Abstract
GSTaxClassifier (Genomic Signature based Taxonomic Classifier) is a program for metagenomics analysis of shotgun DNA sequences. The program includes
a simple but effective algorithm, a modification of the Bayesian method, to predict the most probable genomic origins of sequences at different taxonomical ranks, on the basis of genome databases;
a function to generate genomic profiles of reference sequences with tri-, tetra-, penta-, and hexa-nucleotide motifs for setting a user-defined database;
two different formats (tabular- and tree-based summaries) to display taxonomic predictions with improved analytical methods; and
effective ways to retrieve, search, and summarize results by integrating the predictions into the NCBI tree-based taxonomic information.
GSTaxClassifier takes input nucleotide sequences and using a modified Bayesian model evaluates the genomic signatures between metagenomic query sequences and reference genome databases. The simulation studies of a numerical data sets showed that GSTaxClassifier could serve as a useful program for metagenomics studies, which is freely available at http://helix2.biotech.ufl.edu:26878/metagenomics/.
Keywords: Genomic signature, meta-genomics, taxonomy, Bayesian method
Background
With the advent of whole genome shotgun sequencing (WGS), metagenomics is attracting more and more attention from microbiologists, from characterizing one particular species to understanding the dynamics of a whole microbial community, including assessing the coding potential of environmental organisms, quantifying the relative abundances of known species, and estimating the amount of unknown sequence information. Microbes constitute the vast majority of marine biomass and account for the majority of genetic variations in the oceans. However, the genetic diversity, community composition, relative abundance, and distribution of microbes have challenged microbiologists [1], largely because ˜99% of naturally occurring microbes are not easily isolated in pure culture [2] and analysis of metagenomic data will produce a significant number of sequences that cannot be correctly assigned to known taxa. A practical question emerging from environmental sequencing projects is the extent to which the data are interpretable in the absence of significant individual genome assemblies, especially those uncultivated prokaryotes.
Reference databases, methods for sequence comparison, and a widely accepted taxonomy are three key elements in metagenomics studies. Improvements in cloning and sequencing technologies (e.g., WGS and 454 tag sequencing) have made DNA sequencing feasible for a wide range of microbial communities. Several novel models, based on either sequence homology or composition, were developed to support comprehensive analysis of species composition in complex mixtures such as water, soil, and feces [2–7].
Cloning and sequencing ’marker genes‘ is a common approach to metagenomics classification. Ribosomal RNA (rRNA) genes (e.g., 16S/18S small subunit rRNA) extracted from large-scale environmental shotgun sequencing data are commonly used to quantitatively estimate the community composition, including those uncultivated microbes, such as Crenarchaeota and Acidobacteria from different habitats, by using BLAST alignment [2,3]. However, a homology-based BLAST search depends on the completeness and balance of the reference databases. Many sequences from lowabundance community members cannot be assigned to the most likely phylogenetic neighbors due to lack of homologous marker genes in the databases.
Another approach to characterizing metagenomic sequences is based on the analysis of the genomic composition of oligonucleotides of different lengths (i.e., motifs), called genomic signatures [8,9]. By using a classifier constructed based on genomic signatures, comprehensive analyses of nucleotide frequencies in a wide variety of genomes provides fundamental knowledge of individual genomes [8–10]. For example, GC content or the relative abundance of dinucleotides has been used as a fundamental characteristic of individual genomes [6,8,11,12]. Genomic signatures not only carry phylogenetic information but also are capable of identifying horizontally transferred genes (HGT) in bacteria [12]. Based on a naïve Bayesian [9] or an unsupervised neural network algorithm, a self-organizing map (SOM) [10], di-, tri- and tetra-nucleotide frequencies in a wide variety of prokaryotic and eukaryotic genomes were used to predict the most probably genomic origins of metagenomic sequences [8]. In ’PhyloPythia‘, a composition-based program [6], sequence compositions were used to phylogenetically classify sequence fragments, based on a multiclass support vector machine (SVM). These programs accurately improved the phylogenetic assignment of query sequences to known taxonomic clades. However, either these analyses are based on very complicated algorithms or the predicted results could not be efficiently applied by microbiologists.
In the present study, we introduce a new program, named GSTaxClassifier (Genomic Signature based Taxonomic Classifier), for metagenomics study. Our program includes
a simple but effective model, a modification of the Bayesian method that was proposed by Sandber et al. [9], to predict the most probable genomic origins of metagenomic sequences at different taxonomical levels (i.e., kingdom, phylum order, etc.), on the basis of the reference genome databases;
a function to generate genomic signature profiles of reference sequences with tri-, tetra-, penta-, and hexa-nucleotide motifs for setting a user-defined database;
two different formats (tabular- and tree-based summaries) to display taxonomic predictions with improved analytical methods; and
effective ways to retrieve, search, and summarize results by integrating the predictions into the NCBI treebased taxonomic information.
