Abstract
The complement lectin pathway, an essential component of the innate immune system, is geared for rapid recognition of infections as each C4b deposited via this pathway is capable of forming a C3/C5 convertase. In the present study, role of C4b-binding protein (C4BP) in regulating the lectin pathway C3/C5 convertase assembled on zymosan and sheep erythrocytes coated with mannan (EMan) was examined. While the C4BP concentration for inhibiting 50% (IC50) formation of surface-bound C3 convertase on the two surfaces was similar to that obtained for the soluble C3 convertase (1.05 nM), ∼3- and 41-fold more was required to inhibit assembly of the C5 convertase on zymosan (2.81 nM) and EMan (42.66 nM). No difference in binding interactions between C4BP and surface-bound C4b alone or in complex with C3b was observed. Increasing the C4b density on zymosan (14,000-431,000 C4b/Zym) increased the number of C4b bound per C4BP from 2.87 to 8.23 indicating that at high C4b density all seven α-chains of C4BP are engaged in C4b-binding. In contrast, the number of C4b bound per C4BP remained constant (3.79 ± 0.60) when the C4b density on EMan was increased. The data also show that C4BP regulates assembly and decay of the lectin pathway C3/C5 convertase more stringently than the classical pathway C3/C5 convertase because of a ∼7 to 13-fold greater affinity for C4b deposited via the lectin pathway than the classical pathway. C4BP thus regulates efficiently the four times greater potential of the lectin pathway than the classical pathway in generating the C3/C5 convertase and hence production of pro-inflammatory products, which are required to fight infections but occasionally cause pathological inflammatory reactions.
1. Introduction
Strong similarities between the M1 complex (MBL-MASPs) of the lectin pathway and the C1 complex (C1qrs) of the classical pathway had initially implicated that the overall process of complement activation and regulation of the lectin pathway might be similar to that of the classical pathway. But studies examining different aspects of complement activation have suggested differences between the two pathways (Rossi et al., 2001; Harmat et al., 2004; Rawal et al., 2008). Recent studies by Rawal et al., (2008) have shown that complement activation via the lectin pathway has the potential of generating four times more C3/C5 convertase than the classical pathway and hence more pro-inflammatory products. This is because every C4b deposited via the lectin pathway forms a convertase in contrast to the classical pathway in which only one out of four C4b deposited forms a C3/C5 convertase (Rawal et al., 2003, 2008). In addition, studies employing recombinant forms of mannan-binding lectin (rMBL) have shown that the structural variants of MBL not only form higher order oligomers similar to wild type MBL (MBL/A) but that the structural variant rMBL/D activates the complement lectin pathway to the same extent as rMBL/A while rMBL/B activates weakly and rMBL/C is ineffective (Rajagopalan et al., 2009). Together, these new findings imply that activation of the lectin pathway by MBL or its structural variant can produce significant amounts of anaphylatoxins, C3a and C5a, in a very short time. This can be particularly harmful in infants whose innate immune system relies on activation of the lectin pathway as one of its responses to fight infections (Aittoniemi et al., 1996) or under conditions when unregulated activation of the lectin pathway might be a more prominent contributor to the pathology of inflammatory reactions and tissue damage following oxidative stress (Collard et al., 2000), ischemia and reperfusion injury (Jordan et al., 2001), and in some types of renal diseases (Matsuda et al., 1998; Lhotta et al., 1999; Endo et al., 2000). These reports when considered with the potential therapeutic application of recombinant MBL in MBL-deficient patients emphasize the need to understand the processes of complement activation and regulation via the lectin pathway.
The C3/C5 convertase of the lectin pathway and the classical pathway assembled with C4b as the noncatalytic subunit (C4b,C2a) cleave both C3 and C5 (Rawal et al., 2003, 2008). But C4b,C2a mainly cleaves C3 because it has a weak affinity for C5 and is therefore called a C3 convertase (Rawal et al., 2003, 2008). Deposition of additional C3b molecules by C4b,C2a converts the low affinity C3 convertase to a high affinity C5-binding convertase (C3bC4b,C2a) (Rawal et al., 2003, 2008), which in the present study has been called a C5 convertase. Cleavage products of C3 (C3b and C3a) and C5 (C5b and C5a) have important biological activities that help fight infections. The products recognize targets, mediate inflammatory responses by stimulating neutrophils and phagocytes to the site of injury or infection, and help kill and/or clear microorganisms and altered host cells (Ember et al., 1998). However, these products may also contribute to the pathophysiology of several diseases and conditions such as inflammatory diseases, reperfusion injury and xenotransplantation rejection (Rother, 1998).
Studies analyzing regulation of the lectin pathway of complement have mainly focused on the regulatory role of C1-INH (Wong et al., 1999; Petersen et al., 2000; Nielsen et al., 2007; Kerr et al., 2008). C1-INH has been shown to be a major regulator of MASP-2 than of C1s (Nielsen et al., 2007; Kerr et al., 2008) and based on different sensitivities to various synthetic inhibitors, differences in control of complement activation via the lectin pathway and the classical pathway have been suggested (Petersen et al., 2000). Although the efficiency of formation of the C3/C5 convertase via the two pathways has been reported to be different (Rawal et al., 2008) and regulation of the classical pathway C3/C5 convertase examined by several groups (Parker, 1992; Liszewski et al., 1996; Morgan and Harris, 1999), studies on the regulation of the lectin pathway C3/C5 convertase are limited (Suankratay et al., 1999).
The soluble complement regulator, C4BP is a large plasma protein that exists in many forms with varying subunit composition. The major isoform is made up of seven α-chains (70 kDa) and one β-chain (45 kDa) (α7β1) (Hillarp and Dahlbäck, 1988). The α-chain of C4BP has binding sites for many ligands (Blom et al., 2004a), which include C4b, heparin, serum amyloid protein (SAP), CD40, and CD154 while the β-chain has been reported to bind the vitamin K-dependent anticoagulant protein S. Many pathogens have been reported to bind to the α-chain of C4BP and evade complement-mediated killing (Blom, 2002; Blom et al., 2004b). C4BP controls complement activation by interacting with the noncatalytic subunit C4b of the C3/C5 convertase and thereby inhibits assembly of the convertase, enhances its decay, and exhibits cofactor activity for factor I in inactivating both fluid phase and cell-bound C4b (Fujita et al., 1978; Fujita and Tamura, 1983; Blom et al., 1999; Rawal et al., 2007).
In the present study C4BP regulation of the lectin pathway C3/C5 convertase assembled with purified complement components on zymosan particles, a natural surface rich in mannose residues was examined. For comparison with published studies on the classical pathway C3/C5 convertase that was assembled on sheep erythrocytes (ES) coated with antibody (EA) (Rawal et al., 2007), regulation of the lectin pathway C3/C5 convertase assembled on sheep erythrocytes coated with mannan (EMan) was also examined. The regulatory role of C4BP in inhibiting enzyme formation and enhancing enzyme decay was determined by hemolytic assays measuring the C5 cleavage activity of the lectin pathway C3/C5 convertase. Our findings show that C4BP controls the assembly and decay of the lectin pathway C3/C5 convertase more stringently than the classical pathway C3/C5 convertase. The greater efficiency of C4BP in regulating the C3/C5 convertase of the lectin pathway than the classical pathway is attributed to its higher affinity for C4b deposited via the lectin pathway than the classical pathway. In addition, the data show that on zymosan the polymeric regulator mediates efficient control by utilizing all seven α-chains, each capable of binding one C4b, to regulate the four times greater potential of the lectin pathway than the classical pathway in generating C3/C5 convertase and hence production of proinflammatory products.
2. Materials and Methods
2.1 Reagents
Sheep erythrocytes (ES) and chicken erythrocytes (EC) were isolated from whole blood purchased from Colorado Serum Co. (Denver, CO). Mannan, chromium chloride, NP-40 (nonidet P-40), Tween 20, and EDTA were purchased from Sigma Chemical Co. (St. Louis, MO). Veronal buffered saline (VBS) contained 5 mM barbital, 145 mM NaCl and pH 7.4. Gelatin veronal-buffered saline (GVB) was VBS containing 0.1% gelatin, while GVB++ was GVB containing 0.5 mM MgCl2 and 0.15 mM CaCl2, GVBE was GVB containing 10 mM EDTA.
