UTR | 0.052 | 206696_at | 23 | GPR143 | 0.413 | 7.89E-08 | 0.342 | 1.37E-05 | 0.248 | 9.80E-04 |
| rs888468 | 12 | FGF6 | flanking_3UTR | 0.182 | 211572_s_at | 20 | SLC23A2 | 0.405 | 1.43E-07 | −0.341 | 1.45E-05 | −0.264 | 4.25E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 209756_s_at | 2 | MYCN | 0.421 | 4.04E-08 | 0.328 | 3.15E-05 | 0.312 | 2.73E-05 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 1553755_at | 19 | NXNL1 | 0.395 | 3.14E-07 | 0.328 | 3.15E-05 | 0.312 | 2.82E-05 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 1554782_at | 2 | C2orf19 | 0.453 | 2.64E-09 | 0.328 | 3.15E-05 | 0.304 | 4.58E-05 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 242071_x_at | 10 | ITGA8 | 0.411 | 8.62E-08 | 0.328 | 3.15E-05 | 0.276 | 2.22E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 213417_at | 17 | TBX2 | 0.475 | 3.21E-10 | 0.328 | 3.15E-05 | 0.274 | 2.51E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 240729_at | 3 | C3orf44 | 0.382 | 7.93E-07 | 0.328 | 3.15E-05 | 0.271 | 3.04E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 214018_at | 12 | GRIP1 | 0.490 | 7.06E-11 | 0.328 | 3.15E-05 | 0.270 | 3.23E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 1565812_at | 5 | TRIM36 | 0.461 | 1.25E-09 | 0.328 | 3.15E-05 | 0.268 | 3.53E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 1569002_x_at | 8 | BMP1 | 0.404 | 1.50E-07 | 0.328 | 3.15E-05 | 0.266 | 3.80E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 205947_s_at | 7 | VIPR2 | 0.446 | 4.84E-09 | 0.328 | 3.15E-05 | 0.265 | 4.20E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 205088_at | 23 | MAMLD1 | 0.499 | 2.89E-11 | 0.328 | 3.15E-05 | 0.263 | 4.64E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 235820_at | 12 | LOC100130219 | 0.430 | 1.89E-08 | 0.328 | 3.15E-05 | 0.262 | 4.90E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 215527_at | 6 | KHDRBS2 | 0.428 | 2.17E-08 | 0.328 | 3.15E-05 | 0.259 | 5.71E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 244036_at | NA | --- | 0.480 | 2.01E-10 | 0.328 | 3.15E-05 | 0.257 | 6.26E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 203443_at | 11 | EML3 | 0.438 | 9.32E-09 | 0.328 | 3.15E-05 | 0.255 | 6.84E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 230249_at | 8 | KHDRBS3 | 0.423 | 3.49E-08 | 0.328 | 3.15E-05 | 0.255 | 6.96E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 215953_at | 1 | DKFZP564C196 | 0.387 | 5.72E-07 | 0.328 | 3.15E-05 | 0.252 | 8.12E-04 |
| rs11136070 | 8 | DUSP4 | flanking_5UTR | 0.052 | 206696_at | 23 | GPR143 | 0.455 | 2.19E-09 | 0.328 | 3.15E-05 | 0.248 | 9.80E-04 |
| rs970084 | 20 | SNPH | flanking_5UTR | 0.081 | 222895_s_at | 14 | BCL11B | 0.416 | 5.93E-08 | −0.327 | 3.21E-05 | −0.271 | 3.03E-04 |
| rs970084 | 20 | SNPH | flanking_5UTR | 0.081 | 219528_s_at | 14 | BCL11B | 0.390 | 4.46E-07 | −0.327 | 3.21E-05 | −0.269 | 3.36E-04 |
| rs12412561 | 10 | PCDH15 | flanking_3UTR | 0.069 | 205924_at | 1 | RAB3B | 0.416 | 6.00E-08 | −0.321 | 4.68E-05 | −0.265 | 4.15E-04 |
| rs12128558 | 1 | NPHP4 | flanking_3UTR | 0.055 | 1569419_at | NA | --- | 0.441 | 7.30E-09 | 0.317 | 5.99E-05 | 0.333 | 6.90E-06 |
| rs12128558 | 1 | NPHP4 | flanking_3UTR | 0.055 | 234896_at | NA | --- | 0.465 | 8.89E-10 | 0.317 | 5.99E-05 | 0.259 | 5.49E-04 |
| rs11876487 | 18 | GATA6 | flanking_3UTR | 0.069 | 205947_s_at | 7 | VIPR2 | 0.406 | 1.28E-07 | 0.313 | 7.16E-05 | 0.265 | 4.20E-04 |
| rs11876487 | 18 | GATA6 | flanking_3UTR | 0.069 | 215953_at | 1 | DKFZP564C196 | 0.403 | 1.68E-07 | 0.313 | 7.16E-05 | 0.252 | 8.12E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 1554782_at | 2 | C2orf19 | 0.402 | 1.78E-07 | 0.313 | 7.52E-05 | 0.304 | 4.58E-05 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 206401_s_at | 17 | MAPT | 0.387 | 5.62E-07 | 0.313 | 7.52E-05 | 0.297 | 6.81E-05 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 230666_at | 7 | tcag7.1238 | 0.401 | 1.95E-07 | 0.313 | 7.52E-05 | 0.288 | 1.19E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 242071_x_at | 10 | ITGA8 | 0.385 | 6.44E-07 | 0.313 | 7.52E-05 | 0.276 | 2.22E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 213417_at | 17 | TBX2 | 0.436 | 1.17E-08 | 0.313 | 7.52E-05 | 0.274 | 2.51E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 214018_at | 12 | GRIP1 | 0.451 | 3.20E-09 | 0.313 | 7.52E-05 | 0.270 | 3.23E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 1565812_at | 5 | TRIM36 | 0.421 | 4.11E-08 | 0.313 | 7.52E-05 | 0.268 | 3.53E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 1569002_x_at | 8 | BMP1 | 0.407 | 1.27E-07 | 0.313 | 7.52E-05 | 0.266 | 3.80E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 205947_s_at | 7 | VIPR2 | 0.461 | 1.20E-09 | 0.313 | 7.52E-05 | 0.265 | 4.20E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 205088_at | 23 | MAMLD1 | 0.438 | 1.00E-08 | 0.313 | 7.52E-05 | 0.263 | 4.64E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 235820_at | 12 | LOC100130219 | 0.398 | 2.52E-07 | 0.313 | 7.52E-05 | 0.262 | 4.90E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 215527_at | 6 | KHDRBS2 | 0.403 | 1.62E-07 | 0.313 | 7.52E-05 | 0.259 | 5.71E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 215953_at | 1 | DKFZP564C196 | 0.426 | 2.72E-08 | 0.313 | 7.52E-05 | 0.252 | 8.12E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 224168_at | NA | TXNDC2 | 0.383 | 7.59E-07 | 0.313 | 7.52E-05 | 0.251 | 8.19E-04 |
| rs613359 | 4 | LOC132321 | flanking_3UTR | 0.061 | 206696_at | 23 | GPR143 | 0.379 | 9.67E-07 | 0.313 | 7.52E-05 | 0.248 | 9.80E-04 |
| rs10780183 | 9 | EHMT1 | flanking_5UTR | 0.127 | 225532_at | 18 | CABLES1 | −0.386 | 5.90E-07 | −0.302 | 1.36E-04 | 0.271 | 2.92E-04 |
| rs7612441 | 3 | LRIG1 | flanking_5UTR | 0.145 | 236565_s_at | 15 | LARP6 | 0.445 | 5.28E-09 | −0.298 | 1.69E-04 | −0.274 | 2.49E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 204880_at | 10 | MGMT | −0.407 | 1.22E-07 | −0.296 | 1.85E-04 | 0.332 | 7.44E-06 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 204105_s_at | 7 | NRCAM | 0.432 | 1.65E-08 | −0.296 | 1.85E-04 | −0.297 | 6.80E-05 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 218651_s_at | 15 | LARP6 | 0.398 | 2.39E-07 | −0.296 | 1.85E-04 | −0.289 | 1.11E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 210058_at | 6 | MAPK13 | 0.394 | 3.20E-07 | −0.296 | 1.85E-04 | −0.284 | 1.48E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 218807_at | 1 | VAV3 | 0.488 | 9.16E-11 | −0.296 | 1.85E-04 | −0.279 | 1.95E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 203726_s_at | 18 | LAMA3 | 0.411 | 9.07E-08 | −0.296 | 1.85E-04 | −0.277 | 2.15E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 216959_x_at | 7 | NRCAM | 0.433 | 1.54E-08 | −0.296 | 1.85E-04 | −0.274 | 2.52E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 210059_s_at | 6 | MAPK13 | 0.461 | 1.29E-09 | −0.296 | 1.85E-04 | −0.260 | 5.40E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 213056_at | 3 | FRMD4B | 0.387 | 5.54E-07 | −0.296 | 1.85E-04 | −0.259 | 5.45E-04 |
