Abstract
A Genomic Target Database (GTD) has been developed having putative genomic drug targets for human bacterial pathogens. The selected pathogens are either drug resistant or vaccines are yet to be developed against them. The drug targets have been identified using subtractive genomics approaches and these are subsequently classified into
Drug targets in pathogen specific unique metabolic pathways,
Drug targets in host-pathogen common metabolic pathways, and
Membrane localized drug targets.
HTML code is used to link each target to its various properties and other available public resources. Essential resources and tools for subtractive genomic analysis, sub-cellular localization, vaccine and drug designing are also mentioned. To the best of authors knowledge, no such database (DB) is presently available that has listed metabolic pathways and membrane specific genomic drug targets based on subtractive genomics. Listed targets in GTD are readily available resource in developing drug and vaccine against the respective pathogen, its subtypes, and other family members. Currently GTD contains 58 drug targets for four pathogens. Shortly, drug targets for six more pathogens will be listed.
Availability
GTD is available at IIOAB website http://www.iioab.webs.com/GTD.htm. It can also be accessed at http://www.iioabdgd.webs.com.GTD is free for academic research and non-commercial use only. Commercial use is strictly prohibited without prior permission from IIOAB.
Keywords: Genomic drug targets, database, pathogenic bacteria, metabolic pathway targets, membrane associated targets, candidate vaccine targets
Background
Infectious diseases caused by various pathogenic bacteria are considered to be a major public health problem globally. Several pathogens have been reported to challenge the existing treatment regime by developing drug resistance and in several cases effective vaccines are yet to be developed. Although, researches are going on to develop effective drugs and vaccines, the efforts are not yet successful due to the dynamic adaptability, frequent phase and antigenic modifications, variations in major virulence factors, and adoptive mutations. The advent of several microbial complete genome sequences along with development of various bioinformatics tools, made it faceable for in silico analysis of the genomes and subsequent drug discovery against deadly human pathogen. To date, NCBI genome database has listed approximately 2491 fully sequenced microbial genomes including pathogenic bacteria [1] and computational approaches based on subtractive genomics have successfully been used to identify drug targets in many pathogenic bacteria [2,3,4]. However, structured data for genomic drug targets for any human pathogen do not exist [4]. Therefore, we developed a Genomic Target Database (GTD) to provide putative genomic drug targets categorized into pathogen specific unique metabolic pathways, host-pathogen common metabolic pathways, and membrane/surface localized drug targets for ten most common human pathogenic bacteria. It is hoped that GTD will serve as a readily available resource for both drug and vaccine development for the respective pathogen, its serotypes, family members, and pathogens containing homologous sequences of these drug targets.
Methodology
Data collection
Available drug target data have been collected from various literature sources viz. PubMed [1], ScienceDirect [5], Google Scholar [6] etc. Pathogens, for which no data are available, were identified using subtractive genomics approaches as described elsewhere by Saharkar et al. (2004) [2]. These are based on the assumption that an essential survival gene of a given pathogen that is non-homologous to human host is a candidate drug target [7,8].
Identification of genomic drug targets
Complete genome and proteome sequences of selected pathogens from NCBI [1], BLAST tools, and databases such as Database of Essential Genes (DEG) [9] (http://tubic.tju.edu.cn/deg) and Kyoto Encyclopedia of Genes and Genomes (KEGG) [10] pathway database were used to identify putative drug targets. Each functional gene and corresponding protein sequence of the bacteria were subjected to standard BLAST-X and BLAST-P respectively against DEG. Pathogen homologs that showed significant hits against DEG listed essential genes were selected as putative essential genes for the pathogen under consideration based on the BLAST-P scores [cut off values for bit score (≫100), E-value (≪ E-10), and percentage of identity at amino acid level (≫35%)]. Genes encoding for ≪ 100 amino acids length were purged out. Each identified essential gene and corresponding protein sequence of the pathogen were analyzed for sequence homology with human genome using standard human BLAST-X and BLAST-P in NCBI server. Non-homologous essential genes considered as putative drug targets were selected based on the selection criteria that a drug target should not show any similarity with any human sequence. The function and sub-cellular localization of each drug target was analyzed with Swiss-prot protein database [11] and by using sub-cellular localization prediction tools, CELLO [12], PSORTb [13], PSLpred [14], and SOSUI-GramN [15]. The KEGG database [10] was used for comparative pathway analysis and to identify proteins/enzymes that are involved in host-pathogen common and pathogen specific unique pathways. Targets were listed according to the pathways where they are involved. The membrane or surface proteins (candidate vaccine targets) were grouped separately.