2.2 Purified proteins
Complement proteins, M1, C2, C3, C4, C5, C4b monomer, and C6 were purified from normal human plasma as described (Rawal et al., 2008). Complement components, C4, purified C5b,6, and C4BP were obtained from CompTech (Tyler, TX). All proteins were homogenous by polyacrylamide gel electrophoresis. Functional activity of C2, C4, C5, C6, and C4BP was determined as explained previously (Rawal et al., 2008). Protein concentrations of C2, C3, C4, C5, C6, C5b,6, and C4BP were determined spectrophotometrically as described (Rawal et al., 2008). All purified proteins were stored at −76°C.
2.3 Preparation of ZymM1,C4b, ZymM1,C3bC4b, and ZymC3b cells
ZymM1,C4b was made as described (Rawal et al., 2008). Briefly, zymosan was incubated with M1 in the presence of CaCl2 and MgCl2 to make ZymM1. ZymM1, C4b was made by incubating ZymM1 with radiolabeled (125I) C4. Total number of C4b deposited on washed ZymM1 was determined by the uptake of radiolabeled C4 as C4b. Zymosan having no M1 was employed as control for background C4b binding. ZymM1,C4b was washed and the process repeated until the desired numbers of C4b/Zym were obtained. ZymM1,C3bC4b was made by incubating ZymM1,C4b with C3 and C2 as described (Rawal et al., 2008). Number of C3b molecules bound to ZymM1,C3bC4b was determined by using 125I-Factor B as described (Rawal et al., 1998). The procedure was repeated until the desired numbers of C3b/Zym were obtained. Zymosan having no M1 was employed as controls for background C3b binding. Zymosan bearing only C3b (ZymC3b) was prepared as described (Rawal et al., 2007) and zymosan bearing no C3b was employed as control.
2.4 Preparation of EManM1,C4b and EManM1,C3bC4b cells
Sheep erythrocytes (ES) were coated with mannan (EMan) by the method of Ikeda et al., (Ikeda et al., 1987) as described (Rawal et al., 2008). EManM1 was made by incubating EMan with M1 in the presence of CaCl2 and MgCl2. EManM1 cells were incubated with 125I-C4 to make EManM1,C4b. The number of C4b deposited on EManM1 cells was determined as described above for zymosan. EManM1,C3bC4b cells were made by depositing C3b on EManM1,C4b as described (Rawal et al., 2008).
2.5 Inhibition of formation of the lectin pathway C3/C5 convertase on zymosan and EMan by C4BP measured as the amount of C5 convertase activity remaining
Cleavage of C3 results in C3b generation and deposition of the C3b molecules converts the C3 convertase (C4b,C2a) to a C5 convertase (C3bC4b,C2a). Thus, usage of a C3 cleavage assay would result in analyzing activity of the C5 convertase formed but a C5 cleavage assay would allow analysis of the C3 converatse activity separate from the C5 convertase. Therefore in the present study inhibition of formation of the lectin pathway C3 and C5 convertases by C4BP was measured as the amount of C5 convertase activity in the absence or presence of varying concentrations of C4BP. The C5 cleavage assay was done in two steps. Since formation of enzyme required less than a minute (Rawal et al., 2008), the convertase was assembled in the same reaction mixture in which C5 cleavage assays were done. Surface-bound C3 convertase (ZymM1,C4b,C2a) was assembled on zymosan by incubating ZymM1,C4b with C5 (9 μg, 1.9 μM), saturating concentrations of C2 (0.1 μg, 39 nM) and C6 (0.5 μg, 167 nM), and varying concentrations of C4BP in a total volume of 25 μl of GVB++. Depending on the density of C4b per cell, the concentration of cells was adjusted so as to give ∼1-34 ng of bound C4b in a final volume of 25 μl GVB++ resulting in 0.2-7 nM enzyme concentration. The concentration of enzyme was controlled by using limiting amounts of bound C4b while saturating concentrations of C2 in the reaction mixture maintained a constant amount of enzyme during a 10-min assay even though the lectin pathway C3/C5 convertase was decaying and reforming as has been shown previously (Rawal et al., 2008). After 10 min incubation at 37 °C, the tubes were transferred to an ice bath to prevent further assembly of the enzyme as well as further cleavage of C5. In the second part, an appropriate aliquot of the complete reaction mixture was analyzed for C5b formation as the amount of C5b,6 produced by hemolytic assays using chicken erythrocytes (EC) as described below. Activity determined in the absence of C4BP was considered 100% and the concentration required to inhibit fifty percent activity (IC50) was determined. Surface-bound C5 convertase (ZymM1,C3bC4b,C2a) was assembled on zymosan as described for the C3 convertase (ZymM1,C4b,C2a) above except that ZymM1,C3bC4b and a lower concentration of C5 (0.45 μg, 95 nM) was used.
Inhibition of C3/C5 convertase assembly on EMan cells by C4BP was measured as described above for zymosan except that EManM1,C4b and EManM1,C3bC4b cells were used for assembling the C3 convertase (EManM1,C4b,C2a) and the C5 convertase (EManM1,C3bC4b,C2a), respectively.
2.6 Inhibition of formation of soluble C3 convertase by C4BP
The soluble C3 convertase (C4b,C2a) was assembled by incubating limiting amounts of C4b monomer (0.013-0.026 μg, 2.7-5.3 nM) with C5 (9.6 μg, 2.02 μM), M1 (0.05-0.18 μg) and saturating concentrations of C2 (0.20-0.79 μg, 78-310 nM) and C6 (0.5-1.0 μg, 167-333 nM), and varying concentrations of C4BP in a final volume of 25 μl GVB++. The reaction was started with the addition of C4b. After 10 min incubation at 37 °C, an appropriate aliquot of the complete reaction mixture was analyzed for C5b formation as the amount of C5b,6 produced by hemolytic assays using EC as described below. Enzyme formed in the absence of C4BP was considered 100%.
2.7 Quantitation of reaction products
C5b,6 was measured hemolytically using EC as described (Rawal et al., 1998). Briefly, an aliquot of the complete C5 convertase assay mixture was added to another reaction mixture containing 1.4×106 EC and 5 μ1 of pooled normal human serum in a final volume of 225 μl GVBE. After 10 min incubation at 37 °C, unlysed cells were removed by centrifugation and the amount of hemoglobin released was quantitated spectrophotometrically at 414 nm. 100% lysis was EC lysed in GVBE containing 0.5 % NP-40. Controls containing C5 and C6, but no C2, were subtracted as background. C5b,6 concentration was quantitated from a standard curve using purified C5b,6 as described previously (Rawal et al., 1998).
2.8 Decay acceleration of the lectin pathway C3/C5 convertase assembled on zymosan and EMan by C4BP
Decay accelerating activity of C4BP on the C3 convertase assembled on zymosan and EMan cells bearing 431,000 C4b/Zym and 23,000 C4b/EMan was determined as described (Rawal et al., 2008). Decay accelerating activity of C4BP on the C5 convertase (ZymM1,C3bC4b,C2a and EManM1,C3bC4b,C2a) assembled on zymosan bearing 20,000 C4b and 47,000 C3b per zymosan and EMan cells bearing 450,000 C4b and 1,728,000 C3b was determined as described previously (Rawal et al., 2008).
2.9 Binding assays
Binding of C4BP to zymosan-bound C4b alone, C3b alone, and C3bC4b complexes was measured as the amount of radiolabeled C4BP bound as cpm under equilibrium conditions as described previously (Rawal et al., 2007). ZymM1,C4b or ZymM1,C3bC4b was incubated with varying concentrations of 125I-C4BP at 37 °C for 45 min. Bound and free radiolabeled C4BP were separated by layering the reaction mixture on 250 μl 20% sucrose in GVB++ and centrifuging for 1 min at 10,000×g at 22 °C. The amount of 125I-C4BP bound was calculated from the radioactivity in the pellets after subtracting the contribution of 125I-C4b and nonspecific binding of 125I-C4BP to zymosan having no M1. For ZymC3b cells, non-specific binding of 125I-C4BP to zymosan having no C3b was subtracted as control. The data obtained were fit to one-site binding equation using GraFit version 5.06 Erithacus software to give the apparent dissociation constant Kd. Binding of C4BP to C3bC4b complexes and to C4b alone on sheep erythrocytes coated with mannan (EMan) was determined as described for zymosan except that EManM1,C4b and EManM1,C3bC4b cells were used. Non-specific binding of 125I-C4BP to EMan cells having no M1 was subtracted along with the contribution by 125I-C4b to give the number of C4BP bound per cell.