The simulation results demonstrate that our approach can be applied to metagenomics sequencing projects.
Methodology
The algorithm used in the GSTaxClassifier is a modification of the Bayesian method proposed by Sandberg et al. [9]. Sun et al. [13] mathematically proved that the Bayesian method is equivalent to the nearest neighbor classifier assigning each query sequence to the species whose genomic signature is closest, among the species of interest, to the normalized motif occurrence frequency profile of the query sequence, with respect to the Kullback-Leibler distance. The GSTaxClassifier assigns sequences to the most probable genomic origins by calculating the probabilities of query sequences to belong to all available genomes with Baye's rule. After each sequence is classified into reference species, the number of hits of each species after normalization can be regarded as the relative population density of the species.
To generate genomic signature, a sliding window of a specified motif size was used to scan reference and query sequences and the frequencies of all short oligo-nucleotides (motifs) were counted. If a given motif contains any character other than the characters of A/a, T/t, G/g, and C/c, this motif will not be included. The taxonomic predictions of query sequences were based on the reference genome databases containing 500 bacteria, 40 archaea, and 79 eukaryotes retrieved from NCBI, JGI (DOE Joint Genome Institute), and the Broad Institute of MIT and Harvard. The genomic signature profiles of genome sequences were generated at the motif lengths of tri-, tetra-, penta-, and hexa-nucleotides and can be updated when new genome sequences are available. The GSTaxClassifier was implemented with JSP and the MySQL database and accessible by a web interface. It includes two major functions:
Generating a user-defined database based on reference sequences
The input includes the reference sequences and the motif length. The reference sequences are nucleotide sequences in FASTA format and can be pasted into the input form or uploaded from a local disk. The output of each submission is a genomic signature profile of the reference sequences in text format. This file can be downloaded from the server and uploaded from a local disk as a reference database for sequence prediction.
Predicting taxonomic origins of the query sequences
The processing pipeline for sequence prediction includes several phases. The first step is verifying the input contains valid nucleotide sequences, using a criterion that 80% of the characters of each sequence are ’A‘, ’C‘, ’G‘, ’T‘, ’N‘ or ’X‘. The next step is generating a genomic signature profile of the query sequences with the userdefined motif length. The third step is calculating the probabilities of the query sequences assigned to each genome in the selected reference databases with the modified Bayesian model. The final step is filtering results with a user-defined threshold and saving the results in the relational database with a user-defined project name. The threshold here is defined as the minimum probability of a query sequence assigned to a reference genome to keep the prediction in database.
The input are nucleotide sequences in FASTA format, and can be directly pasted in the input form or uploaded through a plain text file from a local disk. The user is required to create a project name and specify a threshold for retaining results in the database. In addition, GSTaxClassifier provides options for selecting the reference databases, including the databases based on NCBI genomes (e.g., Bacteria, Archaea, and Eukaryota), and a user-defined database, specifying a desired motif length (tri-, tetra-, penta-, and hexa-nucleotides), and choosing an output format. GSTaxClassifier displays results in the context of taxonomic predictions either in a tabular format or in a graphical view. A tabular representation of predictions is displayed on the web page and the results file can be downloaded from the server. In the graphical view, GSTaxClassifier integrates predictions into the tree of life of the NCBI-based taxonomy. The relative sequence abundances at each taxonomic level are represented in terms of the number of the sequences assigned and a total of the prediction probabilities of those assigned sequences. Also, the server provides advanced options, such as searching and summarizing the predictions for a specific taxon. In addition, two other functions were implemented in GSTaxClassifier for dealing with prediction results. The function of ’Retrieve Results‘ enables user to retrieve and search the predictions with different formats, while the function of ’Delete Project‘ enables user to remove a project and its results.
Discussion
In order to validate our algorithm we conducted simulation studies using six data sets, each set containing 1,000 known genomic DNA sequence segments. The test sequences were taxonomically assigned using GSTaxClassifier as a test of prediction accuracy, based on the reference databases in the server. The prediction accuracy is defined in terms of true positive and false positive rates. Sequences assigned to correct taxa are considered true positives, while incorrect assignments are false positives. A threshold of 40% minimum identity (or probability) between query and hit genome is used in our simulation studies.
