Abstract
Background
The clinical characteristics of sporadic brachial plexus neuropathy (S-BPN) and hereditary brachial plexus neuropathy (H-BPN) are similar. At times of attack inflammation in brachial plexus nerves has been identified in both conditions. SEPT-9 mutations (Arg88Trp, Ser93Phe, 5UTR-131G to C) occur in some families with H-BPN. These mutations were not found in American H-BPN kindreds with a conserved 500 Kb sequence of DNA at 17q25 (the location of SEPT-9) where a founder mutation has been suggested.
Objective
To study 17q25 and SEPT-9 in S-BPN (56 patients) and H-BPN (13 kindreds).
Methods
Allele analysis at 17q25, SEPT-9 DNA sequencing and mRNA analysis from lymphoblast cultures.
Results
A conserved 17q25 sequence was found in 5 of 13 H-BPN kindreds and one S-BPN patient. This conserved sequence was not found in the family with a SEPT-9 mutation (Arg88Trp) or controls (182). SEPT-9 mRNA expression did not differ between forms of H-BPN and controls. No known mutations of SEPT-9 were found in S-BPN.
Conclusions/Relevance
Rare S-BPN patients have the same conserved 17q25 sequence found in many American H-BPN kindreds. BPN patients with this conserved sequence do not appear to have SEPT-9 mutations or alterations of its mRNA expression levels in lymphoblast cultures. BPN patients with this conserved sequence may have the most common genetic cause in the Americas by a founder effect mutation.
Keywords: sporadic brachial plexus neuropathy (S-BPN), hereditary brachial plexus neuropathy (H-BPN), hereditary neuralgic amyotrophy, SEPT-9, Parsonage-Turner syndrome
Introduction
Hereditary brachial plexus neuropathy (H-BPN) [a.k.a. hereditary neuralgic amyotrophy; OMIM #162100] is an autosomal dominant, variably penetrant disease in which affected persons suffer periodic attacks of unilateral or asymmetric pain, weakness, atrophy, and sensory alterations of the shoulder girdle and upper limb [1, 2]. It is readily distinguished from hereditary neuropathy with pressure palsy (HNPP), which tends to be painless, affects nerves at usual sites of compression, and is associated with mild generalized neuropathy [3]. In H-BPN the symptoms, distribution of neurological findings, and course of the attacks are difficult to distinguish from those of sporadic brachial plexus neuropathy (S-BPN). Both H-BPN and S-BPN attacks may associate with antecedent immunization or infection. Combined with descriptions of brachial plexus nerve inflammation at times of attack altered immunity has been suggested as a potential causative mechanism in BPN attacks [4, 5].
Recently, mutations of the septin-like molecule SEPT-9 (Arg88Trp, Ser93Phe, 5UTR-131G to C) have been reported as molecular genetic abnormalities associated with H-BPN, but they were found in only some kindreds [6]. The septin protein belongs to a class of GTP-binding scaffold proteins thought to be important in membrane dynamics, vesicle trafficking, apoptosis, and cytoskeletal remodeling. The protein is ubiquitously expressed with greater levels of expression in lymphocytes compared to neural tissues [7]. Cell culture transfection experiments using Arg88Trp and Ser93Phe constructs within mouse mammary gland cells have suggested possible disruption of septin interactions or the Rho family of GTPases [8] Why, however, these mutations lead to intermittent attack and possible inflammatory immune mechanism is not understood, and a clear model for BPN has not been established.
Mutations of SEPT-9 have not been identified in American kindreds who share a conserved 500 Kb region of DNA at 17q25 where SEPT-9 is found [6]. Watts et al. [9] predicted that among American H-BPN families with a conserved DNA region at 17q25 a common mutation would be found originating from a shared ancestor, i.e. a founder mutation. Because no mutations in SEPT-9 have been identified in the expressed protein in these families, a mutation that alters expression regulation of SEPT-9 could still be causative, and was not studied by earlier investigations.
Here we test whether 1) a conserved DNA region at 17q25 occurs in S-BPN attacks 2) whether known mutations of SEPT-9 are present in S-BPN and 3) whether SEPT-9 mRNA from lymphocyte cultures differs between American kindreds with and those without a conserved region at 17q25.