| rs3797418 | 5 | IQGAP2 | intron | 0.101 | 218806_s_at | 1 | VAV3 | 0.412 | 8.14E-08 | −0.296 | 1.85E-04 | −0.248 | 9.46E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 204880_at | 10 | MGMT | −0.382 | 8.06E-07 | −0.296 | 1.88E-04 | 0.332 | 7.44E-06 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 204866_at | 23 | PHF16 | 0.392 | 3.94E-07 | −0.296 | 1.88E-04 | −0.311 | 3.00E-05 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 204105_s_at | 7 | NRCAM | 0.421 | 4.15E-08 | −0.296 | 1.88E-04 | −0.297 | 6.80E-05 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 218807_at | 1 | VAV3 | 0.451 | 3.04E-09 | −0.296 | 1.88E-04 | −0.279 | 1.95E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 203726_s_at | 18 | LAMA3 | 0.404 | 1.54E-07 | −0.296 | 1.88E-04 | −0.277 | 2.15E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 216959_x_at | 7 | NRCAM | 0.416 | 5.89E-08 | −0.296 | 1.88E-04 | −0.274 | 2.52E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 222548_s_at | 2 | MAP4K4 | 0.382 | 7.91E-07 | −0.296 | 1.88E-04 | −0.273 | 2.63E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 218181_s_at | 2 | MAP4K4 | 0.381 | 8.73E-07 | −0.296 | 1.88E-04 | −0.271 | 2.94E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 203922_s_at | 23 | CYBB | −0.393 | 3.45E-07 | −0.296 | 1.88E-04 | 0.265 | 4.12E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 210059_s_at | 6 | MAPK13 | 0.382 | 7.87E-07 | −0.296 | 1.88E-04 | −0.260 | 5.40E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 213056_at | 3 | FRMD4B | 0.379 | 9.66E-07 | −0.296 | 1.88E-04 | −0.259 | 5.45E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 239202_at | NA | --- | 0.414 | 7.17E-08 | −0.296 | 1.88E-04 | −0.250 | 8.71E-04 |
| rs17637119 | 3 | CADPS | intron | 0.069 | 218806_s_at | 1 | VAV3 | 0.431 | 1.81E-08 | −0.296 | 1.88E-04 | −0.248 | 9.46E-04 |
| rs11796743 | 23 | GSPT2 | flanking_5UTR | 0.061 | 1553755_at | 19 | NXNL1 | 0.420 | 4.75E-08 | 0.288 | 2.90E-04 | 0.312 | 2.82E-05 |
| rs1408077 | 1 | CR1 | intron | 0.195 | 1567320_at | NA | LOC57802 | 0.403 | 1.76E-07 | 0.283 | 3.85E-04 | 0.249 | 9.14E-04 |
| rs5962901 | 23 | MID2 | intron | 0.067 | 226529_at | 7 | TMEM106B | −0.380 | 9.95E-07 | 0.280 | 4.38E-04 | −0.276 | 2.23E-04 |
| rs5962901 | 23 | MID2 | intron | 0.067 | 213417_at | 17 | TBX2 | 0.408 | 1.26E-07 | 0.280 | 4.38E-04 | 0.274 | 2.51E-04 |
| rs5962901 | 23 | MID2 | intron | 0.067 | 214018_at | 12 | GRIP1 | 0.403 | 1.78E-07 | 0.280 | 4.38E-04 | 0.270 | 3.23E-04 |
| rs5962901 | 23 | MID2 | intron | 0.067 | 1565812_at | 5 | TRIM36 | 0.413 | 8.17E-08 | 0.280 | 4.38E-04 | 0.268 | 3.53E-04 |
| rs5962901 | 23 | MID2 | intron | 0.067 | 205088_at | 23 | MAMLD1 | 0.389 | 5.18E-07 | 0.280 | 4.38E-04 | 0.263 | 4.64E-04 |
| rs5962901 | 23 | MID2 | intron | 0.067 | 215527_at | 6 | KHDRBS2 | 0.385 | 6.82E-07 | 0.280 | 4.38E-04 | 0.259 | 5.71E-04 |
| rs5962901 | 23 | MID2 | intron | 0.067 | 206696_at | 23 | GPR143 | 0.432 | 1.73E-08 | 0.280 | 4.38E-04 | 0.248 | 9.80E-04 |
| rs17183491 | 15 | WDR72 | flanking_5UTR | 0.263 | 227067_x_at | 1 | NOTCH2NL | −0.382 | 9.55E-07 | 0.280 | 4.59E-04 | −0.256 | 6.48E-04 |
| rs6491526 | 13 | CLYBL | intron | 0.075 | 214018_at | 12 | GRIP1 | 0.398 | 2.46E-07 | 0.277 | 4.84E-04 | 0.270 | 3.23E-04 |
| rs6491526 | 13 | CLYBL | intron | 0.075 | 1569002_x_at | 8 | BMP1 | 0.382 | 7.76E-07 | 0.277 | 4.84E-04 | 0.266 | 3.80E-04 |
| rs6491526 | 13 | CLYBL | intron | 0.075 | 210961_s_at | 20 | ADRA1D | 0.380 | 9.07E-07 | 0.277 | 4.84E-04 | 0.254 | 7.25E-04 |
| rs1669748 | 5 | ADAMTS16 | flanking_5UTR | 0.462 | 239202_at | NA | --- | 0.394 | 3.21E-07 | −0.277 | 4.93E-04 | −0.250 | 8.71E-04 |
| rs11706227 | 3 | WNT5A | flanking_3UTR | 0.061 | 1560397_s_at | NA | KLHL6 | −0.384 | 6.71E-07 | 0.274 | 5.61E-04 | −0.286 | 1.33E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 1554782_at | 2 | C2orf19 | 0.488 | 9.19E-11 | 0.272 | 6.23E-04 | 0.304 | 4.58E-05 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 1570330_at | NA | --- | 0.386 | 5.97E-07 | 0.272 | 6.23E-04 | 0.284 | 1.42E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 214018_at | 12 | GRIP1 | 0.424 | 3.25E-08 | 0.272 | 6.23E-04 | 0.270 | 3.23E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 1565812_at | 5 | TRIM36 | 0.423 | 3.44E-08 | 0.272 | 6.23E-04 | 0.268 | 3.53E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 205947_s_at | 7 | VIPR2 | 0.384 | 6.71E-07 | 0.272 | 6.23E-04 | 0.265 | 4.20E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 205088_at | 23 | MAMLD1 | 0.379 | 9.83E-07 | 0.272 | 6.23E-04 | 0.263 | 4.64E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 235820_at | 12 | LOC100130219 | 0.446 | 4.75E-09 | 0.272 | 6.23E-04 | 0.262 | 4.90E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 215527_at | 6 | KHDRBS2 | 0.463 | 9.78E-10 | 0.272 | 6.23E-04 | 0.259 | 5.71E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 203443_at | 11 | EML3 | 0.401 | 1.89E-07 | 0.272 | 6.23E-04 | 0.255 | 6.84E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 230249_at | 8 | KHDRBS3 | 0.381 | 8.25E-07 | 0.272 | 6.23E-04 | 0.255 | 6.96E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 215953_at | 1 | DKFZP564C196 | 0.401 | 1.99E-07 | 0.272 | 6.23E-04 | 0.252 | 8.12E-04 |
| rs9833533 | 3 | FHIT | intron | 0.058 | 206696_at | 23 | GPR143 | 0.446 | 4.87E-09 | 0.272 | 6.23E-04 | 0.248 | 9.80E-04 |
| rs13197839 | 6 | EGFL11 | flanking_3UTR | 0.119 | 200972_at | 15 | TSPAN3 | −0.382 | 8.52E-07 | 0.269 | 7.47E-04 | −0.266 | 3.94E-04 |
| rs5984367 | 23 | TGIF2LX | flanking_5UTR | 0.098 | 207278_s_at | 19 | CD209 | 0.424 | 3.09E-08 | 0.268 | 7.48E-04 | 0.289 | 1.10E-04 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 209756_s_at | 2 | MYCN | 0.441 | 7.77E-09 | 0.267 | 7.95E-04 | 0.312 | 2.73E-05 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 230666_at | 7 | tcag7.1238 | 0.400 | 2.12E-07 | 0.267 | 7.95E-04 | 0.288 | 1.19E-04 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 214454_at | 5 | ADAMTS2 | 0.395 | 3.13E-07 | 0.267 | 7.95E-04 | 0.274 | 2.55E-04 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 214018_at | 12 | GRIP1 | 0.426 | 2.70E-08 | 0.267 | 7.95E-04 | 0.270 | 3.23E-04 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 1565812_at | 5 | TRIM36 | 0.456 | 1.98E-09 | 0.267 | 7.95E-04 | 0.268 | 3.53E-04 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 205088_at | 23 | MAMLD1 | 0.446 | 4.60E-09 | 0.267 | 7.95E-04 | 0.263 | 4.64E-04 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 235820_at | 12 | LOC100130219 | 0.442 | 6.92E-09 | 0.267 | 7.95E-04 | 0.262 | 4.90E-04 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 203443_at | 11 | EML3 | 0.424 | 3.05E-08 | 0.267 | 7.95E-04 | 0.255 | 6.84E-04 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 244522_at | 11 | SYVN1 | 0.407 | 1.19E-07 | 0.267 | 7.95E-04 | 0.248 | 9.64E-04 |
| rs11770570 | 7 | LHFPL3 | intron | 0.055 | 206696_at | 23 | GPR143 | 0.422 | 3.65E-08 | 0.267 | 7.95E-04 | 0.248 | 9.80E-04 |
| rs11063679 | 12 | NTF3 | flanking_5UTR | 0.075 | 205088_at | 23 | MAMLD1 | 0.461 | 1.19E-09 | 0.265 | 8.56E-04 | 0.263 | 4.64E-04 |
| rs5768857 | 22 | CELSR1 | intron | 0.127 | 209310_s_at | 11 | CASP4 | 0.381 | 8.40E-07 | 0.265 | 8.57E-04 | 0.267 | 3.65E-04 |
| rs4730550 | 7 | IFRD1 | intron | 0.064 | 218229_s_at | 1 | POGK | −0.401 | 2.18E-07 | 0.266 | 8.73E-04 | −0.256 | 6.58E-04 |
| rs8180101 | 3 | GBE1 | flanking_5UTR | 0.081 | 205982_x_at | 8 | SFTPC | 0.393 | 3.64E-07 | 0.264 | 8.91E-04 | 0.248 | 9.83E-04 |
| rs772492 | 4 | LOC285513 | flanking_5UTR | 0.173 | 213056_at | 3 | FRMD4B | 0.379 | 9.81E-07 | −0.264 | 8.98E-04 | −0.259 | 5.45E-04 |
| rs549467 | 4 | RAP1GDS1 | flanking_5UTR | 0.069 | 216030_s_at | 20 | SEMG2 | 0.436 | 1.15E-08 | 0.263 | 9.43E-04 | 0.332 | 7.74E-06 |
60 probe sets (54 unique genes) associated with 30 SNPs (30 unique SNP-nearby genes) were significantly correlated with AraC IC50. Genes that were associated with SNPs in IQGAP2 are bolded for AraC, too. p-value for each association are indicated. R values represent the correlation coefficient for each association.