2.10 Data analysis
Binding data obtained were fit by nonlinear regression analysis using Grafit version 5.0 software (Erithacus software, Staines, England) to a one-site binding equation: y = [L]Cap/Kd + [L], where y is the amount of radiolabeled ligand bound, [L] the concentration of the free ligand, Cap the capacity for binding the ligand, and Kd is the apparent dissociation constant. Inhibition data were fit by nonlinear regression analysis to the following equation: % radiolabeled protein bound = 100%/(l + [inhibitor concentration/IC50]s), where s is the slope factor.
2.11 Preparation of radiolabeled proteins
Factor B, C4 and, C4BP (100 μg) were radiolabeled with 125I for 30 min at 0°C in a glass tube coated with Iodogen (Pierce Chemical Co., USA). Free 125I was removed by centrifugal desalting (Christopherson, 1983). Specific activities of radiolabeled factor B ranged from 2.09-4.16 μCi/μg. Radiolabeled C4, and C4BP were diluted with cold C4 and C4BP, respectively, to give specific activities of 0.05-0.85 μCi/μg.
3. Results
3.1 Inhibition of formation of the lectin pathway C3/C5 convertase on zymosan by C4BP
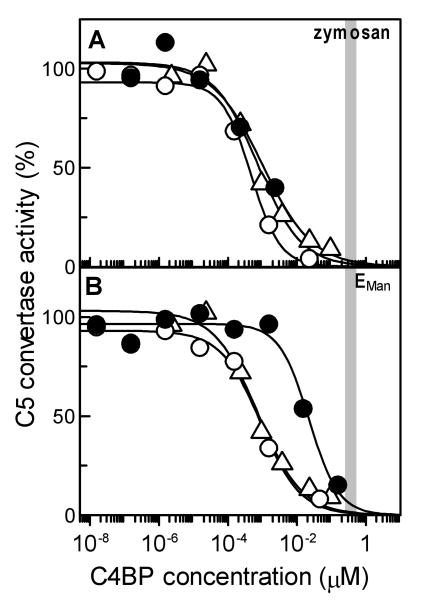
Role of C4BP in regulating assembly of the lectin pathway soluble C3 convertase and the surface-bound C3 and C5 convertases on zymosan particles was determined by measuring the amount of C5 convertase formed in the presence of varying concentrations of C4BP. A comparison of the extent of inhibition at normal serum concentration of C4BP (250 to 400 nM shown as the grey shaded area, Fig. 1A) revealed that greater than 98% of the soluble (C4b,C2a) and surface-bound (ZymM1,C4b,C2a) forms of the C3 convertase were inhibited from assembling on zymosan. The IC50 measured for inhibiting formation of the surface-bound C3 convertase (1.27 ± 1.09 nM) was similar to that obtained for the soluble form of enzyme (1.05 ± 0.36 nM) indicating similar sensitivities for the two forms of the lectin pathway C3 convertase to C4BP regulation (Table 1). The IC50 measured for inhibiting assembly of the C5 convertase (ZymM1,C3bC4b,C2a, 2.81 ± 2.46 nM) was 2- and 3-fold higher than that obtained respectively for the surface-bound and soluble forms of the C3 convertase suggesting that C4BP regulates formation of the C3 convertase more effectively than the C5 convertase. Nevertheless, at normal serum concentration of C4BP (250 to 400 nM shown as the grey shaded area) more than 95% of the C5 convertase was inhibited from assembling on zymosan.
Fig. 1.
Inhibition of formation of the lectin pathway C3/C5 convertase by C4BP on zymosan and EMan cells measured as the amount of C5 convertase activity remaining. (A) The C3 convertase was allowed to form on zymosan particles bearing 346,000 C4b/Zym while the C5 convertase was assembled on zymosan bearing 54,000 C4b and 362,000 C3b per zymosan in the presence of indicated concentration of C4BP. Inhibition of enzyme formation was measured as the amount of C5 convertase activity remaining by determining the amount of C5b,6 formed using hemolytic assay with EC. C5b,6 formed in the absence of C4BP was considered 100%. To obtain IC50, the data were fit by nonlinear regression using Grafit version 5.0 Erithacus software. Vertical bar indicates the normal concentration range of C4BP in plasma (250-400 nM). Symbols: soluble C3 convertase C4b,C2a (Δ); surface-bound C3 convertase, ZymM1,C4b,C2a (○); surface-bound C5 convertase, ZymM1,C3bC4b,C2a (●). (B). Inhibition of formation of the lectin pathway C3/C5 convertase on EMan cells by C4BP. Experimental details are as described above in A except that EMan cells bearing 471,000 C4b/EMan were employed to assemble the C3 convertase while cells bearing 138,000 C4b and 110,000 C3b per EMan were used for assembling the C5 convertase. Symbols: soluble C3 convertase, C4b,C2a(Δ); surface-bound C3 convertase, EManM1,C4b,C2a (○); surface-bound C5 convertase, EManM1,C3bC4b,C2a (●).
Table 1.
Inhibition of formation of C3/C5 convertases by C4BP
| Enzyme | IC50a (nM) |
|---|---|
| Lectin pathway C3/C5 convertase | |
| Soluble C3 convertase (C4b,C2a) | 1.05 ± 0.36 |
| Surface-bound C3 convertase (ZymM1,C4b,C2a) | 1.27 ± 1.09 |
| Surface-bound C3 convertase (EManM1,C4b,C2a) | 2.63 ± 1.69 |
| High Affinity C5 convertase (ZymM1,C3bC4b,C2a) | 2.81 ± 2.46 |
| High Affinity C5 convertase (EManM1,C3bC4b,C2a) | 42.66 ± 42.33 |
| Classical pathway C3/C5 convertasesb | |
| Soluble C3 convertase (C4b,C2a) | 5 ± 3 |
| Surface-bound C3 convertase (EAC1,C4b,C2a) | 35 ± 12 |
| High affinity C5 convertase (EAC1,C3bC4b,C2a) | 72 ± 16 |
IC50, C4BP concentration required to inhibit formation of C3 convertases and high affinity C5 convertases by 50% as described in Materials and Methods and shown in Figs. 1 and 2. The lectin pathway C3/C5 convertases were assembled on zymosan or EMan cells bearing different densities of C4b (21,000-346,000 C4b/Zym and 8,000-471,000 C4b/EMan). Values are mean ± S.D. (n = 3 or more)
IC50 values for classical pathway C3/C5 convertases that were assembled on EAC1 cells bearing different densities of C4b (9,300-462,000 C4b/EA) and have been reported previously (Rawal et al., 2007).
3.2 Inhibition of formation of the lectin pathway C3/C5 convertase on EMan cells by C4BP
C4BP inhibited formation of the C3 and C5 convertases on EMan but the extent varied as seen in Fig. 1B. A comparison of the extent of inhibition at normal serum concentration of C4BP (250-400 nM shown as the grey shaded area) revealed greater than 98% inhibition of the surface-bound C3 convertase (EManM1,C4b,C2a) whereas about 90% of the C5 convertase (EManM1,C3bC4b,C2a) was inhibited. The IC50 for inhibiting assembly of the C3 convertase on EMan (EManM1,C4b,C2a, 2.63 ± 1.69 nM) was 2-fold while that for the C5 convertase (EManM1,C3bC4b,C2a, 42.66 ± 42.33 nM) was 41 times more than the IC50 obtained for the soluble C3 convertase (1.05 ± 0.36 nM) (Table 1). These results show that C4BP regulates formation of the C3 convertase more effectively than the C5 convertase on EMan.