Methods
Patients and controls
With institutional review board approval, we studied 13 persons with H-BPN, 56 persons with S-BPN, and 182 non-disease controls. Previously obtained transformed lymphoblast cell lines were available for our studies. Subjects had undergone clinical examination and nerve conduction studies in epidemiologic studies of healthy subjects in Olmstead County. We reported the disease features of four of these H-BPN kindreds who previously had undergone nerve or brachial plexus biopsies during attacks [5]. Probands of each family and sporadic cases had all been examined by us during attacks and had been studied (i.e., nerve conductions, needle electromyography, brachial plexus and or cervical spine imaging, and other laboratory studies from which we judged this disorder to be H-BPN). Confirmation that the disorder was inherited was based either on information from the probands or their families having undergone kindred evaluation.
Studies in the S-BPN case with conserved allele
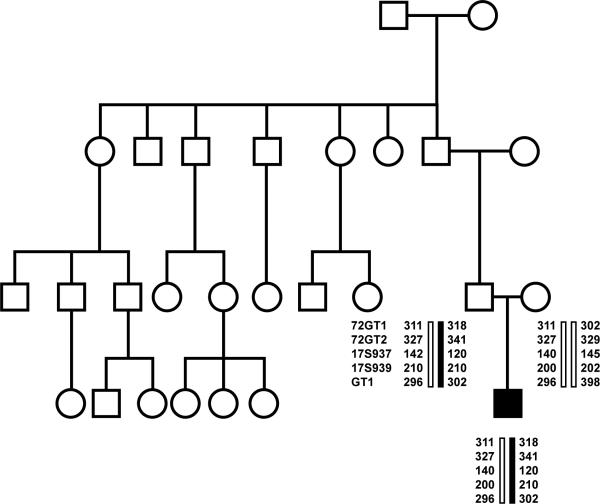
Additional clinical attention was paid to the S-BPN individual with the identified conserved alleles at 17q25 (Fig. 1). That patient had BPN attacks at 6 and 24 years of age. He was from a large family with no other members having known attacks of upper extremity pain or weakness. He was without clear dysmorphic features (i.e. hypotelorism or short stature). His parents were examined and underwent DNA collection. Multiple other family members underwent questioning and medical history review and were without known attacks. We inquired about mimickers of neuritic attacks, i.e., rotator cuff disease (shoulder pain), anterior interosseous syndrome (index and thumb weakness), radial neuropathy (wrist drop), and recurrent pneumonia (diaphragm paralysis), but none were elicited from family members.
Figure 1.
The constructed kindred of one S-BPN individual with the conserved set of alleles at 17q25 also found in our H-BPN kindreds, see Figure 3. No attacks were identified in his other family members including in his father who shared the conserved set of alleles, i.e. asymptomatic carrier (black bar).
SEPT-9 mutation analysis
The 13 probands of kindreds with H-BPN, 56 S-BPN patients and 182 controls all underwent DNA sequencing analysis (pyrosequencing) of the codons previously identified with alterations of [Arg88Trp, Ser93Phe, 5 UTR-131G to C]. For all probands of H-BPN we sequenced the entire open reading frame of SEPT-9, utilizing the PCR primers previously published [6].
Allele studies at 17q25
Based on previously published data [9], fluorescent-labeled oligonucleotide primers 72GT1, 72GT2, D17S937, D17S939, and GT1 were created within a 500 kb region at 17q25. Utilizing these allele markers, linkage and haplotype analysis was performed by conventional fragment analysis methods using a 3730 DNA analyzer and Genotyper software (Applied Biosystems, Forester City, CA). Two-point LOD scores were calculated in 6 of 13 kindreds who were of sufficient size to permit such analysis. Autosomal dominant inheritance was assumed and penetrance was set at 99%. High penetrance was set to allow for discovery of those not localizing to 17q25, i.e. exclusionary linkage. The frequency of mutant alleles was designated at 0.0001. Haplotypes were generated assuming least numbers of crossovers, and prior knowledge of a disease associated set of alleles between affected families [9]. One-hundred-and-eight-two normal individuals also underwent allele analysis of the above polymorphic markers.