Cytidine Analogue Pathway Gene Analysis
Gemcitabine and AraC share the same metabolism pathway that activates these drugs in cells. There is a total of 19 genes within this pathway (Table S6). Many previous pharmacogenetic studies have been focused on genetic variation in genes within this pathway, and there is evidence showing that variation in the expression of genes within this pathway can influence response to cytidine analogues [7], [29], [30]. Therefore, we took the advantage of the genome-wide SNP data and selected all SNPs within pathway genes that were present on the Illumina 550K chip to perform an analysis among SNP, expression and gemcitabine and AraC IC50 values for these pathway genes. A total of 749 SNPs for cytidine analogue pathway genes were present on the Illumina 550K chip. Association of gemcitabine IC50 with SNPs in pathway genes identified 38 SNPs within or close to 8 genes with p-values <0.05, while 23 SNPs close to 9 genes were significantly associated with AraC IC50 values with p-values <0.05 ( Tables 3 and 4 ). To determine the association of SNPs with cis-regulation of pathway gene expression, we associated all of these SNPs with expression for all genes within the pathway. That analysis identified 24 cis-acting SNPs that regulate the expression of pathway genes with p-values <0.0001 ( Table 5 ). The top three SNPs (p = 7.54×10−33, 1.54×10−32 and 2.69×10−10, respectively) were all mapped to the NT5C3L1 gene, a nucleotidase family member ( Table 5 ).
Table 3. Top 38 significant SNPs within or close to 8 pathway genes associated with gemcitabine IC50 values (p<0.05).
| SNP | r-value | p-value | q-value | N | MAF | Chr | Position | Genome Build | Gene Symbol | Gene | Location | Location Relative to Gene |
| rs2037067 | 0.269 | 0.001 | 0.82119 | 170 | 0.471 | 4 | 1.84E+08 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −237660 |
| rs3775289 | 0.227 | 0.005 | 0.82119 | 170 | 0.203 | 4 | 72081538 | 36.1 | DCK | NM_000788.1 | Intron | −1110 |
| rs4308342 | 0.227 | 0.005 | 0.82119 | 170 | 0.203 | 4 | 72103069 | 36.1 | DCK | NM_000788.1 | Intron | −3879 |
| rs956727 | −0.218 | 0.007 | 0.82119 | 171 | 0.117 | 9 | 86036753 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −46159 |
| rs515583 | 0.211 | 0.009 | 0.82119 | 171 | 0.19 | 6 | 85933010 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −283518 |
| rs13144500 | −0.205 | 0.011 | 0.82119 | 171 | 0.123 | 4 | 1.84E+08 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −335310 |
| rs12500335 | −0.202 | 0.013 | 0.82119 | 170 | 0.091 | 4 | 1.84E+08 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −103463 |
| rs3966882 | 0.202 | 0.013 | 0.82119 | 168 | 0.318 | 6 | 85938190 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −278338 |
| rs10006225 | 0.200 | 0.013 | 0.82119 | 171 | 0.064 | 4 | 1.84E+08 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −102248 |
| rs1932660 | 0.200 | 0.013 | 0.82119 | 171 | 0.234 | 9 | 86017079 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −65833 |
| rs4877822 | 0.200 | 0.013 | 0.82119 | 171 | 0.234 | 9 | 86021002 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −61910 |
| rs7874528 | −0.198 | 0.014 | 0.82119 | 171 | 0.17 | 9 | 86020319 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −62593 |
| rs9472236 | 0.196 | 0.015 | 0.82119 | 171 | 0.225 | 6 | 44314728 | 36.1 | SLC29A1 | NM_004955.1 | flanking_3UTR | −4872 |
| rs1570933 | 0.196 | 0.015 | 0.82119 | 171 | 0.292 | 6 | 85937335 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −279193 |
| rs17087049 | −0.194 | 0.017 | 0.82119 | 171 | 0.208 | 9 | 86104192 | 36.1 | SLC28A3 | NM_022127.1 | Intron | −124 |
| rs10780659 | −0.193 | 0.017 | 0.82119 | 171 | 0.146 | 9 | 86071310 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −11602 |
| rs7173860 | 0.192 | 0.017 | 0.82119 | 171 | 0.465 | 15 | 83230545 | 36.1 | SLC28A1 | NM_004213.3 | Intron | −362 |
| rs7597224 | 0.189 | 0.019 | 0.85358 | 171 | 0.342 | 2 | 18829103 | 36.1 | NT5C1B | NM_001002006.1 | flanking_5UTR | −194784 |
| rs12683783 | −0.188 | 0.020 | 0.82119 | 171 | 0.126 | 9 | 86060628 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −22284 |
| rs10122651 | −0.188 | 0.020 | 0.82119 | 171 | 0.468 | 9 | 86123704 | 36.1 | SLC28A3 | NM_022127.1 | Intron | −5515 |
| rs9294329 | −0.187 | 0.021 | 0.82119 | 171 | 0.295 | 6 | 85958337 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −258191 |
| rs10780663 | −0.186 | 0.022 | 0.82119 | 170 | 0.465 | 9 | 86120882 | 36.1 | SLC28A3 | NM_022127.1 | Intron | −2693 |
| rs9362176 | −0.181 | 0.025 | 0.82119 | 171 | 0.152 | 6 | 86036174 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −180354 |
| rs4585823 | −0.179 | 0.026 | 0.82119 | 171 | 0.132 | 9 | 86094005 | 36.1 | SLC28A3 | NM_022127.1 | Intron | −884 |
| rs12684950 | −0.178 | 0.027 | 0.82119 | 171 | 0.155 | 9 | 86066751 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −16161 |
| rs556388 | 0.178 | 0.028 | 0.82119 | 171 | 0.447 | 8 | 1.03E+08 | 36.1 | RRM2B | NM_015713.3 | flanking_3UTR | −110177 |
| rs10955282 | 0.179 | 0.028 | 0.82119 | 170 | 0.488 | 8 | 1.03E+08 | 36.1 | RRM2B | NM_015713.3 | flanking_3UTR | −39785 |
| rs10780656 | 0.178 | 0.028 | 0.82998 | 171 | 0.196 | 9 | 86031222 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −51690 |
| rs1321742 | −0.175 | 0.032 | 0.82119 | 170 | 0.444 | 6 | 85937908 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −278620 |
| rs501344 | 0.172 | 0.034 | 0.83550 | 171 | 0.327 | 8 | 1.03E+08 | 36.1 | RRM2B | NM_015713.3 | flanking_3UTR | −102158 |
| rs3743162 | 0.171 | 0.035 | 0.82119 | 171 | 0.129 | 15 | 83231973 | 36.1 | SLC28A1 | NM_201651.1 | Intron | −7 |
| rs11885014 | −0.173 | 0.035 | 0.82119 | 167 | 0.272 | 2 | 18810796 | 36.1 | NT5C1B | NM_001002006.1 | flanking_5UTR | −176477 |
| rs9990999 | 0.170 | 0.037 | 0.83550 | 169 | 0.352 | 4 | 1.84E+08 | 36.1 | DCTD | NM_001921.2 | Intron | −8215 |
| rs944664 | −0.166 | 0.041 | 0.82119 | 171 | 0.444 | 6 | 85945694 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −270834 |
| rs4877824 | 0.165 | 0.042 | 0.82119 | 171 | 0.38 | 9 | 86045679 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −37233 |
| rs2251530 | −0.163 | 0.044 | 0.82119 | 171 | 0.386 | 9 | 86002760 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −80152 |
| rs4507403 | 0.164 | 0.046 | 0.82119 | 166 | 0.41 | 4 | 1.84E+08 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −251819 |
| rs6850978 | 0.159 | 0.050 | 0.82119 | 171 | 0.424 | 4 | 1.84E+08 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −238854 |
Table 4. Top 23 significant SNPs within or close to 9 pathway genes associated with AraC IC50 values (p<0.05).