3.3 Effect of C4b density on C4BP regulation of surface-bound C3 convertase assembly on zymosan and EMan cells
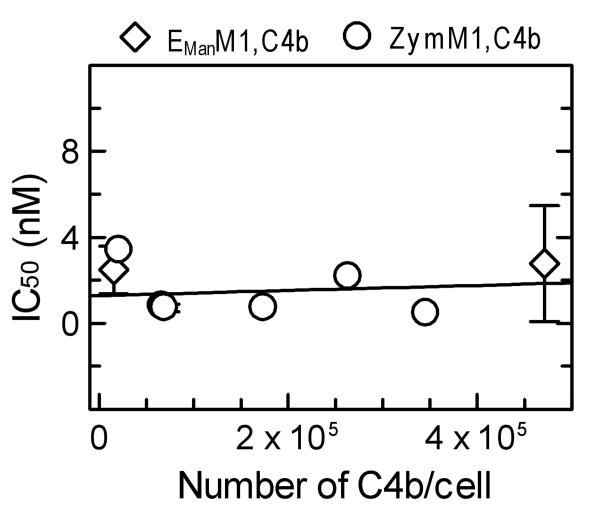
The effect of C4b density on C4BP regulation of the lectin pathway C3 convertase was examined by using zymosan bearing different densities of C4b (22,000-346,000 C4b/Zym) for assembling the lectin pathway C3 convertase (ZymM1,C4b,C2a). Inhibition of formation of ZymM1,C4b,C2a was determined by measuring its C5 convertase activity (Materials and Methods). As seen in Fig. 2, the IC50 obtained did not vary even though the C4b density on the cell surface was increased by 16-fold (22,000-346,000 C4b/Zym). The average IC50 was 1.27 ± 1.09 nM (Table I). Similar results were obtained when the C4b density on EMan cells was increased by 31-fold (15,000-471,000 C4b/EMan, Fig. 2). The average IC50 was 2.63 ± 1.69 nM (Table I). Together, the results show that under the conditions employed the inhibition efficiency of C4BP for the lectin pathway C3 convertase assembled on zymosan or EMan cells is not influenced by the number of C4b molecules deposited via the lectin pathway.
Fig. 2.
Effect of C4b density on C4BP regulation of surface-bound C3 convertase assembly on zymosan and EMan cells. Inhibition of C3 convertase (ZymM1,C4b,C2a and EManM1,C4b,C2a) assembly on zymosan and EMan cells bearing different amounts of C4b/cell ranging from 22,000-346,000 C4b/Zym and 15,000-471,000 C4b/EMan. The number of C4b/cell was measured using 125I-labeled C4. Inhibition of enzyme formation was measured as the amount of C5 convertase activity remaining as described in Fig. 1. The IC50 for each density of C4b/cell employed in the assays was calculated from individual inhibition plots. These IC50 values were considered as one set of data and were fit by linear regression using Grafit version 5.0 Erithacus software. Symbols: ZymM1,C4b,C2a (○); EManM1,C4b,C2a (◊).
3.4 Equilibrium binding interactions between C4BP and surface-bound C4b
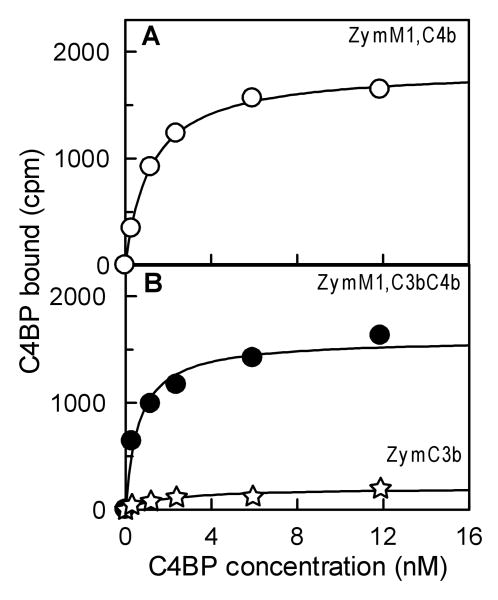
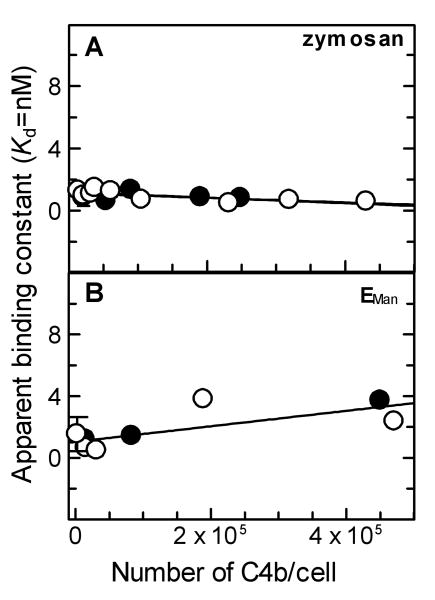
Binding affinity of C4BP for surface-bound C4b deposited via the lectin pathway was determined by measuring binding of radio-labeled C4BP to ZymM1,C4b bearing different densities of C4b/cell under equilibrium conditions. Fig. 3A shows the data obtained upon the binding of radiolabeled C4BP to ZymM1,C4b bearing 59,000 C4b/Zym. Non-linear regression analysis of the data using a one-site binding equation indicated an apparent dissociation constant (Kd) of 1.19 nM, suggesting a high affinity binding interaction between C4BP and surface-bound C4b. Increasing the density of C4b/Zym by 31-fold (14,000 to 431,000 C4b/Zym) did not change the binding affinity of C4BP for surface-bound C4b (Fig. 4A). The average apparent Kd was 0.95 ± 0.35 nM. These results indicate that the already high affinity binding interaction between C4BP and C4b did not become stronger with increasing C4b density.
Fig. 3.
Equilibrium binding of C4BP to surface-bound C4b alone, C3b alone, and C4b-C3b complexes on zymosan. (A). C4BP binding to surface-bound C4b was measured as the amount of 125I-C4BP bound as cpm to ZymM1,C4b (○) under equilibrium conditions. ZymM1,C4b (0.44×106) bearing 59,000 C4b/Zym was incubated for 45 min at 37 °C with indicated concentration of 125I-C4BP (4.1×105 cpm/μg). The y axis represents the amount of radiolabeled C4BP bound to ZymM1C4b as cpm. The data were fit by nonlinear regression to a one-site binding equation to obtain the apparent binding constant (Kd). (B). C4BP binding to surface-bound C4b-C3b complexes was analyzed as described in Fig. 3(A) except that 0.44×106 ZymM1,C3bC4b bearing 54,000 C4b and 362,000 C3b/Zym was incubated with radiolabeled 125I-C4BP (4.1×105 cpm/μg). C4BP binding to zymosan bearing only C3b was also analyzed by incubating ZymC3b bearing 181,000 C3b/Zym with radiolabeled 125I-C4BP (7.65×105 cpm/μg). Data obtained with 0.84×106 ZymC3b was normalized for 0.44×106 ZymM1,C4b as well as for the specific activity of 125I-C4BP used and are shown in the figure. Symbols: ZymM1,C4bC3b (●); ZymC3b (✯).
Fig. 4.
Effect of C4b density on the binding of C4BP to surface-bound C4b alone and C4b-C3b complexes on zymosan and EMan cells. (A). Effect of C4b density on the binding of C4BP to ZymM1,C4b and ZymM1,C3bC4b. The apparent binding constant (Kd) obtained for binding interactions of C4BP with ZymM1,C4b (○) or ZymM1,C3bC4b (●) assembled with different amounts of C4b/Zym (ranging from 14,000-431,000 C4b/Zym) was measured under equilibrium conditions and calculated by nonlinear regression analysis of individual binding curves as described in Fig. 3. The apparent Kd value thus obtained from each binding curve were considered as one data set and was fit by linear regression using Grafit version 5.0 Erithacus software as shown in the figure. (B). Effect of C4b density on the binding of C4BP to EManM1,C4b and EManM1,C3bC4b cells. The apparent binding constant (Kd) obtained for binding interactions of C4BP with EManM1,C4b (○) and EManM1,C3bC4b (●) assembled with different densities of C4b/EMan (ranging from 15,000-471,000 C4b/EMan) was measured under equilibrium conditions and calculated by nonlinear regression analysis of individual binding curves as described in Fig. 3. The apparent Kd thus obtained from each binding curve was considered as one data set and was fit by linear regression using Grafit version 5.0 Erithacus software as shown in the figure.