Real-time PCR analysis of SEPT-9
Total RNA was extracted from transformed lymphoblasts using RNeay Mini Kit (Qiagen, CA) from 13 patient cell lines with H-BPN, 1 sporadic individual with conserved haplotype at 17q25, and 14 normal controls. Their total RNA was subject to reverse transcription per instruction of Taqman Reverse Transcription Reagents (Applied Biosystems, CA). The amplification cycle is as follows: 25°C for 10 minutes, 48°C for 30 minutes, 95°C for 5 minutes, then maintained at 4°C. Real-time PCR was performed using TaqMan Gene Expression Assays mixes (Applied Biosystems, CA, Part No.: 4331182) labeled with fluorescent color FAM for targeting endogenous control GAPDH (Assay ID: Hs00266705_g1) and target gene SEPT-9 (Assay ID: Hs00246396_m1), respectively. Reverse transcription product (5 ul) was used for real-time PCR setup per instruction of Taqman Universal PCR Mastermix, 2X (Appliedbiosystems, Part No.: 4324018). Amplification Cycle program is as follows: 50°C for 2 minutes, 95°C for 10 minutes, followed by 95°C for 15 seconds, and 60°C for 1 minute for 40 cycles. All real-time PCR data were analyzed by standard comparative ΔΔCT method and compared to normal control samples. Fisher's exact test was used to compare the expression levels between those with and without the conserved 17q25 haplotype and between H-BPN and normal controls.
Results
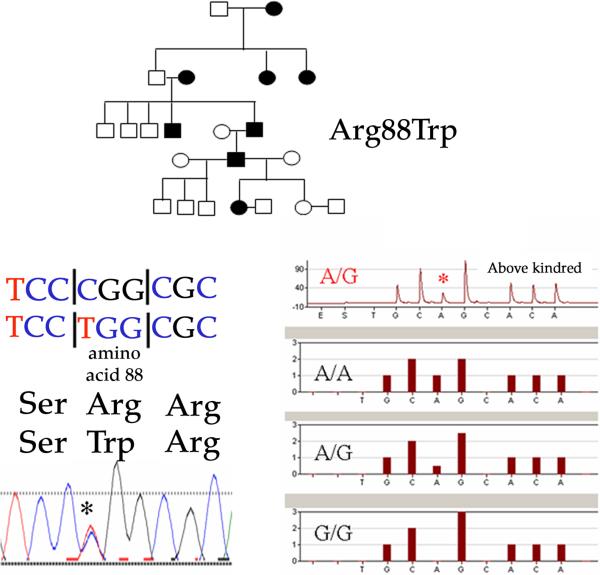
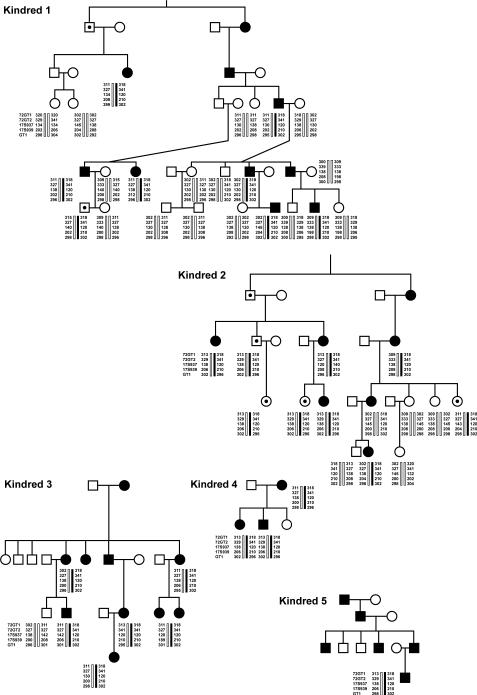
Of the 13 probands of our American kindreds with H-BPN, 1 was identified with a SEPT-9 mutation (Arg88Trp[262C→T]). This family was of German-Austrian, Pennsylvania-Dutch, Irish, and Native-American descent (Fig. 2). Redundant skin folds and short stature were not identified. However, interpupillary distance was short at the 2nd percentile in both affected and unaffected individuals. Haplotype analysis identified a conserved set of alleles [72GT1-318, 72GT2-341, 17S937-120, 17S939-210, GT1-296] in 5 of the 13 kindreds (Fig. 3), although not in this family identified with a SEPT-9 mutation (Arg88Trp) or the 182 normal controls. Additionally, these same alleles were found in one S-BPN individual and his unaffected father described above and shown in Figure 1. The maximal 2-point LOD scores in 5 of 6 kindreds favored linkage to 17q25, with values ranging from 0.49 to 3.7 LOD with juxtaposed markers consistent with linkage to this region. Five of these kindreds were those with the conserved haplotype (Fig. 3). None of the 56 S-BPN cases or the 182 control subjects had known mutations of SEPT-9, based on the results from complete lymphoblast open reading frame DNA sequencing analysis of all probands. Specifically, the S-BPN individual with the conserved haplotype but without family history was not identified with any mutations. SEPT-9 RNA levels by RT-PCR comparing the H-BPN patients with the conserved 17q25 haplotype and without were not statistically different. Additionally, no significant difference occurred between normal control SEPT-9 mRNA levels and H-BPN.