| SNP | r-value | p-value | q-value | N | MAF | Chr | Position | Genome Build | Gene Symbol | Gene | Location | Location Relative To Gene |
| rs9472236 | 0.240 | 0.003 | 0.99971 | 173 | 0.225 | 6 | 44314728 | 36.1 | SLC29A1 | NM_004955.1 | flanking_3UTR | −4872 |
| rs2037067 | 0.222 | 0.006 | 0.99971 | 172 | 0.471 | 4 | 183810578 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −237660 |
| rs11999726 | −0.219 | 0.006 | 0.99971 | 173 | 0.073 | 9 | 86104976 | 36.1 | SLC28A3 | NM_022127.1 | intron | −516 |
| rs7830150 | −0.213 | 0.008 | 0.99971 | 173 | 0.187 | 8 | 103162269 | 36.1 | RRM2B | NM_015713.3 | flanking_3UTR | −123638 |
| rs1614627 | −0.196 | 0.015 | 0.99971 | 173 | 0.17 | 1 | 20819343 | 36.1 | CDA | NM_001785.1 | flanking_3UTR | −1360 |
| rs2224211 | 0.192 | 0.017 | 0.99971 | 173 | 0.196 | 6 | 85619788 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −596740 |
| rs7874528 | −0.183 | 0.023 | 0.99971 | 173 | 0.17 | 9 | 86020319 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −62593 |
| rs17414857 | −0.182 | 0.023 | 0.99971 | 173 | 0.094 | 8 | 103139477 | 36.1 | RRM2B | NM_015713.3 | flanking_3UTR | −146430 |
| rs4877839 | −0.182 | 0.024 | 0.99971 | 173 | 0.12 | 9 | 86118829 | 36.1 | SLC28A3 | NM_022127.1 | intron | −640 |
| rs17343066 | 0.181 | 0.024 | 0.99971 | 173 | 0.36 | 9 | 86135893 | 36.1 | SLC28A3 | NM_022127.1 | intron | −9416 |
| rs12526113 | −0.181 | 0.024 | 0.99971 | 173 | 0.33 | 6 | 85612351 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −604177 |
| rs2324753 | −0.181 | 0.024 | 0.99971 | 173 | 0.33 | 6 | 85615181 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −601347 |
| rs12554292 | −0.180 | 0.025 | 0.99971 | 173 | 0.135 | 9 | 86256385 | 36.1 | SLC28A3 | NM_022127.1 | flanking_5UTR | −83152 |
| rs1570311 | −0.175 | 0.029 | 0.99971 | 173 | 0.161 | 6 | 86211870 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −4658 |
| rs4487548 | −0.175 | 0.029 | 0.99971 | 173 | 0.161 | 6 | 86199130 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −17398 |
| rs9344525 | −0.175 | 0.029 | 0.99971 | 173 | 0.161 | 6 | 86208157 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −8371 |
| rs6850978 | 0.171 | 0.034 | 0.99971 | 173 | 0.424 | 4 | 183809384 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −238854 |
| rs9450270 | −0.170 | 0.034 | 0.99971 | 173 | 0.187 | 6 | 86171485 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −45043 |
| rs3743162 | 0.170 | 0.034 | 0.99971 | 173 | 0.129 | 15 | 83231973 | 36.1 | SLC28A1 | NM_201651.1 | intron | −7 |
| rs1265138 | 0.165 | 0.042 | 0.99971 | 171 | 0.148 | 8 | 103269212 | 36.1 | RRM2B | NM_015713.3 | flanking_3UTR | −16695 |
| rs1569040 | 0.163 | 0.043 | 0.99971 | 172 | 0.147 | 17 | 17246812 | 36.1 | NT5M | NM_020201.3 | flanking_3UTR | −55110 |
| rs12500335 | −0.162 | 0.044 | 0.99971 | 172 | 0.091 | 4 | 183944775 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −103463 |
| rs11598702 | −0.163 | 0.045 | 0.99971 | 171 | 0.246 | 10 | 104887975 | 36.1 | NT5C2 | NM_012229.2 | intron | −1178 |
Table 5. 24 candidate SNPs with cis-regulation of pathway genes (p<0.0001).
| SNP | Probeset | r-value (SNP vs Exp) | p-value (SNP vs Exp) | SNP | MAF | Chr | Position | Genome Build | Gene Symbol | Gene | Location | Location Relative to Gene |
| rs1319763 | 225044_at | 0.6435 | 7.54E-33 | rs1319763 | 0.22 | 17 | 37245316 | 36.1 | NT5C3L | NM_052935.2 | intron | −266 |
| rs1046403 | 225044_at | 0.6420 | 1.54E-32 | rs1046403 | 0.219 | 17 | 37237346 | 36.1 | NT5C3L | NM_052935.2 | coding | [142/58] |
| rs4796712 | 225044_at | 0.3728 | 2.69E-10 | rs4796712 | 0.099 | 17 | 37240656 | 36.1 | NT5C3L | NM_052935.2 | coding | [77/12] |
| rs9344525 | 227486_at | −0.3034 | 3.91E-07 | rs9344525 | 0.167 | 6 | 86208157 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −8371 |
| rs1570311 | 227486_at | −0.3034 | 3.91E-07 | rs1570311 | 0.167 | 6 | 86211870 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −4658 |
| rs4487548 | 227486_at | −0.3034 | 3.91E-07 | rs4487548 | 0.167 | 6 | 86199130 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −17398 |
| rs9344525 | 203939_at | −0.2821 | 2.59E-06 | rs9344525 | 0.167 | 6 | 86208157 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −8371 |
| rs1570311 | 203939_at | −0.2821 | 2.59E-06 | rs1570311 | 0.167 | 6 | 86211870 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −4658 |
| rs4487548 | 203939_at | −0.2821 | 2.59E-06 | rs4487548 | 0.167 | 6 | 86199130 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −17398 |
| rs7277 | 201572_x_at | −0.2768 | 4.20E-06 | rs7277 | 0.341 | 4 | 184048507 | 36.1 | DCTD | NM_001012732.1 | 3UTR | [269/1038] |
| rs9344525 | 1553995_a_at | −0.2709 | 6.56E-06 | rs9344525 | 0.167 | 6 | 86208157 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −8371 |
| rs1570311 | 1553995_a_at | −0.2709 | 6.56E-06 | rs1570311 | 0.167 | 6 | 86211870 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −4658 |
| rs4487548 | 1553995_a_at | −0.2709 | 6.56E-06 | rs4487548 | 0.167 | 6 | 86199130 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −17398 |
| rs4910907 | 201477_s_at | −0.2705 | 6.81E-06 | rs4910907 | 0.167 | 11 | 4125427 | 36.1 | RRM1 | NM_001033.2 | flanking_3UTR | −8745 |
| rs9907244 | 225044_at | −0.2650 | 1.05E-05 | rs9907244 | 0.455 | 17 | 37234426 | 36.1 | NT5C3L | NM_052935.2 | flanking_3UTR | −558 |
| rs9344525 | 1553994_at | −0.2621 | 1.33E-05 | rs9344525 | 0.167 | 6 | 86208157 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −8371 |
| rs1570311 | 1553994_at | −0.2621 | 1.33E-05 | rs1570311 | 0.167 | 6 | 86211870 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −4658 |
| rs4487548 | 1553994_at | −0.2621 | 1.33E-05 | rs4487548 | 0.167 | 6 | 86199130 | 36.1 | NT5E | NM_002526.1 | flanking_5UTR | −17398 |
| rs2250159 | 203302_at | −0.2613 | 1.46E-05 | rs2250159 | 0.369 | 9 | 85966866 | 36.1 | SLC28A3 | NM_022127.1 | flanking_3UTR | −116046 |
| rs1474500 | 201477_s_at | −0.2561 | 2.11E-05 | rs1474500 | 0.166 | 11 | 4080688 | 36.1 | RRM1 | NM_001033.2 | intron | −801 |
| rs12990630 | 1553540_a_at | −0.2518 | 2.94E-05 | rs12990630 | 0.082 | 2 | 18973412 | 36.1 | NT5C1B | NM_001002006.1 | flanking_5UTR | −339093 |
| rs4910896 | 201477_s_at | −0.2492 | 3.58E-05 | rs4910896 | 0.172 | 11 | 4118132 | 36.1 | RRM1 | NM_001033.2 | flanking_3UTR | −1450 |
| rs16845804 | 236703_at | 0.2410 | 6.54E-05 | rs16845804 | 0.221 | 4 | 72208285 | 36.1 | DCK | NM_000788.1 | flanking_3UTR | −92808 |
| rs17271644 | 201572_x_at | −0.2376 | 8.30E-05 | rs17271644 | 0.495 | 4 | 184046266 | 36.1 | DCTD | NM_001012732.1 | flanking_3UTR | −1972 |
Functional Characterization of Candidate Genes
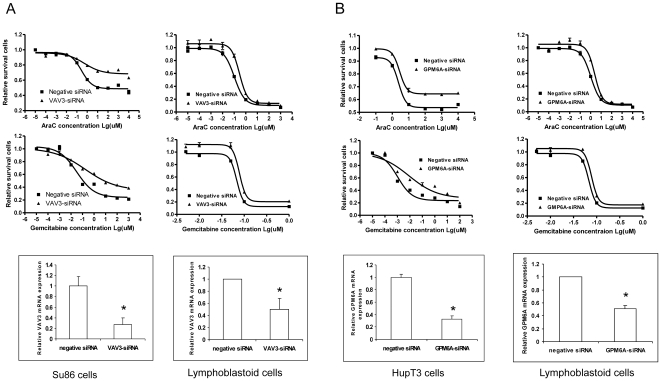
Our association studies were performed with human lymphoblastoid cell lines but the gene regulation is tissue specific [31]. Therefore, to investigate the impact of the SNPs that we had identified on gene expression and on gemcitabine and AraC cytotoxicity and to functionally validate our association results, we also studied tumor cell lines, in this case, two pancreatic cancer cell lines, to validate our association results. We performed siRNA knock down for three SNP-associated genes, including VAV3, MGMT that are associated with the SNP in IQGAP2 and GPM6A which is associated with the SNP in TGM3, to determine whether genes that had expression associated with the two SNPs might influence drug IC50. We selected VAV3 and GPM6A because each of these genes had two probesets associated with rs3797418 and rs6082527, respectively. VAV3 was also highly associated with gemcitabine and AraC IC50 while GPM6A gene expression was associated with gemcitabine IC50 ( Figure 4A and 4B ). As with TGM3, we also performed knock down of GPM6A for AraC. However, the association was most significant in the CA group, which could be due to differences in allele frequencies among the three ethnic groups studied. Expression of the third gene, MGMT, was also associated with both SNPs (rs3797418 with p = 1.22×10−7 and rs6082527 with p = 8.06×10−8) and with gemcitabine and AraC IC50 values (p = 5.49×10−8 and 1.85×10−4, respectively). We also included IQGAP2 and TGM3 in the siRNA knock down studies to determine whether the genes harboring these two SNPs might themselves have functional effects on gemcitabine or AraC cytotoxicity. Finally, we included a positive control siRNA, FKBP5, a gene that we had previously shown to be important for gemcitabine response [25].