Binding between C4BP and surface-bound C4b deposited on EMan cells was also found to be a high affinity interaction. As seen in Fig. 4B, increasing the density of C4b/EMan by 31-fold from 15,000 to 471,000 C4b/EMan resulted in a ∼4-fold increase in the apparent Kd from 0.64 to 2.36 nM indicating that on EMan cells ∼4-fold weaker binding interactions occurred between C4BP and surface-bound C4b with increasing C4b density. Nevertheless, the data show that the interaction between C4BP and surface-bound C4b is still a high affinity binding interaction with an average apparent Kd = 1.82 ± 1.57 nM.
3.5 Equilibrium binding of C4BP to surface-bound C4b-C3b complexes and C3b alone
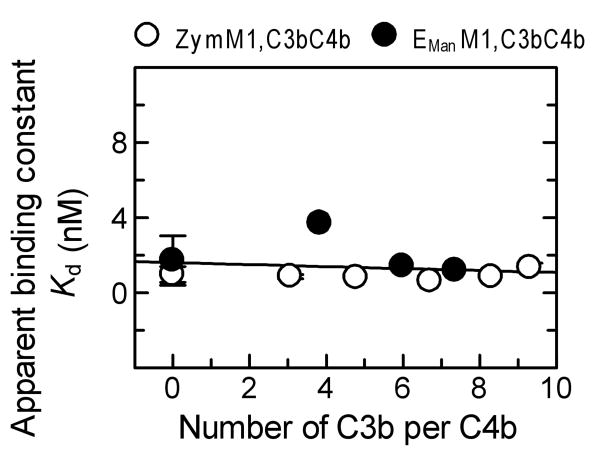
Since deposition of additional C3b molecules is necessary to convert the C3 convertase to a C5 convertase, the role of additional C3b molecules in modifying the high affinity binding interaction of C4BP with surface-bound C4b in complex with C3b was examined. This was done by measuring binding of radio-labeled C4BP to ZymM1,C3bC4b bearing 54,000 C4b and 362,000 C3b/Zym under equilibrium conditions. As seen in Fig. 3B non-linear regression analysis of the equilibrium binding data indicated an apparent Kd of 0.62 nM, similar to that obtained with ZymM1,C4b (average apparent Kd = 0.95 ± 0.35 nM). These results indicate a high affinity binding interaction between C4BP and surface-bound C4b even when in complex with C3b. The binding of C4BP to ZymM1,C3bC4b was observed to be a high affinity interaction not only over a wide range of C4b molecules per cell (21,000-249,000 C4b/Zym, Fig. 4A) but also over a wide range of C3b molecules deposited per C4b (Fig. 5). This was indicated by an average apparent Kd = 0.90 ± 0.27 nM that did not change significantly, even though the number of C3b molecules deposited on ZymM1,C4b was increased from zero to nine C3b molecules deposited per C4b (Fig. 5). Binding experiments between C4BP and zymosan particles that had only C3b on them (ZymC3b) detected no interaction of C4BP with surface-bound C3b (Fig. 3B). Considered together, the data suggest that the high affinity binding interaction of C4BP to surface-bound C4b is not altered when C4b is in complex with C3b.
Fig. 5.
Effect of C3b density on the binding of C4BP to ZymM1,C3bC4b and EManM1,C3bC4b cells. The apparent binding constants (Kd) obtained for binding interactions of C4BP with ZymM1,C3bC4b (○) and EManM1,C3bC4b cells (●) assembled with different ratio of number of C3b:C4b was measured under equilibrium conditions. The number of C4b/cell was measured using 125I-C4 and the number of C3b/cell was measured using 125I-Factor B as described (Rawal et al., 1998). The data were fit by nonlinear regression to a one-site binding equation to obtain the apparent dissociation constant (Kd) as described in Fig. 3. The apparent Kd obtained from these individual plots was considered as one set of data and was fit by linear regression using Grafit version 5.0 Erithacus software as shown in the figure.
Binding of C4BP to EManM1,C3bC4b cells was also observed to be a high affinity interaction over a wide range of C3b molecules deposited per C4b (Fig. 5). This was indicated by an average apparent Kd = 2.10 ± 1.39 nM that did not change significantly, even though the number of C3b molecules deposited on EManM1 cells was increased from zero to seven C3b molecules deposited per C4b (Fig. 5). Data in Fig. 5 show that the presence of additional C3b molecules does not influence the high affinity binding interaction of the regulator with surface-bound C4b on EMan cells as also observed for zymosan.
3.6 Effect of C4b density on the number of C4b molecules bound per C4BP at saturation
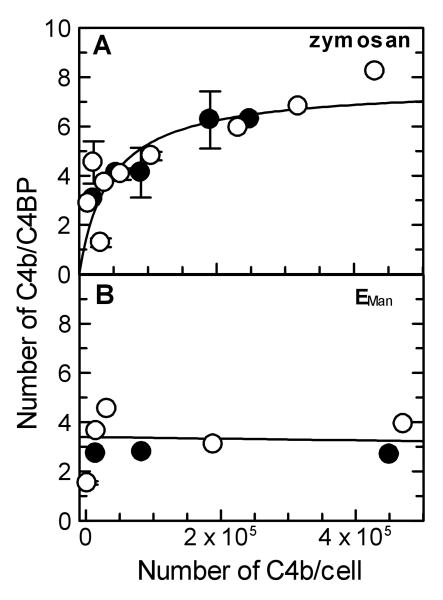
The number of C4BP molecules that could bind to surface-bound C4b was determined from saturating binding curves obtained by incubating varying amounts of 125I-C4BP with ZymM1,C4b. As seen in Figs. 3A and 3B, the binding of 125I-C4BP to ZymM1,C4b and ZymM1,C3bC4b was saturated. The number of C4BP molecules bound per C4b was determined from the maximum amount of C4BP bound. The number of C4b molecules bound per C4BP was found to increase from 2.87 to 8.23 when C4b density was increased from 14,000 to 431,000 C4b/zymosan (Fig. 6A). The ratio was also observed to increase from 3.06 to 6.28 when ZymM1,C3bC4b was used instead of ZymM1,C4b (Fig. 6A). And as seen in the figure, at C4b densities ≥ about 300,000 C4b/Zym a maximum of seven C4b molecules are bound per C4BP even in the presence of C3b.
Fig. 6.
Comparison of the effect of C4b density on the number of C4b molecules bound per C4BP at saturation on zymosan and EMan cells. (A). The effect of C4b density on the number of C4b molecules bound per C4BP at saturation on zymosan. The number of C4b/cell was measured using 125I-C4. The number of C4BP molecules bound to ZymM1,C4b (○) and ZymM1,C3bC4b (●) was calculated from the maximum amount of 125I-C4BP bound to the cells under saturating conditions and from the specific activity of C4BP after subtracting the contribution from 125I-C4b and correcting for nonspecific binding of both. The number of surface-bound C4b molecules bound to C4BP, determined from individual binding plots, was considered as one set of data and was fit by nonlinear regression using Grafit version 5.0 Erithacus software and is shown in the figure. (B). Effect of C4b density on the number of C4b molecules bound per C4BP at saturation on EMan cells. Details are as described in Fig. 6A for zymosan except that EManM1,C4b (○) and EManM1,C3bC4b (●) cells were used. The number of surface-bound C4b molecules bound to C4BP, determined from individual binding plots, was considered as one set of data and was fit by linear regression using Grafit version 5.0 Erithacus software and is shown in the figure. Data obtained with EManM1,C4b cells bearing 2,300 C4b/EMan were not included in the fit.
In contrast, the ratio of the number of C4b molecules bound per C4BP did not change when C4b density on EManM1,C4b cells was increased 31 times from 15,000 to 471,000 C4b/EMan (Fig. 6B). The ratio was constant (3.79 ± 0.60) indicating that a maximum of four C4b molecules bind C4BP on EMan cells. The ratio was observed to be constant even in the presence of C3b although the average number of C4b molecules bound per C4BP decreased from four to three when EManM1,C3bC4b cells were employed (2.73 ± 0.05). These findings indicate that C4BP binds a maximum of three to four surface-bound C4b molecules on EMan.