Figure 2.
One of 13 kindreds studied with hereditary brachial plexus neuropathy (H-BPN) identified with SEPT-9 mutation. Top, 5-generation kindred affected by recurrent attacks of brachial plexus neuropathy with apparent dominant inheritance. (bottom, left) Sequencing analysis identifying 262C→T transition leading to Arg88Trp. (bottom, right) Complimentary strand confirmatory pyrosequencing assay showing A and G heterozygous occurrence at base position 262, with kindred evaluation on top and possible combination below.
Figure 3.
Five kindreds of 13 with H-BPN and identified with a conserved set of alleles (black bar) at 17q25 when analyzed by polymorphic markers [72GT1, 72GT2, 17S937, 17S939, GT1]. Haplotypes were generated assuming least numbers of crossovers, and prior knowledge of a disease associated set of alleles between affected families with H-BPN [9].
Discussion
The clinical and histopathologic similarities between S-BPN and H-BPN [1, 2, 5] suggest the possibility that a shared underlying mechanism is important in both disorders. Several different genetic or epigenetic mechanisms are likely operational [10]. Previously, intensive questioning and examination of family members for a history of symptoms and signs of brachial plexopathy was the mainstay of identifying hereditary predisposition to BPN. Now, genetic discoveries in H-BPN [6, 10] permit addressing whether similar alterations at SEPT-9 and 17q25 might explain the similarities between S-BPN and H-BPN patients. The current molecular information suggests at least five genetic classifications for BPN attacks: 1) H-BPN with SEPT-9 mutation; 2) H-BPN with conserved 17q25 haplotype; 3) H-BPN not localizing to 17q25; 4) S-BPN with conserved 17q25 haplotype; 5) S-BPN without a known genetic abnormality.
The evidence for genetic predisposition in some S-BPN cases in our study is not definitive, rather suggestive. Specifically, only one S-BPN patient of 56 was found with the same conserved sequence seen at 17q25 among American H-BPN kindreds. No known mutations of SEPT-9 were identified in that patient or other S-BPN patients. Genetically, it appears this presumed S-BPN individual's family is at risk for attacks of brachial plexopathy. The fact that this patient had his first attack at 6 and experienced recurrent attacks at older years supports this possibility [2]. The absence of attacks in other family members including his father who carries the same conserved chromosome region might suggest low penetrance in this family for unclear reason. Alternatively, we considered whether this conserved set of alleles could be found in normal individuals not at risk, but did not find similar allele combination in 182 normal examined controls, nor prior report of such phenomena in 50 additional controls [9].
Our data supports the earlier work by Watts et al. [9] that a shared mutation which localizes to 17q25 has yet to be identified in many families with H-BPN. Specifically, among our kindreds, 5 of 13 had a conserved region of DNA at 17q25, suggesting a common mutation inherited from a shared ancestor in each of these families, (i.e. a founder mutation). In those 5 families, 88% of affected individuals had the conserved region of DNA, suggesting high penetrance in most families intensely studied. The percentage of H-BPN kindreds with the conserved 17q25 region is less than previously reported by Watts et al. [9], where 6 of 7 kindreds shared the conserved region. Their study, however, is different from ours in that all families were pre-selected by linkage to have causative genetic localization at 17q25. In our study, only 6 of 13 kindreds were of sufficient size to allow for linkage analysis. Of these, 5 had LOD scores suggestive of chromosomal localization to 17q25, and all but 1 shared the conserved haplotype at 17q25; that family had the SEPT-9 Arg88Trp mutation. Overall, our observations are consistent with genetic diversity as a cause for H-BPN by finding kindreds without SEPT-9 gene abnormality or conserved 17q25 sequence.[10]
Whether some of the genetic diversity identified in H-BPN relates to occult SEPT-9 distant promotor or alternative gene mutation(s) resulting in variable SEPT-9 mRNA expression levels was also addressed. The level of mRNA expression between our kindreds with and without the conserved 17q25 haplotype was not significantly different by RT-PCR analysis from lymphoblast cell lines. There was also no significant difference between expression levels in affected individuals compared to normal controls. The relevance of this data depends on whether SEPT-9 is still responsible in those with the conserved 17q25 sequence despite normal conventional DNA sequencing of that gene. Also the relevance of these mRNA studies depends on whether the primary pathogenesis of H-BPN is humeral based i.e. an inflammatory mechanism. Our data suggests no obvious expression level difference in lymphoblasts. Additional research into SEPT-9 mRNA evaluation looking for aberrant splicing or transcript variance in nerve could be the next step in evaluation of whether such mechanisms could represent the founder mutation in American H-BPN kindreds.