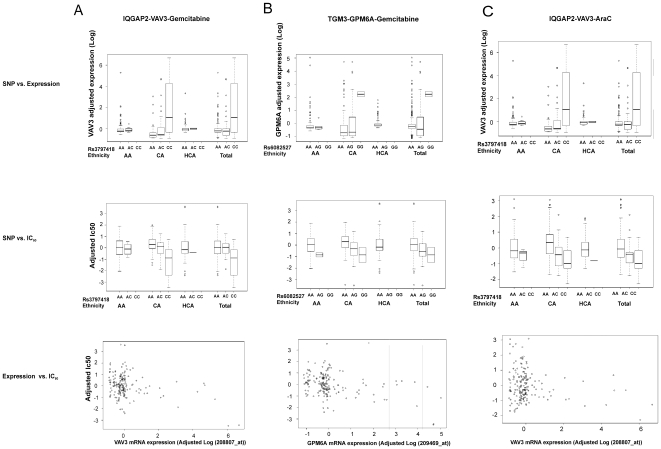
Figure 4. Association among SNPs in IQGAP2 and TGM3 with expression and gemcitabine or AraC cytotoxicity.
(A) SNP rs3797418 (A/C) association with VAV3 expression, gemcitabine IC50 values and VAV3 expression association with gemcitabine IC50 in individual ethnic groups and all of the cell lines. (B) SNP rs6082527 (A/G) association with GPM6A gene expression, gemcitabine IC50 values and GPM6A gene expression association with gemcitabine IC50 values. (C). SNP rs3797418 (A/C) association with VAV3 expression, AraC IC50 values and VAV3 expression association with AraC IC50 in individual ethnic groups and all of the cell lines. Each dot represents one sample. Genotypes for each SNP are plotted against gene expression levels (upper panels) as well as gemcitabine or AraC IC50 values (middle panels). In the lower panel, correlations were determined between gene expression levels and gemcitabine or AraC IC50 values.
Specific and negative control siRNAs were transiently transfected into human SU86 or Hup-T3 pancreatic cancer cell lines, cell lines selected on the basis of endogenous expression level of genes of interest, followed by gemcitabine or AraC cytotoxicity assays. We selected pancreatic cancer cell lines because gemcitabine is the standard of care for the therapy of pancreatic cancer. Down regulation of VAV3 in SU86 cells significantly desensitized the cells to gemcitabine and AraC ( Figure 5A ), consistent with the result of the association study (gemcitabine p = 9.14E-10 and AraC p = 1.24E-11). Down regulation of GPM6A in HupT3 cell line also resulted in desensitization to gemcitabine (p = 1.35E-07), an observation consistent with our association study results for GPM6A gene expression and gemcitabine IC50 ( Figure 5B ). Interestingly, although GPM6A expression was not significantly associated with AraC response, the knock down experiments showed a similar effect for AraC (p = 3.66E-12) ( Figure 5B ). We also performed similar experiments of knock down of these two genes using selected lymphoblastoid cell lines and it showed similar results as in the tumor cell lines ( Figure 5A right panel, AraC p = 0.0074, gemcitabine p = 0.0085; Figure 5B right panel, AraC p = 0.001, gemcitabine p = 0.002). However, down-regulation of MGMT did not alter either gemcitabine or AraC cytotoxicity. Furthermore, knock down of IQGAP2 and TGM3 themselves, did not have significant effect on gemcitabine or AraC cytotoxicity, an observation indicating that the genes harboring these two SNPs might not have a direct effect on cytotoxicity for the two drugs, and that rs3797418 and rs6082527 might just be markers for gemcitabine and/or AraC cytotoxicity.
Figure 5. Functional characterization of candidate genes with siRNA knock down of VAV3 (A) and GPM6A (B).
Knock down of VAV3 and GPM6A expression in human SU86 and Hup-T3 pancreatic cancer cell lines as well as lymphoblastoid cell lines showed increased resistance to gemcitabine and AraC after siRNA knock down as determined by MTS assays. Error bars represent SEM values for 3 independent experiments. Quantitative RT-PCR was performed to assess VAV3 and GPM6A gene expression levels after knock down with specific siRNAs. Results are expressed as % of control. Error bars represent SEM values for 3 independent experiments. * = p<0.05.
Characterization of SNPs in IQGAP2 and TGM3
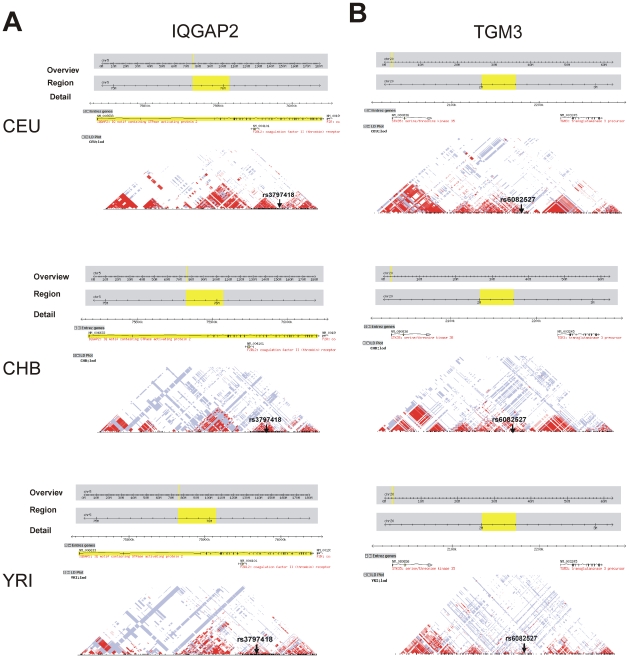
In order to further characterize the two SNPs (rs3797418 and rs6082527) in IQGAP2 and TGM3, we also performed genotype-phenotype correlation studies with the expression of these two genes, i.e., we determined the possible cis-association of the SNPs with IQGAP2 and TGM3 expression. However, neither SNP was associated with the expression of either IQGAP2 or TGM3, indirectly supporting the results of the IQGAP2 and TGM3 knock down experiments. We then determined LD patterns for 500 kb regions around each of the two SNPs using the HapMap data for each ethnic group. As shown in Figure 6 . LD patterns surrounding the two SNPs differed among the three ethnic groups. No SNPs within the 500 kb region around rs3797418 were in high LD with rs3797418. One SNP, rs6047696 in an intergenic region, was in moderate LD with rs6082527 in the CEPH population (r2 = 0.682, D′ = 0.826). In addition, rs12479538, located in an intron of STK35, a serine/threonine kinase 35, was in complete linkage disequilibrium with rs6082527 in the Han Chinese population (r2 = 1, D′ = 1). The p-value for the association between the rs6082527 SNP and gemcitabine IC50 was 0.006, which was not significant after correction for multiple comparisons. Unfortunately, the rs12479538 SNP was not on the 550K SNP array. Obviously, the association of the two SNPs in IQGAP2 and TGM3 with gemcitabine or AraC response and with the expression of other genes might result from the effects of either known or unknown polymorphisms in tight linkage disequilibrium with these two SNPs.
Figure 6. Linkage disequilibrium within ∼300 Kb surrounding the rs3797418 and rs6082527 SNPs.
Red indicates combinations where D′ = 1 and linkage of disequilibrium (LOD) ≥2; light red, combinations where D′<1 and LOD ≥2; red squares, combinations where D′ = 1 and LOD <2; white squares, combinations with D′<1 and LOD <2. SNPs are arranged in order from 5′ to 3′ in each gene, as shown in the gene structure above each plot.
Since these two SNPs, rs3797418 (A/C) and rs6082527 (A/G), were located in the intron of IQGAP2 and the 5′-FR of TGM3, respectively, we determined the relationship among genes associated with these two SNPs. Ingenuity Pathway analysis was performed by mapping the 10 and 8 unique genes that were significantly associated with rs3797418 in IQGAP2 for gemcitabine and AraC, respectively. Results of the Ingenuity Pathway Analysis showed a network that connected all of the 10 genes centered on ERK and TNF for gemcitabine and all of the 8 genes centered on TNF, ERK, TP53 for AraC (Figure S1). However, since only 5 genes were identified to be associated with rs6082527 through our analysis, no network was identified when we analyzed genes that were associated with this SNP.
Discussion
Variation in response to chemotherapeutic agents is a common phenomenon in the clinic. Many factors can contribute to this variation. However, genetic variation is one of the major factors that play an important role in determining response to drugs. Therefore, it is important to identify SNPs that might be used as predictive markers for drug response. In a previous study, we identified candidate genes for which variation in expression levels were associated with variation in response to gemcitabine and AraC using a lymphoblastoid cell line model system [25]. In the current set of experiments, we expanded our study to include genome-wide SNPs to test the hypothesis that SNPs across the genome might contribute significantly to variation in response to two cytidine analogues, gemcitabine and AraC. Lymphoblastoid cell lines with genome-wide SNP and expression data have been used successfully to identify pharmacogenomic candidates associated with response to other chemotherapeutic agents [26], [32]. The advantage of using this cell-based model system is that we have obtained different types of high throughput genomic and transcriptomic data, which makes it possible to test the hypothesis that genetic variation might contribute to variation in drug response, in this case, two cytidine analogues [18], [19], [33], [34]. Many previous pharmacogenetic studies for these two drugs were focused on the gemcitabine bioactivation and metabolism pathway [35], [36]. For example, SNPs identified in genes encoding ribonucleotide reductase (RRM1) and cytidine deaminase (CDA) were found to be associated with gemcitabine chemosensitivity in the NCI60 cell lines or with active gemcitabine plasma metabolites levels [37]–[39]. Those findings provided the initial evidence that genetic variation might contribute to the variation in cytidine analogue response.