3.7 Decay acceleration of the lectin pathway C3/C5 convertase by C4BP
As seen in Table 2, C4BP enhanced the decay of the C3 convertase and the C5 convertase assembled on zymosan or EMan to the same extent. The nearly similar IC50 obtained for accelerating the decay of the C3 and C5 convertases on zymosan (0.54 ± 0.19 and 0.81 ± 0.11 nM) or on EMan (1.03 ± 1.00 and 3.47 ± 2.15 nM) suggest that the surface or the presence of additional C3b molecules play no role in influencing the decay enhancing activity of C4BP towards the C5 convertase. And because the IC50 is far below the normal physiological concentration of C4BP (200-450 nM) more than 95% of the C3 and C5 convertases formed on the surface of zymosan or EMan were subjected to the decay accelerating activity of C4BP.
Table 2.
Decay acceleration of C3/C5 convertases by C4BP
| Enzyme | IC50a (nM) |
|---|---|
| Lectin pathway C3/C5 convertases | |
| Soluble C3 convertase (C4b,C2a) | NDb |
| Surface-bound C3 convertase (ZymM1,C4b,C2a) | 0.54 ± 0.19 |
| Surface-bound C3 convertase (EManM1,C4b,C2a) | 1.03 ± 1.00 |
| High affinity C5 convertase (ZymM1,C3bC4b,C2a) | 0.81 ± 0.11 |
| High affinity C5 convertase (EManM1,C3bC4b,C2a) | 3.47 ± 2.15 |
| Classical pathway C3/C5 convertasesc | |
| Soluble C3 convertase (C4b,C2a) | 17 ± 6 |
| Surface bound C3 convertase (EAC1,C4b,C2a) | 11 ± 8 |
| High Affinity C5 convertase (EAC1,C3bC4b,C2a) | 20 ± 11 |
IC50, C4BP concentration required to accelerate the decay of C3 convertase and C5 convertase by 50% as described in Materials and Methods. The lectin pathway C3 convertase was assembled on zymosan and EMan cells bearing 20,000-431,000 C4b/Zym and 23,000-189,000 C4b/EMan. The lectin pathway C5 convertase was assembled on zymosan bearing 11,000-25,000 C4b/Zym and 450,000 C4b/EMan. Values are mean ± S.D. (n = 3).
ND, not determined.
IC50 for classical pathway C3 convertase that was assembled on EA bearing 60,000 C4b/EA and the C5 convertase assembled on EA bearing 11,200 C4b/EA have been reported previously (Rawal et al., 2007).
4. Discussion
The present study shows that the formation and decay of the lectin pathway C3/C5 convertase on two surfaces; zymosan and EMan exhibit greater sensitivity to C4BP regulation when compared to data published for the respective classical pathway C3/C5 convertase assembled on EA (Rawal et al., 2007). Although many groups have analyzed C4BP regulation of the classical pathway C3/C5 convertase (Gigli et al., 1979, 1985; Blom et al., 1999, 2001) and the lectin pathway has been examined by Suankratay et al., (1999), these studies did not report IC50 values. In the present study, a quantitative comparison of the IC50 values obtained for inhibiting formation of the lectin pathway C3 and C5 convertases is made to that published from our laboratory on the classical pathway C3/C5 convertase (Table 1). The analysis indicates that on zymosan 28- and 26-fold less of C4BP was required than reported for the classical pathway C3 and C5 convertases (35 and 72 nM, Table 1) (Rawal et al., 2007). Inhibiting the assembly of the lectin pathway C3 and C5 convertases on EMan also required less C4BP (13- and ∼2-fold) than the respective classical pathway convertase (Rawal et al., 2007). In addition, IC50 obtained for accelerating the decay of the C3 and C5 convertases on zymosan was 20- and 25- while that on EMan cells was 11- and 6-fold less than reported for the classical pathway C3 and C5 convertases (11 and 20 nM) (Table 2). Moreover, the C3 convertase is the precursor for the C5 convertase and the IC50 value for inhibiting assembly of the surface-bound C3 convertase on zymosan and EMan cells is 197-315 and 95-152 while that for accelerating their decay is 463-741 and 243-388 times below the normal serum concentration of C4BP (250 to 400 nM, shaded area in Fig. 1). Considered together, these findings suggest that at normal serum concentration of C4BP by effectively regulating the C3 convertase and hence the C5 convertase, C4BP controls the four times greater potential of the lectin pathway than the classical pathway in generating pro-inflammatory products.
Our studies show that on EMan the number of C4b molecules bound per C4BP remained constant at about four but on zymosan it increased from about three to nearly seven with increasing C4b density (Fig. 6A). Since one α-chain of C4BP has been reported to bind one C4b (Blom. 2002), the increase in the number of C4b molecules bound per C4BP indicates that the number of α-chains engaged in C4b-binding increases from three to seven with increasing C4b density on the surface of zymosan. And at C4b densities ≥ ∼300,000 C4b/Zym all seven α-chains are engaged in C4b-binding. In contrast, on EMan at densities ≥ 15,000 C4b/EMan, each C4BP molecule had reached its maximum capacity of binding three to four C4b molecules i.e. only three to four α-chains were engaged in C4b-binding. It is likely that the heterogeneous cell surface of zymosan might contribute to deposition of C4b molecules that are spread out but yet near enough for all seven α-chains of C4BP to engage in C4b-binding at high C4b density. Whereas the homogeneous cell surface of EMan may result in C4b clusters that are made up of only three to four C4b molecules or more, but the additional C4b molecules may not be near enough for C4BP to engage in binding more than four C4b molecules. These results suggest that the C4b-C4b and C3b-C4b complexes deposited via the lectin pathway on the two surfaces are different.
Studies analyzing regulation of the classical pathway C3/C5 convertase have reported one C4BP binds an average of two to three C4b molecules when EA cells bearing 19,000 C4b/cell were used (Fujita and Tamura, 1983) while four C4b molecules have been reported to bind one C4BP at densities ≥ 19,000 C4b/EA (Rawal et al., 2007). The unoccupied α-chains of C4BP have been proposed to be free and available for binding to other ligands such as heparin-like molecules (Blom et al., 1999). Studies have also reported that although some α-chains may interact with the pathogen, others are free to inhibit complement (Meri et al, 2004, 2006; Jarva et al., 2005; Hallstrom et al, 2007; Potempa et al., 2008). Our studies on regulation of the lectin pathway C3/C5 convertase show that on EMan one C4BP binds a maximum of three to four C4b molecules but on zymosan up to seven C4b molecules are bound. The findings suggest that depending on the surface, the potential to engage all seven α-chains of C4BP exists to control the innate immune response via the lectin pathway of complement.
The inhibition efficiency of C4BP has been reported to increase with increasing C4b density when EA cells with a very low density of C4b/cell were employed for assembling the classical pathway C3 convertase (Blom et al., 2001). Fujita and Tamura (1983) showed that EA cells bearing 19,000 or more C4b/cell were about 6-fold more efficient in binding C4BP than cells bearing 3,000 C4b/cell while Rawal et al., (2007) employing EA cells bearing high C4b density (23,000-178,000 C4b/EA) reported no change in the inhibition efficiency of C4BP. Our studies on the lectin pathway show that even though the number of α-chains engaged in C4b-binding increased from three to seven with increasing C4b density on zymosan (14,000 to 431,000 C4b/Zym, Fig. 6A), the apparent binding affinity (Kd) between C4BP and C4b deposited on zymosan did not change (Kd range = 1.29 to 0.58 nM) (Fig. 4A). These findings explain why no change in the inhibition efficiency of C4BP was observed in Fig. 2 when the number of C4b molecules deposited on zymosan was increased from 22,000-346,000 C4b/Zym.
Comparison of the average apparent Kd for C4BP binding to C4b deposited via the lectin pathway and the classical pathway shows a ∼13- and 7-fold greater affinity for C4b deposited on zymosan and EMan than on EA. This was indicated by an average apparent Kd = 0.95 ± 0.35 and 1.82 ± 1.57 nM obtained for C4b deposited on zymosan and EMan (Fig. 5) versus 12 nM reported for C4b deposited on EA (Rawal et al., 2007). The binding interaction of C4BP for surface-bound C4b immobilized on a microtiter plate has been reported to be high (Kd = 2.2 and 5 nM) (Blom et al., 2001; Fujita and Tamura, 1983) while that for soluble C4b has been reported to be weak (Kd = 80 nM) (Ziccardi et al., 1984). Our findings when considered with the published reports indicate that C4BP has a greater affinity for C4b deposited via the lectin pathway than the classical pathway.