Three simulation experiments were performed to validate our approach. The first experiment was based on six test data sets, each containing a total of 1,000 sequences that were randomly generated from individual genomes of bacteria, archaea, or eukaryotes. Three data sets were generated with sequence length of 400 bps (base pairs), and three additional data sets contained sequences of 1,000 bps each. The prediction accuracy was estimated by comparing each of the six test data sets against the reference genome databases, based on the motif length of hexa-nucleotides. Overall, GSTaxClassifier showed robust accuracies in bacteria and archaea, with accuracies of 79.8%- 89% input sequences assigned at the Order level, 82%-92% at the Phylum level, and 89%-95% at the Kingdom level. We also observed that sequences as short as 400 bps in eukaryotes can be correctly classified with 66.6%-82.1% accuracy from Order to Kingdom. Analysis of all data sets indicated that the amount of correctly assigned sequences correlated strongly with the sequence length. The prediction accuracy of 1,000 bp sequence segments is relatively higher than that of 400 bp sequences.
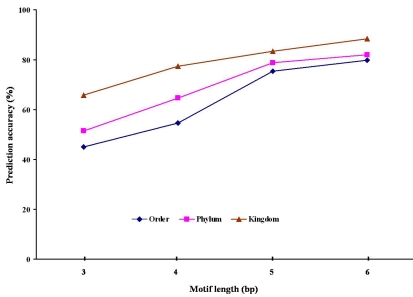
To further demonstrate how well our method assigns query sequences to the correct taxonomic units, we did cross-validation analyses. In the second experiment, we compared each of the six test data sets against the specially constructed reference genome databases that excluded the genomes used to generate the test data sets. Each of the test data set query sequences could be assigned to either one of the ancestral or sisterhood nodes of the genome species (the true positives), another node (the false positives), or not be assigned to any node (unknown) if the best prediction probability was below the threshold. The estimated accuracies of the test data sets based on a hexa-nucleotide motif length revealed that, even when the genomes were excluded from the reference database, the classifier is still sensitive for detection as well as specific for discrimination between the species and their near relatives (Table 1 insupplementary material). For the ranks from Order to Kingdom, the assignment accuracies were significantly increased across all test data sets, by 63.2%-95.3 % in bacteria, 61.9%- 96.4% in archaea, and 54.3%-76.5% in eukaryotes. The lower prediction accuracy in eukaryotes could be attributed to few eukaryotic genomes in the reference database and a loss in sensitivity across more distantly related species. The third validation experiment was to estimate the influence of tri-, tetra-, penta-, and hexa-nucleotide motiflength on estimation accuracy, by using the six test data sets. The results suggested that longer motifs produced more accurate estimates of taxonomic contents (Figure 1). Genomic signatures based on hexanucleotides exhibited the best performance, coinciding with early reports [6,9]. We demonstrated that genomic signature allows accurate characterization of genomic DNA sequence fragments based on the reference genome databases, but it is important to note that the simulated data sets were explicitly intended as examples for a benchmarking study.
Figure 1.
Influence of motif lengths on prediction accuracy of sequences from bacteria.
Conclusion
We present a web server capable of assigning metagenomic sequences to taxonomic clades. The simple but efficient algorithm used allows fast and reliable identification of metagenomic sequences. The key characteristics of the server are the ability to accurately identify the likely origin of query sequences at high phylogenetic ranks as well as integrating predictions into the NCBI tree of life based on taxonomic information. The web interface is simple and easy to use.
Supplementary material
Acknowledgments
We would like to acknowledge A. Gardner and J. Warfield for help in server design and support. This research has been supported by the ICBR (Interdisciplinary Center for Biotechnology Research) of the University of Florida.
Footnotes
Citation:Yu et al, Bioinformation 4(1): 46-49 (2009)
References
- 1.Curtis TP, Sloan WT. Science. 2005;309:1331. doi: 10.1126/science.1118176. [DOI] [PubMed] [Google Scholar]
- 2.Tringe SG, Rubin EM. Nature Review Genetics. 2005;6:805. doi: 10.1038/nrg1709. [DOI] [PubMed] [Google Scholar]
- 3.Huson DH, et al. Genome Res. 2007;17:377. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin HG, et al. Nat Biotechnol. 2006;24:1263. [Google Scholar]
- 5.Tsirigos A, Rigoutsos I. Nucleic Acids Res. 2005;33:922. doi: 10.1093/nar/gki187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McHardy AC, et al. Nature Methods. 2007;4:63. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 7.Venter JC, et al. Science. 2004;304:66. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 8.Karlin S, Burge C. Trends Genet. 1995;11:283. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 9.Sandberg R, et al. Genome Res. 2001;11:1404. doi: 10.1101/gr.186401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abe T, et al. DNA Res. 2005;12:281. doi: 10.1093/dnares/dsi015. [DOI] [PubMed] [Google Scholar]
- 11.Karlin S, et al. J Bacteriol. 1997;179:3899. doi: 10.1128/jb.179.12.3899-3913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalevi D, et al. Bioinformatics. 2006;22:517. doi: 10.1093/bioinformatics/btk029. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y, et al. BIOCOMP. 2007;1:163. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.