In the present study, we performed a genome-wide association study with 171 lymphoblastoid cell lines for which we obtained genome-wide SNPs by use of Illumina 550K SNP chips, as well as, expression array data obtained with Affymetrix U133 Plus 2.0 GeneChips. Those data made it possible for us to take a comprehensive approach in which we combined genotype, expression and cytotoxicity data for the two cytidine analogues ( Figure 3 ) to identify SNPs that might contribute to drug sensitivity through their impact on the regulation of gene expression ( Tables 1 and 2 ). However, there were only 49 SNPs among the top significant SNPs for gemcitabine and AraC that were common for both drugs, indicating even though the two drugs share common mechanisms and the same metabolic pathway, there are differences between these two drugs. This stepwise analysis helped us to narrow our focus to two SNPs associated with both expression and drug cytotoxicity. Those two SNPs, rs3797418 and rs6082527, were located, respectively, in an intron of IQGAP2 and the 5′-FR of TGM3. To further understand the biology responsible for the association of rs3797418 and rs6082527 with gene expression and drug cytotoxicity, we performed knock down of selected genes with expression that was associated with these two SNPs. Knock down of VAV3 and GPM6A in pancreatic cancer cell lines desensitized the cells to gemcitabine and AraC ( Figure 5A and 5B ). VAV3 is a member of the VAV family that has guanine nucleotide exchange activity toward small GTP-binding proteins. VAV3 is a known proto-oncogene that can be involved in tumorgenesis [40]–[42]. As shown in our network analysis (Figure S1), VAV3 can be involved in multiple signaling pathways. Although the mechanism responsible for VAV3 involvement in gemcitabine and AraC response is unclear, given the multiple roles of VAV3 in cancer development, it was not surprising that this gene might contribute to variation in response to chemotherapeutic agents. GPM6A, also known as M6a, is a transmembrane glycoprotein that belongs to the myelin protolipid protein (PLP) family. This gene is expressed mainly in nervous system, and recent studies have suggested the importance of GPM6A in processes involved in neural development such as neurite extension, survival and differentiation [43]–[45]. GPM6A also interacts with a number of G protein coupled receptors and downstream signaling pathways [46]. One recent report suggested that GPM6A might function as a chaperone in cancer cells as part of global profiling of the cell surface proteome for cancer cells [47]. Although there is limited information with regard to the involvement of this gene in tumorgenesis and drug resistance, our results suggest a possible role for GPM6A in response to cytidine analogue antineoplastic agents.
Although the two SNPs in IQGAP2 and TGM3 that we studied were associated with multiple downstream genes with expression that was also associated with gemcitabine or AraC cytotoxicity, we did not identify an association between these two SNPs and either IQGAP2 and TGM3 gene expression, nor did we observe an influence of IQGAP2 and TGM3 on gemcitabine or AraC cytotoxicity after knock down of these two genes.
IQGAP2 is approximately 300 kb and TGM3 is about 45 kb in length. Figure 6 shows the haplotype structure within ∼300 kb surrounding the rs3797418 and rs6082527 SNPs. Linkage analysis was also performed with HapMap data sets in different ethnic groups. However, one of the SNPs linked to rs6082527 was not associated with gemcitabine IC50, and the other was not present on the 550K SNP array. Therefore, the effect of the two SNPs in IQGAP2 and TGM3 on drug response and other gene expression might result from the effects of either known or unknown polymorphisms linked to those two SNPs.
In summary, we have used a data-rich cell-based model system to perform association studies of gemcitabine and AraC cytotoxicity with genome-wide SNP and expression array data. Use of this approach enabled us to identify SNPs that might be markers for cytidine analogue sensitivity and genes that might play an important role in variation in response to these drugs. There are also limitations associated with the use of these cell lines, as described by Choy et. al [48]. The potential confounding factors besides genetic effect could include cell growth rate, EBV transformation and ATP condition. However, that same paper stated that “RNA levels are predominantly correlated to inter-individual differences in EC50. Much less of the correlation between RNA and EC50s reflects intra-individual variation”. That conclusion supports the hypothesis that inter-individual variation in RNA levels due to genetic variation may contribute to variation in drug response phenotype. One limitation of the use of these cell lines is the possibility that EBV transformation might influence drug sensitivity and/or expression profiles, so we might miss some genes of importance for the drug response phenotype, either because they are not expressed in these cell lines or, after transformation, are down-regulated. There is also evidence that gemcitabine and doxorubicin can induce the lytic form of EBV transformed cells [49], [50]. However, we and other groups have demonstrated success with the use of this type of cell line in previous pharmacogenomic studies [25], [26], [51], and functional characterization of the genes identified during the present study supports the feasibility of this approach. The results of the present study enhance our understanding of the role of genetic variation in variation in gemcitabine and AraC sensitivity and resistance and may help to identify mechanisms involved in the actions of these drugs. In addition, these SNPs and genes can now potentially be tested in the clinical setting and, if confirmed, they could significantly enhance our ability to individualize treatment with these two drugs.
Materials and Methods
Cell Line
174 lymphoblastoid cell lines from 60 Caucasian-American (CA), 54 African-American (AA) and 60 Han Chinese-American (HCA) (sample sets HD100CAU, HD100AA, and HD100CHI) subjects were purchased from the Coriell Cell Repository (Camden, NJ). All of these cell lines had been obtained and anonymized by the National Institute of General Medical Sciences prior to deposit, and all subjects had provided written consent for the use of their DNA and cells for experimental purposes. Human SU86 and Hup-T3 pancreatic cancer cell lines were gifts from Dr. Daniel D. Billadeau, Mayo Clinic.
siRNA
The siRNA duplexes used in the knock down studies were purchased from the QIAGEN Inc. (Valencia, CA).
Drugs and Cell Proliferation Assays
Gemcitabine was provided by Eli Lilly (Indianapolis, IN). AraC was purchased from Sigma-Aldrich (St. Louis, MO). Cytotoxicity assays with the lymphoblastoid cell and tumor cell lines were performed with the CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Promega Corporation, Madison, WI) as previously described [25]. Estimation of the IC50 phenotype (effective dose that kills 50% of the cells) was calculated using a four parameter logistic model. Of the 174 cell lines, cytotoxicity curves for two cell lines were removed from further analysis due to possible experimental error.
Gene Expression
Expression array data were obtained for all 174 lymphoblastoid cell lines as previously described [25]. Specifically, biotin-labeled cRNA, produced by in vitro transcription, was hybridized to Affymetrix Human Genome U133 Plus 2.0 GeneChips. Each of these GeneChip contains 54,613 probesets. Normalization of the expression array data was performed using GCRMA [52], [53].
Genome-Wide SNP Analysis
Illumina HumanHap 550K BeadChips were used to obtain genome-wide SNP data for all 174 cell lines. Specifically, each DNA sample was genotyped using the Illumina version 3 BeadChip, which assays 561,278 single nucleotide polymorphisms (SNPs). All genotyping was conducted at the Genotype Shared Resource (GSR) at the Mayo Clinic, Rochester, MN. SNPs with call rates <0.95 were excluded, as were DNA samples with call rates <0.95. The call rate for rs3797418 and rs6082527 SNPs were 100% for all cell lines. One African American sample was removed due to a call rate <0.95, resulting in 173 cell lines with genotypic data and 171 cell lines with both genotypic data and phenotypic data (gemcitabine IC50 values)
Transient Transfection and RNA Interference
Human SU86 and HupT3 pancreatic cancer cell lines were used to perform the siRNA studies. The Hiperfect transfection reagent (QIAGEN) was used for siRNA reverse transfection. Specifically, cells were seeded into 96-well plates and were mixed with siRNA-complex, consisting of 5 nM of specific or negative control siRNA (QIAGEN), and the Hiperfect transfection reagent. The lymphoblastoid cell lines were transfected with control siRNA and specific siRNA for GPM6A and VAV3 using the electroporation (Nucleofector® Lonza Walkersville Inc., Walkersville, MD). Specifically, 1.25×106 cells suspended in Cell Line Nucleofector Solution V (Lonza Walkersville Inc.) were mixed with 2 µM control or specific siRNA before the electroporation.
Quantitative Real-Time Reverse Transcription-PCR
Total RNA was isolated from cultured cells with the Qiagen RNeasy kit (QIAGEN Inc. Valencia, CA), followed by QRT-PCR performed with the 1-step, Brilliant SYBR Green QRT-PCR master mix kit (Stratagene, La Jolla, CA). Specifically, primers purchased from Qiagen were used to perform QRT-PCR using the Stratagene Mx3005P™ Real-Time PCR detection system (Stratagene). All experiments were performed in triplicate with β-actin as an internal control. Reverse transcribed Universal Human reference RNA (Stratagene) was used to generate a standard curve. Control reactions lacked RNA template.