In addition to the C4b binding site mapped on the α-chain of C4BP (Blom et al., 2001, 2003), the binding site for cofactor activity of C4BP toward C3b, which overlaps and extends beyond the C4b-binding site, has been localized to CCP1-4 on the α-chain (Fujita and Nussenzweig, 1979; Fukui et al., 2002; Blom et al., 2003). In the present study, deposition of C3b molecules on ZymM1,C4b or EManM1,C4b cells had no effect on the binding of C4BP (Fig. 5). C4BP binding to ZymM1,C3bC4b or EManM1,C3bC4b cells exhibited average apparent Kd values (0.90 ± 0.27 and 2.10 ± 1.39 nM) similar to that obtained with ZymM1,C4b and EManM1,C4b cells (0.95 ± 0.35 and 1.82 ± 1.57 nM). Increasing the number of C3b molecules from 0 to 9 and 0 to 7 C3b molecules per C4b on zymosan and EMan had no effect on the binding of C4BP to ZymM1,C3bC4b and EManM1,C3bC4b cells, respectively (Fig. 5). The findings are in contrast to the enhancing effect reported for the additional C3b molecule on the cofactor activity of the membrane-bound regulator CR1 in inactivating C4b (Medof and Nussenzweig, 1984) and in enhancing the decay of the C5 convertase (Krych-Goldberg et al., 2005). Our results suggest that the C3b site on C4BP plays no role in C4BP regulation of the lectin pathway C3/C5 converatse as has been reported for the classical pathway C3/C5 convertase (Rawal et al., 2007).
Acknowledgments
We thank Prof. M. K. Pangburn for providing some of the complement reagents used in the present study.
This research was supported by National Institutes of Health Grant [HL-073804] from the National Heart, Lung, And Blood Institute to N.R. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, And Blood Institute of the National Institutes of Health.
Abbreviations
- C4BP
C4b-binding protein
- C3b, C4b and C5b
the proteolytically activated form of C3, C4, and C5 respectively
- EA
antibody coated sheep erythrocytes
- EC
chicken erythrocytes
- MBL
mannan-binding lectin
- pMBL/A
MBL purified from human plasma
- M1 complex
MBL-MASPs complex
- MASP
MBL-associated serine proteases
- C1 complex, consists of three components
C1q and the serine proteases C1r and C1s in the ratio 1:2:2
- EMan
mannan coated sheep erythrocytes
- C4b
C2a, soluble C3 convertase
- EManM1
C4b,C2a and ZymM1,C4b,C2a, surface-bound lectin pathway C3 convertases
- EManM1
C3bC4b,C2a and ZymM1,C3bC4b,C2a, surface-bound lectin pathway C5 convertases
- ZymC3b
C3b bound to zymosan particles
- IC50
concentration required to inhibit fifty percent
Footnotes
Disclosures: Dr. N. Rawal is an officer of and has financial interest in CompTech, a supplier of complement reagents.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aittoniemi J, Miettinen A, Laippala P, Isolauri E, Viikari J, Ruuska T, Soppi E. Age-dependent variation in the serum concentration of mannan-binding protein. Acta Paediatr. 1996;85:906–909. doi: 10.1111/j.1651-2227.1996.tb14182.x. [DOI] [PubMed] [Google Scholar]
- Blom AM, Webb J, Villoutreix BO, Dahlbäck B. A cluster of positively charged amino acids in the C4BP α-chain is crucial for C4b binding and factor I cofactor function. J Biol Chem. 1999;274:19237–19245. doi: 10.1074/jbc.274.27.19237. [DOI] [PubMed] [Google Scholar]
- Blom AM, Kask L, Dahlbäck B. Structural requirements for the complement regulatory activities of C4BP. J Biol Chem. 2001;276:27136–27144. doi: 10.1074/jbc.M102445200. [DOI] [PubMed] [Google Scholar]
- Blom AM. Structural and functional studies of complement inhibitor C4b-binding protein. Biochem Soc Trans. 2002;30:978–982. doi: 10.1042/bst0300978. [DOI] [PubMed] [Google Scholar]
- Blom AM, Kask L, Dahlbäck B. CCP1-4 of the C4b-binding protein α-chain are required for factor I mediated cleavage of complement factor C3b. Mol Immunol. 2003;39:547–556. doi: 10.1016/s0161-5890(02)00213-4. [DOI] [PubMed] [Google Scholar]
- Blom AM, Villoutreix BO, Dahlbäck B. Complement inhibitor C4b-binding protein-friend or foe in innate immune system? Mol Immunol. 2004a;40:1333–1346. doi: 10.1016/j.molimm.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Blom AM, Villoutreix BO, Dahlbäck B. Functions of human complement inhibitor C4b-binding protein in relation to its structure. Arch Immunol Ther Exp. 2004b;52:83–95. [PubMed] [Google Scholar]
- Christopherson RI. Desalting protein solutions in a centrifuge column. Methods Enzymol. 1983;91:278–281. doi: 10.1016/s0076-6879(83)91025-x. [DOI] [PubMed] [Google Scholar]
- Chung LP, Reid KB. Structural and functional studies on C4b-binding protein, a regulatory component of the human complement system. Biosci Rep. 1985;5:855–865. doi: 10.1007/BF01119897. [DOI] [PubMed] [Google Scholar]
- Collard CD, Väkevä A, Morrissey MA, Agah A, Rollins SA, Reenstra WR, Buras JA, Meri S. Complement activation after oxidative stress. Role of the lectin complement pathway. Am J Pathol. 2000;156:1549–1556. doi: 10.1016/S0002-9440(10)65026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ember JA, Jagels MA, Hugli TE. Characterization of complement anaphylatoxins and their biological responses. In: Volanakis JE, Frank MM, editors. The Human Complement System in Health and Disease. Marcel Dekker; New York: 1998. pp. 241–284. [Google Scholar]
- Endo M, Ohi H, Ohsawa I, Fujita T, Matsushita M. Complement activation through the lectin pathway in patients with Henoch-Schönlein purpura nephritis. Am J Kidney Dis. 2000;35:401–407. doi: 10.1016/s0272-6386(00)70192-2. [DOI] [PubMed] [Google Scholar]
- Fujita T, Gigli I, Nussenzweig V. The role of C4-binding protein: II Role in proteolysis of C4b by C3b-inactivator. J Exp Med. 1978;148:1044–1051. doi: 10.1084/jem.148.4.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Nussenzweig V. The role of C4-binding protein and beta 1H in proteolysis of C4b and C3b. J Exp Med. 1979;150:267–276. doi: 10.1084/jem.150.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Tamura N. Interaction of C4-binding protein with cell-bound C4b: A quantitative analysis of binding and the role of C4-binding protein in proteolysis of cell-bound C4b. J Exp Med. 1983;157:1239–1251. doi: 10.1084/jem.157.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui A, Yuasa-Nakagawa T, Murakami Y, Funami K, Kishi N, Matsuda T, Fujita T, Seya T, Nagasawa S. Mapping of the sites responsible for factor I-cofactor activity of C3b cleavage of C3b and C4b on human C4b-binding protein (C4bp) by deletion mutagenesis. J Biochem. 2002;132:719–728. doi: 10.1093/oxfordjournals.jbchem.a003279. [DOI] [PubMed] [Google Scholar]
- Gigli I, Fujita T, Nussenzweig V. Modulation of the classical pathway C3 convertase by plasma proteins C4 binding protein and C3b inactivator. Proc Natl Acad Sci U S A. 1979;76:6596–6600. doi: 10.1073/pnas.76.12.6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigli I, Sorvillo J, Halbwachs-Mecarelli L. Regulation and deregulation of the fluid-phase classical pathway C3 convertase. J Immunol. 1985;135:440–444. [PubMed] [Google Scholar]
- Hallstrom T, Jarva H, Riesback K, Blom AM. Interaction with C4b-binding protein contributes to nontypeable Haemophilus influenza serum resistance. J Immunol. 2007;178:6359–6366. doi: 10.4049/jimmunol.178.10.6359. [DOI] [PubMed] [Google Scholar]