Molecular abnormalities other than mutations of SEPT-9 exons or the exon-intron boundaries sequenced here are likely responsible for many cases of H-BPN. A single genetic mutation of H-BPN in American kindreds with the conserved 17q25 sequence is supported by this work (i.e., founder mutation), and appears the most common identified genetic cause in the Americas. Regardless of the identity of additional molecular markers in H-BPN interrogation of S-BPN patients with discovered abnormalities will be important in consideration of additional genetic influence among presumed S-BPN.
Acknowledgment
: This work was supported in part from grants received from the National Institute of Neurologic Disorders and Strokes (NS36797), a previous grant from the Muscular Dystrophy Association, and by CA15083 for the Mayo Gene Discovery Shared Resource (Mayo Comprehensive Cancer Center Grant) from the National Cancer Institute. We would like to thank the affected individuals for their invaluable help.
References
- 1.Klein CJ, Windebank AJ. Hereditary Brachial Plexus Neuropathy. In: Dyck PJ, Thomas PK, editors. Peripheral Neuropathy. Fourth Edition Elsevier; Philadelphia: 2005. pp. 1753–67. [Google Scholar]
- 2.van Alfen N, van Engelen BG. The clinical spectrum of neuralgic amyotrophy in 246 cases. Brain. 2006;129(Pt 2):438–50. doi: 10.1093/brain/awh722. [DOI] [PubMed] [Google Scholar]
- 3.Windebank AJ, Schenone A, Dewald GW. Hereditary neuropathy with liability to pressure palsies and inherited brachial plexus neuropathy--two genetically distinct disorders. Mayo Clin. Proc. 1995;70(8):743–6. doi: 10.4065/70.8.743. [DOI] [PubMed] [Google Scholar]
- 4.Suarez GA, Giannini C, Bosch EP, Barohn RJ, Wodak J, Ebeling P, et al. Immune brachial plexus neuropathy: Suggestive evidence for an inflammatory immune pathogenesis. Neurology. 1996;46(2):559–61. doi: 10.1212/wnl.46.2.559. [DOI] [PubMed] [Google Scholar]
- 5.Klein CJ, Dyck PJ, Friedenberg SM, Burns TM, Windebank AJ. Inflammation and neuropathic attacks in hereditary brachial plexus neuropathy. J. Neurol. Neurosurg. Psychiatry. 2002;73(1):45–50. doi: 10.1136/jnnp.73.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhlenbaumer G, Hannibal MC, Nelis E, Schirmacher A, Verpoorten N, Meuleman J, et al. Mutations in SEPT9 cause hereditary neuralgic amyotrophy. Nat. Genet. 2005;37(10):1044–46. doi: 10.1038/ng1649. [DOI] [PubMed] [Google Scholar]
- 7.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. Updated http://genome.ucsc.edu (2006) [DOI] [PMC free article] [PubMed]
- 8.Sudo K, Ito H, Iwamoto I, Morishita R, Asano T, Nagata K. SEPT9 sequence alternations causing hereditary neuralgic amyotrophy are associated with altered interactions with SEPT4/SEPT11 and resistance to Rho/Rhotekin-signaling. Hum. Mutat. 2007;28(10):1005–13. doi: 10.1002/humu.20554. [DOI] [PubMed] [Google Scholar]
- 9.Watts GD, O'Briant KC, Chance PF. Evidence of a founder effect and refinement of the hereditary neuralgic amyotrophy (HNA) locus on 17q25 in American families. Hum. Genet. 2002;110(2):166–72. doi: 10.1007/s00439-001-0647-5. [DOI] [PubMed] [Google Scholar]
- 10.Watts GD, O'Briant KC, Borreson TE, Windebank AJ, Chance PF. Evidence for genetic heterogeneity in hereditary neuralgic amyotrophy. Neurology. 2001;56(5):675–78. doi: 10.1212/wnl.56.5.675. [DOI] [PubMed] [Google Scholar]