Statistical Methods
Quality control for the Illumina genotypes was performed before performing the association study. SNPs that deviated from HWE (minimum exact test for HWE [54], [55] and stratified test for HWE [56] p-values <0.001), with call rates <95% or SNPs with MAFs <5% were removed from the analysis. Subjects with call rates <95% were also removed from all SNP analyses. No effects for SNPs were seen with the Illumina platforms. The plot for the most significant SNPs are shown in Figure S2. Next, population stratification was assessed. Because we studied cell lines chosen to represent common human genetic diversity, multiple races/ethnic groups were represented in the lymphoblastoid cell lines. Therefore, we used the method developed by Price et al. [57] to adjust for population stratification. This method uses an eigen analysis to detect and adjust for population stratification. Within each race, eigen analysis was completed, with the top five eigenvectors saved. Using the five eigenvectors within each race, individual genotypes were adjusted using the model 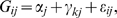 with
with 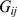 representing the genotype for the ith cell line in racial group j (j = 1, 2, 3),
representing the genotype for the ith cell line in racial group j (j = 1, 2, 3), 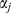 the race effect for race j, and
the race effect for race j, and 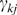 the k
th eigenvector, k = 1, 5, effect for race j. Similarly, the log transformed IC50 values, were also adjusted for race using the five eigenvectors, in addition to gender. Pearson correlation coefficients were then computed using the adjusted genotypes and IC50 variables. False Discovery q-values [58], [59] were also computed for each test. Similar sets of analyses were also performed for the association of IC50 with mRNA expression and SNPs with mRNA expression. Pairwise LD was estimated using r-squared statistics and were displayed graphically using the Haploview software [60].
the k
th eigenvector, k = 1, 5, effect for race j. Similarly, the log transformed IC50 values, were also adjusted for race using the five eigenvectors, in addition to gender. Pearson correlation coefficients were then computed using the adjusted genotypes and IC50 variables. False Discovery q-values [58], [59] were also computed for each test. Similar sets of analyses were also performed for the association of IC50 with mRNA expression and SNPs with mRNA expression. Pairwise LD was estimated using r-squared statistics and were displayed graphically using the Haploview software [60].
Integrative Genomic Analysis
To integrate genotype, expression and drug cytotoxicity data, we used an approach similar to “genomic convergence” [61]–[63], as depicted graphically in Figure 2 . We used a less stringent cut off for p-values used to identify the initial set of SNPs that were associated with IC50 to avoid missing possible candidates with biological significance. First, we identify SNPs associated with IC50 (p-values <0.001). One of the functional mechanisms by which SNPs might influence cytotoxicity is by regulating transcription in either a cis (SNP within or near the gene) or trans- (SNP not within or near the gene) manner. Therefore, we determined the expression probesets that were associated with these identified SNPs (p-values <10−6). Next, to determine whether the expression probesets associated with these SNPs were also associated with IC50 values for gemcitabine and AraC, we correlated these expression probesets with IC50 for both drugs with a p<0.001. A set of genes was then selected to test functionally for their possible biological relevance with regard to drug response.
Bioinformatics Pathway Analysis
To further understand biological interactions between the downstream genes, pathway analysis was performed on genes that had significant associations with the two SNPs and with phenotype. In the case of gemcitabine, SNP rs3797418 present in the intronic region of IQGAP2 showed significant associations with 14 probesets during the SNP-expression analysis; and these same 14 probesets also had significant correlations with IC50 phenotype. Therefore, these14 probesets that mapped to 10 unique genes were used to perform pathway analysis. A similar analysis was performed with AraC using the 8 genes that were significantly associated with the SNP rs3797418 to perform the pathway analysis. SNP rs6082527 close to TGM3 gene was associated with 6 probesets that showed significant correlations with IC50 phenotype. Therefore, pathway analysis was also performed for the 6 probesets that mapped to 5 genes to identify major pathways that might be involved with the downstream effects of this SNP. Ingenuity software was used to perform the pathway analysis. This software consists of a curated database and several analysis tools to obtain pathways associated with a set of genes. Ingenuity calculates p-value for the probability of finding a set of genes within a given pathway. Fisher's exact test is used to calculate the p-values.
Supporting Information
Top polymorphism associated with gemcitabine cytotoxicity (IC50 values). 492 significant SNPs were associated with gemcitabine IC50 values with p-values <0.001.
(0.09 MB XLS)
Top candidates for association between 550K genome-wide SNPs and AraC cytotoxicity. 553 significant SNPs were associated with AraC IC50 values with p-values <0.001.
(0.10 MB XLS)
Top SNPs that were significantly associated with both gemcitabine and AraC (p<0.001).
(0.05 MB XLS)
Top candidates for association between 492 SNPs and whole gene expression array data (54,000 probe sets). 100 SNPs among 492 located within or close to 78 genes were associated with 296 expression probe sets (220 unique genes) with p-values <10−6.
(0.19 MB XLS)
Top candidates for association between 553 SNPs and whole gene expression data (54,000 probesets). 140 SNPs among 553 SNPs located within or close to 843 probe sets (563 unique genes) with p-values <10−6.
(0.33 MB XLS)
Genes within the gemcitabine/AraC transport, metabolism and target pathway.
(0.02 MB PDF)
Network analysis. Genes that were associated with rs3797418, an intron SNP in IQGAP2, were used to perform network analysis using Ingenuity Pathway Analysis. Dotted line indicates an indirect connection and solid lines indicate a direction interaction between genes.
(0.32 MB PDF)
Illumina intensity plot for top candidate SNPs.
(0.13 MB PDF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Supported in part by National Institutes of Health (NIH) grants GM61388 (The Pharmacogenetics Research Network) (B.F., G.J, L.L., R.K. and L.W.), CA102701 (The Pancreatic Cancer SPORE) (L.W.), R01 CA132780 (L.L. and R.M.W.), R01 GM28157 (R.M.W), K22 CA130828 (L.W.) and R01 CA138461 (L.W.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 2.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 3.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morley M, Molony CM, Weber TM, Devlin JL, Ewens KG, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smid K, Bergman AM, Eijk PP, Veerman G, van Haperen VW, et al. Micro-array analysis of resistance for gemcitabine results in increased expression of ribonucleotide reductase subunits. Nucleosides Nucleotides Nucleic Acids. 2006;25:1001–1007. doi: 10.1080/15257770600890269. [DOI] [PubMed] [Google Scholar]
- 8.Bergman AM, Eijk PP, Ruiz van Haperen VW, Smid K, Veerman G, et al. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as the major determinant. Cancer Res. 2005;65:9510–9516. doi: 10.1158/0008-5472.CAN-05-0989. [DOI] [PubMed] [Google Scholar]
- 9.Gwee PC, Tang K, Sew PH, Lee EJ, Chong SS, et al. Strong linkage disequilibrium at the nucleotide analogue transporter ABCC5 gene locus. Pharmacogenet Genomics. 2005;15:91–104. doi: 10.1097/01213011-200502000-00005. [DOI] [PubMed] [Google Scholar]
- 10.Half E, Tang XM, Gwyn K, Sahin A, Wathen K, et al. Cyclooxygenase-2 expression in human breast cancers and adjacent ductal carcinoma in situ. Cancer Res. 2002;62:1676–1681. [PubMed] [Google Scholar]
- 11.Wu X, Gwyn K, Amos CI, Makan N, Hong WK, et al. The association of microsomal epoxide hydrolase polymorphisms and lung cancer risk in African-Americans and Mexican-Americans. Carcinogenesis. 2001;22:923–928. doi: 10.1093/carcin/22.6.923. [DOI] [PubMed] [Google Scholar]
- 12.Campbell TB, Borok M, Gwanzura L, MaWhinney S, White IE, et al. Relationship of human herpesvirus 8 peripheral blood virus load and Kaposi's sarcoma clinical stage. Aids. 2000;14:2109–2116. doi: 10.1097/00002030-200009290-00006. [DOI] [PubMed] [Google Scholar]
- 13.Bussey KJ, Chin K, Lababidi S, Reimers M, Reinhold WC, et al. Integrating data on DNA copy number with gene expression levels and drug sensitivities in the NCI-60 cell line panel. Mol Cancer Ther. 2006;5:853–867. doi: 10.1158/1535-7163.MCT-05-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gullans SR. Connecting the dots using gene-expression profiles. N Engl J Med. 2006;355:2042–2044. doi: 10.1056/NEJMcibr065953. [DOI] [PubMed] [Google Scholar]
- 16.Heinemann V, Hertel LW, Grindey GB, Plunkett W. Comparison of the cellular pharmacokinetics and toxicity of 2′,2′-difluorodeoxycytidine and 1-beta-D-arabinofuranosylcytosine. Cancer Res. 1988;48:4024–4031. [PubMed] [Google Scholar]
- 17.Heinemann V, Xu YZ, Chubb S, Sen A, Hertel LW, et al. Cellular elimination of 2′,2′-difluorodeoxycytidine 5′-triphosphate: a mechanism of self-potentiation. Cancer Res. 1992;52:533–539. [PubMed] [Google Scholar]
- 18.Kern W, Estey EH. High-dose cytosine arabinoside in the treatment of acute myeloid leukemia: Review of three randomized trials. Cancer. 2006;107:116–124. doi: 10.1002/cncr.21543. [DOI] [PubMed] [Google Scholar]
- 19.Kindler HL. In focus: advanced pancreatic cancer. Clin Adv Hematol Oncol. 2005;3:420–422. [PubMed] [Google Scholar]