- Harmat V, Gál P, József K, Szilágyi K, Ambrus G, Végh B, Náray-Szabó G, Závodszky P. The structure of MBL-associated serine protease-2 reveals that identical substrate specificities of C1s and MASP-2 are realized through different sites of enzyme-substrate interactions. J Mol Biol. 2004;342:1533–1546. doi: 10.1016/j.jmb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Hessing M, Kanters D, Takeya H, van't Veer C, Hackeng TM, Iwanaga S, Bouma BN. The region Ser333-Arg356 of the α-chain of human C4b-binding protein is involved in the binding of complement C4b. FEBS Lett. 1993;317:228–232. doi: 10.1016/0014-5793(93)81281-4. [DOI] [PubMed] [Google Scholar]
- Hillarp A, Dahlbäck B. Novel subunit in C4b-binding protein required for protein S binding. J Biol Chem. 1988;263:12759–12764. [PubMed] [Google Scholar]
- Ikeda K, Sannoh T, Kawasaki N, Kawasaki T, Yamashina I. Serum Lectin with known structure activates complement through the classical pathway. J Biol Chem. 1987;262:7451–7454. [PubMed] [Google Scholar]
- Jarva H, Ram S, Vogel U, Blom AM, Meri S. Binding of the complement inhibitor C4bp to serogroup B Neisseria meningitidis. J Immunol. 2005;174:6299–6307. doi: 10.4049/jimmunol.174.10.6299. [DOI] [PubMed] [Google Scholar]
- Jordan JE, Montalto MC, Stahl GL. Inhibition of mannose-binding lectin reduces postischemic myocardial reperfusion injury. Circulation. 2001;104:1413–1418. doi: 10.1161/hc3601.095578. [DOI] [PubMed] [Google Scholar]
- Kerr FK, Thomas AR, Wijeyewickrema LC, Whisstock JC, Boyd SE, Kaiserman D, Matthews AY, Bird PI, Thielens NM, Rossi V, Pike RN. Elucidation of the substrate specificity of the MASP-2 protease of the lectin pathway and identification of the enzyme as a major physiological target of the serpin, C1-inhibitor. Mol Immunol. 2008;45:670–677. doi: 10.1016/j.molimm.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Krych-Goldberg M, Hauhart RE, Porzukowiak T, Atkinson JP. Synergy between two active sites of human complement receptor type 1 (CD35) in complement regulation: Implications for the strructure of the classical pathway C3 convertase and generation of more potent inhibitors. J Immunol. 2005;175:4528–4535. doi: 10.4049/jimmunol.175.7.4528. [DOI] [PubMed] [Google Scholar]
- Lhotta K, Würzner R, König P. Glomerular deposition of mannose-binding lectin in human glomerulonephritis. Nephrol Dial Transplant. 1999;14:881–886. doi: 10.1093/ndt/14.4.881. [DOI] [PubMed] [Google Scholar]
- Liszewski MK, Farries TC, Lublin DM, Rooney IA, Atkinson JP. Control of the complement system. Adv Immunol. 1996;61:201–283. doi: 10.1016/s0065-2776(08)60868-8. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Shikata K, Wada J, Sugimoto H, Shikata Y, Kawasaki T, Makino H. Deposition of mannan binding protein and mannan binding protein-mediated complement activation in the glomeruli of patients with IgA nephropathy. Nephron. 1998;80:408–413. doi: 10.1159/000045212. [DOI] [PubMed] [Google Scholar]
- Medof N, Nussenzweig V. Control of the function of substrate-bound C4b-C3b by the complement receptor CR1. J Exp Med. 1984;159:1669–1685. doi: 10.1084/jem.159.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meri T, Blom AM, Hartmann A, Lenk D, Meri S, Zipfel PF. The yeast and hyphal forms of Candida albicans bind complement regulator C4b-binding protein. Infect Immun. 2004;11:6633–6641. doi: 10.1128/IAI.72.11.6633-6641.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meri T, Cutler SJ, Blom AM, Meri S, Jokiranta TS. Relapsing fever spirochetes Borrelia recurrentis and B. duttonii acquire complement regulators C4b-binding protein and factor H. Infect Immun. 2006;74:4157–4163. doi: 10.1128/IAI.00007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan PB, Harris CL. Regulation in the activation pathways. In: Morgan BP, Harris CL, editors. Complement Regulatory proteins. Academia; San Diego: 1999. pp. 41–136. [Google Scholar]
- Nielsen EW, Waage C, Fure H, Brekke OL, Sfyoera G, Lambris JD, Mollnes TE. Effect of supraphysiologic levels of C1-inhibitor on the classical, lectin and alternative pathways of complement. Mol Immunol. 2007;44:1819–1826. doi: 10.1016/j.molimm.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Parker CJ. Regulation of complement by membrane proteins: an overview. Curr Top Microbiol Immunol. 1992;178:1–6. doi: 10.1007/978-3-642-77014-2_1. [DOI] [PubMed] [Google Scholar]
- Petersen SV, Thiel S, Jensen L, Vorup-Jensen T, Koch C, Jensenius JC. Control of the classical and the MBL pathway of complement activation. Mol Immunol. 2000;37:803–811. doi: 10.1016/s0161-5890(01)00004-9. [DOI] [PubMed] [Google Scholar]
- Potempa M, Potempa J, Okroj M, Popadiak K, Eick S, Nguyen KA, Riesback K, Blom AM. Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J Immunol. 2008;181:5537–5544. doi: 10.4049/jimmunol.181.8.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Salvi VP, Jensenius JC, Rawal N. New insights on the structural/functional properties of recombinant human mannan-binding lectin and its variants. Immunol Lett. 2009;123:114–124. doi: 10.1016/j.imlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Rawal N, Pangburn MK. C5 convertase of the alternative pathway of complement. Kinetic analysis of the free and surface-bound forms of the enzyme. J Biol Chem. 1998;273:16828–16835. doi: 10.1074/jbc.273.27.16828. [DOI] [PubMed] [Google Scholar]
- Rawal N, Pangburn MK. Formation of high-affinity C5 convertase of the classical pathway of complement. J Biol Chem. 2003;278:38476–38483. doi: 10.1074/jbc.M307017200. [DOI] [PubMed] [Google Scholar]
- Rawal N, Pangburn MK. Role of the C3b-binding site on C4b-binding protein in regulating classical pathway C5 convertase. Mol Immunol. 2007;44:1105–1114. doi: 10.1016/j.molimm.2006.07.282. [DOI] [PubMed] [Google Scholar]
- Rawal N, Rajagopalan R, Salvi VP. Activation of Complement Component C5: Comparison of C5 convertases of the lectin pathway and the classical pathway of complement. J Biol Chem. 2008;283:7853–7863. doi: 10.1074/jbc.M707591200. [DOI] [PubMed] [Google Scholar]
- Rossi V, Cseh S, Bally I, Thielens NM, Jensenius JC, Arlaud GJ. Substrate specificities of recombinant mannan-binding-lectin-associated serine proteases-1 and -2. J Biol Chem. 2001;276:40880–40887. doi: 10.1074/jbc.M105934200. [DOI] [PubMed] [Google Scholar]
- Rother K. Complement in Inflammation. In: Rother K, Till GO, Hänsch GM, editors. The Complement System. Springer; New York: 1998. pp. 462–471. [Google Scholar]
- Suankratay C, Mold C, Zhang Y, Lint TF, Gewurz H. Mechanism of complement-dependent haemolysis via the lectin pathway: role of the complement regulatory proteins. Clin Exp Immunol. 1999;117:442–448. doi: 10.1046/j.1365-2249.1999.00998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong NKH, Kojima M, Dobó J, Ambrus G, Sim RB. Activities of the MBL-associated serine proteases (MASPs) and their regulation by natural inhibitors. Mol Immunol. 1999;36:853–861. doi: 10.1016/s0161-5890(99)00106-6. [DOI] [PubMed] [Google Scholar]
- Ziccardi RJ, Dahlbäck B, Müller-Eberhard HJ. Characterization of the interaction of human C4b-binding protein with physiological ligands. J Biol Chem. 1984;259:13674–13679. [PubMed] [Google Scholar]