Features, design, and contents of GTD
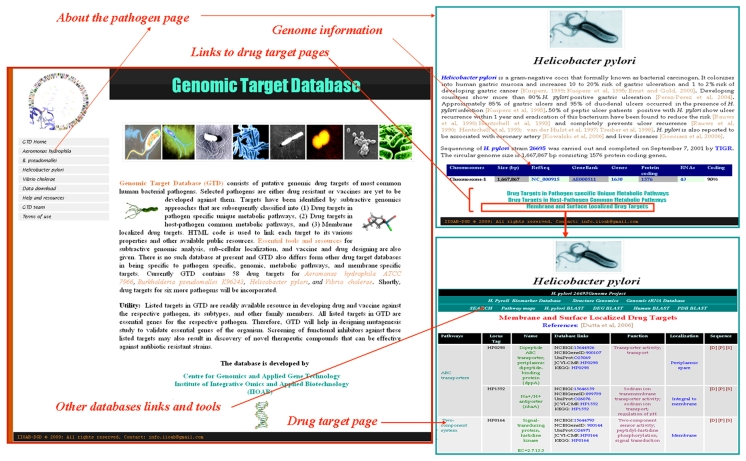
The GTD is a HTML based database and is represented in table format. The screenshot of the database is shown in Figure 1. For each genome, four pages are there. The first page contains the brief description of the pathogen, its taxonomy, virulence, and genome information etc. At the end of this page, three links (Drug targets in pathogen specific unique metabolic pathways, Drug targets in host pathogen common metabolic pathways, and Membrane and surface localized drug targets) are given. Each corresponding link page contains list of identified gene/protein targets for that particular category. At the top of this page pathogen specific links to the main genome project, other publicly available DBs, search option, pathway links, and various BLASTs are given for subtractive genomics analysis. Below that literature references are mentioned (if available) for corresponding drug targets. Each entry in GTD is a potential drug target for the corresponding pathogen and links are provided for their various properties and other publicly accessible databases. In silico predicted sub-cellular localization is denoted as (P). Two separate pages have been allotted that contains downloadable data of GTD and links to various resources for drug and vaccine designing.
Figure 1.
Screenshot image of Genomic Target Database (GTD)
Data statistics and future development
The objective to develop GTD is to list out drug targets of 10 most common human pathogenic bacteria. Currently GTD contains 58 drug targets for four pathogens. There are targets, specific to unique metabolic pathways in Burkholderia pseudomallei K96243 (27), Aeromonas hydrophila ATCC 7966 (18), and Vibrio cholerae (3). Ten membrane/surface localized drug targets have been listed for Helicobacter pylori. Listing of targets for other pathogens is in progress.
Utility
The GTD is designed to provide a readily available resource of putative genomic drug targets in most common human pathogenic bacteria.
All listed targets in GTD are essential genes for the pathogen. Therefore, GTD will help in designing mutagenesis study to validate essential genes of the organism.
The data can be used for both the drug and vaccine development for the respective pathogen, its sub-types, family members, and other similar species.
Screening of functional inhibitors against these listed targets may also result in discovery of novel therapeutic compounds that can be effective against antibiotic resistant strains.
Author contributions:
DB: Conceived collected, analyzed data, designed GTD, entered data and prepared the manuscript. AK and ANM provided inputs and reviewed datasets, analysis and database.
Acknowledgments
We acknowledge www.webs.com for the free server and all the database providers whose links are taken to develop GTD. We also thank all members of IIOAB for their continuous support and encouragement in developing the database. AK and ANM acknowledge the facilities of DBT's Bioinformatics sub-centers hosted at respective University Departments for review, analysis and feedback on database development.
Footnotes
Citation:Barh et al, Bioinformation 4(1): 50-51 (2009)
References
- 1. http://www.ncbi.nlm.nih.gov.
- 2.Sakharkar KR, et al. In Silico Biol. 2004;4:355. [PubMed] [Google Scholar]
- 3.Dutta A, et al. In Silico Biol. 2006;6:43. [PubMed] [Google Scholar]
- 4.Sharma V, et al. In Silico Biol. 2008;8:331. [PubMed] [Google Scholar]
- 5. http://www.sciencedirect.com/
- 6. http://scholar.google.com/
- 7.Allsop AE. Curr Opin Microbiol. 1998;1:530. doi: 10.1016/s1369-5274(98)80085-4. [DOI] [PubMed] [Google Scholar]
- 8.Schmid MB. Curr Opin Chem Biol. 1998;2:529. doi: 10.1016/s1367-5931(98)80130-0. [DOI] [PubMed] [Google Scholar]
- 9.Zhang R, et al. Nucleic Acids Res. 2004;32:D271. doi: 10.1093/nar/gkh024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogata H, et al. Nucleic Acids Res. 1999;27:29. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. http://www.expasy.org/sprot.
- 12.Yu CS, et al. Protein Science. 2004;13:1402. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardy JL, et al. Bioinformatics. 2005;21:617. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 14.Bhasin M, et al. Bioinformatics. 2005;21:2522. doi: 10.1093/bioinformatics/bti309. [DOI] [PubMed] [Google Scholar]
- 15.Imai K, et al. Bioinformation. 2008;2:417. doi: 10.6026/97320630002417. [DOI] [PMC free article] [PubMed] [Google Scholar]