- 20.Tomasson MH, Xiang Z, Walgren R, Zhao Y, Kasai Y, et al. Somatic mutations and germline sequence variants in the expressed tyrosine kinase genes of patients with de novo acute myeloid leukemia. Blood. 2008;111:4797–4808. doi: 10.1182/blood-2007-09-113027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bleibel WK, Duan S, Huang RS, Kistner EO, Shukla SJ, et al. Identification of genomic regions contributing to etoposide-induced cytotoxicity. Hum Genet. 2009;125:173–180. doi: 10.1007/s00439-008-0607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla SJ, Duan S, Badner JA, Wu X, Dolan ME. Susceptibility loci involved in cisplatin-induced cytotoxicity and apoptosis. Pharmacogenet Genomics. 2008;18:253–262. doi: 10.1097/FPC.0b013e3282f5e605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan S, Bleibel WK, Huang RS, Shukla SJ, Wu X, et al. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Fridley B, Kalari K, Jenkins G, Batzler A, et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res. 2008;68:7050–7058. doi: 10.1158/0008-5472.CAN-08-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang RS, Duan S, Kistner EO, Hartford CM, Dolan ME. Genetic variants associated with carboplatin-induced cytotoxicity in cell lines derived from Africans. Mol Cancer Ther. 2008;7:3038–3046. doi: 10.1158/1535-7163.MCT-08-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Burmeister M. Genetical genomics: combining genetics with gene expression analysis. Hum Mol Genet. 2005;14 Spec No. 2:R163–169. doi: 10.1093/hmg/ddi267. [DOI] [PubMed] [Google Scholar]
- 28.Bystrykh L, Weersing E, Dontje B, Sutton S, Pletcher MT, et al. Uncovering regulatory pathways that affect hematopoietic stem cell function using ‘genetical genomics’. Nat Genet. 2005;37:225–232. doi: 10.1038/ng1497. [DOI] [PubMed] [Google Scholar]
- 29.Schoch C, Haferlach T, Haase D, Fonatsch C, Loffler H, et al. Patients with de novo acute myeloid leukaemia and complex karyotype aberrations show a poor prognosis despite intensive treatment: a study of 90 patients. Br J Haematol. 2001;112:118–126. doi: 10.1046/j.1365-2141.2001.02511.x. [DOI] [PubMed] [Google Scholar]
- 30.Seve P, Mackey JR, Isaac S, Tredan O, Souquet PJ, et al. cN-II expression predicts survival in patients receiving gemcitabine for advanced non-small cell lung cancer. Lung Cancer. 2005;49:363–370. doi: 10.1016/j.lungcan.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hertel LW, Boder GB, Kroin JS, Rinzel SM, Poore GA, et al. Evaluation of the antitumor activity of gemcitabine (2′,2′-difluoro-2′-deoxycytidine). 50:4417-22. Cancer Res. 1990;50:4417–4422. [PubMed] [Google Scholar]
- 34.Lokiec F, Lansiaux A. [Gemcitabine: from preclinic to clinic passing by pharmacokinetics]. Bull Cancer. 2007;94 Spec No Actualites:S85–89. [PubMed] [Google Scholar]
- 35.Kim SR, Saito Y, Maekawa K, Sugiyama E, Kaniwa N, et al. Thirty novel genetic variations in the SLC29A1 gene encoding human equilibrative nucleoside transporter 1 (hENT1). Drug Metab Pharmacokinet. 2006;21:248–256. doi: 10.2133/dmpk.21.248. [DOI] [PubMed] [Google Scholar]
- 36.Kim SR, Saito Y, Maekawa K, Sugiyama E, Kaniwa N, et al. Twenty novel genetic variations and haplotype structures of the DCK gene encoding human deoxycytidine kinase (dCK). Drug Metab Pharmacokinet. 2008;23:379–384. doi: 10.2133/dmpk.23.379. [DOI] [PubMed] [Google Scholar]
- 37.Kwon WS, Rha SY, Choi YH, Lee JO, Park KH, et al. Ribonucleotide reductase M1 (RRM1) 2464G>A polymorphism shows an association with gemcitabine chemosensitivity in cancer cell lines. Pharmacogenet Genomics. 2006;16:429–438. doi: 10.1097/01.fpc.0000204999.29924.da. [DOI] [PubMed] [Google Scholar]
- 38.Yonemori K, Ueno H, Okusaka T, Yamamoto N, Ikeda M, et al. Severe drug toxicity associated with a single-nucleotide polymorphism of the cytidine deaminase gene in a Japanese cancer patient treated with gemcitabine plus cisplatin. Clin Cancer Res. 2005;11:2620–2624. doi: 10.1158/1078-0432.CCR-04-1497. [DOI] [PubMed] [Google Scholar]
- 39.Sugiyama E, Kaniwa N, Kim SR, Kikura-Hanajiri R, Hasegawa R, et al. Pharmacokinetics of gemcitabine in Japanese cancer patients: the impact of a cytidine deaminase polymorphism. J Clin Oncol. 2007;25:32–42. doi: 10.1200/JCO.2006.06.7405. [DOI] [PubMed] [Google Scholar]
- 40.Hornstein I, Alcover A, Katzav S. Vav proteins, masters of the world of cytoskeleton organization. Cell Signal. 2004;16:1–11. doi: 10.1016/s0898-6568(03)00110-4. [DOI] [PubMed] [Google Scholar]
- 41.Trenkle T, McClelland M, Adlkofer K, Welsh J. Major transcript variants of VAV3, a new member of the VAV family of guanine nucleotide exchange factors. Gene. 2000;245:139–149. doi: 10.1016/s0378-1119(00)00026-3. [DOI] [PubMed] [Google Scholar]
- 42.Turner M, Billadeau DD. VAV proteins as signal integrators for multi-subunit immune-recognition receptors. Nat Rev Immunol. 2002;2:476–486. doi: 10.1038/nri840. [DOI] [PubMed] [Google Scholar]
- 43.Lagenaur C, Kunemund V, Fischer G, Fushiki S, Schachner M. Monoclonal M6 antibody interferes with neurite extension of cultured neurons. J Neurobiol. 1992;23:71–88. doi: 10.1002/neu.480230108. [DOI] [PubMed] [Google Scholar]
- 44.Mukobata S, Hibino T, Sugiyama A, Urano Y, Inatomi A, et al. M6a acts as a nerve growth factor-gated Ca(2+) channel in neuronal differentiation. Biochem Biophys Res Commun. 2002;297:722–728. doi: 10.1016/s0006-291x(02)02284-2. [DOI] [PubMed] [Google Scholar]
- 45.Alfonso J, Fernandez ME, Cooper B, Flugge G, Frasch AC. The stress-regulated protein M6a is a key modulator for neurite outgrowth and filopodium/spine formation. Proc Natl Acad Sci U S A. 2005;102:17196–17201. doi: 10.1073/pnas.0504262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu DF, Koch T, Liang YJ, Stumm R, Schulz S, et al. Membrane glycoprotein M6a interacts with the micro-opioid receptor and facilitates receptor endocytosis and recycling. J Biol Chem. 2007;282:22239–22247. doi: 10.1074/jbc.M700941200. [DOI] [PubMed] [Google Scholar]
- 47.Shin BK, Wang H, Yim AM, Le Naour F, Brichory F, et al. Global profiling of the cell surface proteome of cancer cells uncovers an abundance of proteins with chaperone function. J Biol Chem. 2003;278:7607–7616. doi: 10.1074/jbc.M210455200. [DOI] [PubMed] [Google Scholar]
- 48.Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2009;4:1–16. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng WH, Israel B, Raab-Traub N, Busson P, Kenney SC. Chemotherapy induces lytic EBV replication and confers ganciclovir susceptibility to EBV-positive epithelial cell tumors. Cancer Res. 2002;62:1920–1926. [PubMed] [Google Scholar]
- 50.Feng WH, Hong G, Delecluse HJ, Kenney SC. Lytic induction therapy for Epstein-Barr virus-positive B-cell lymphomas. J Virol. 2004;78:1893–1902. doi: 10.1128/JVI.78.4.1893-1902.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang RS, Duan S, Bleibel WK, Kistner EO, Zhang W, et al. A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity. Proc Natl Acad Sci U S A. 2007;104:9758–9763. doi: 10.1073/pnas.0703736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 53.Wu Z, Irizarry R, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligobucleotide expression arrays. J Am Stat Assn. 2004;99:909–917. [Google Scholar]
- 54.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 55.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–893. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schaid DJ, Batzler AJ, Jenkins GD, Hildebrandt MA. Exact tests of Hardy-Weinberg equilibrium and homogeneity of disequilibrium across strata. Am J Hum Genet. 2006;79:1071–1080. doi: 10.1086/510257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 58.Storey JD. A direct approach to false discovery rates. J Royal Stat Soc. 2002;B 64:479–498. [Google Scholar]
- 59.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 61.Hauser MA, Li YJ, Takeuchi S, Walters R, Noureddine M, et al. Genomic convergence: identifying candidate genes for Parkinson's disease by combining serial analysis of gene expression and genetic linkage. Hum Mol Genet. 2003;12:671–677. [PubMed] [Google Scholar]
- 62.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. Markov Model for Rapid Haplotyping and Genotype Imputation in Genome Wide Studies. 2008 University of Michigan. [Google Scholar]
- 63.Schadt EE. Exploiting naturally occurring DNA variation and molecular profiling data to dissect disease and drug response traits. Curr Opin Biotechnol. 2005;16:647–654. doi: 10.1016/j.copbio.2005.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top polymorphism associated with gemcitabine cytotoxicity (IC50 values). 492 significant SNPs were associated with gemcitabine IC50 values with p-values <0.001.
(0.09 MB XLS)
Top candidates for association between 550K genome-wide SNPs and AraC cytotoxicity. 553 significant SNPs were associated with AraC IC50 values with p-values <0.001.
(0.10 MB XLS)
Top SNPs that were significantly associated with both gemcitabine and AraC (p<0.001).
(0.05 MB XLS)
Top candidates for association between 492 SNPs and whole gene expression array data (54,000 probe sets). 100 SNPs among 492 located within or close to 78 genes were associated with 296 expression probe sets (220 unique genes) with p-values <10−6.
(0.19 MB XLS)
Top candidates for association between 553 SNPs and whole gene expression data (54,000 probesets). 140 SNPs among 553 SNPs located within or close to 843 probe sets (563 unique genes) with p-values <10−6.
(0.33 MB XLS)
Genes within the gemcitabine/AraC transport, metabolism and target pathway.
(0.02 MB PDF)
Network analysis. Genes that were associated with rs3797418, an intron SNP in IQGAP2, were used to perform network analysis using Ingenuity Pathway Analysis. Dotted line indicates an indirect connection and solid lines indicate a direction interaction between genes.
(0.32 MB PDF)
Illumina intensity plot for top candidate SNPs.
(0.13 MB PDF)