Summary
Bacillus anthracis the causative agent of anthrax, is a dangerous biological weapon, as spores derived from drug-resistant strains cause infections for which antibiotic therapy is no longer effective. We sought to develop an antiinfective therapy for anthrax and targeted CapD, an enzyme that cleaves poly-γ-D-glutamate capsule and generates amide bonds with peptidoglycan crossbridges to deposit capsular material into the envelope of B. anthracis. In agreement with the model that capsule confers protection from phagocytic clearance, B. anthracis capD variants failed to deposit capsule into the envelope and displayed defects in anthrax pathogenesis. By screening chemical libraries, we identified the CapD inhibitor capsidin, 4-[(4-bromophenyl)thio]-3-(diacetylamino)benzoic acid), which covalently modifies the active site threonine of the transpeptidase. Capsidin treatment blocked capsular assembly by B. anthracis and enabled phagocytic killing of non-encapsulated vegetative forms.
Keywords: PDGA, anchor structure, GGT, transpeptidation, therapeutic
Introduction
Antiinfectives target molecular mechanisms essential for the replication of microbial pathogens during infection (Ehrlich and Bertheim, 1907), whereas antibiotics block all bacterial growth even outside of a human host (Chain et al., 1940; Domagk, 1935). Antibiotics inhibit growth of many different microbial species and offer a considerable advantage over the narrowly focused efficacy spectrum of antiinfectives – antibiotic therapy can be effective even without microbiological diagnosis of the disease causing pathogen (Chain, 1971). This overarching principle established antibiotics as powerful drugs for the therapy of infectious diseases over the past six decades, but also inadvertently accelerated the evolution of drug-resistant microbes (Walsh, 2000). Today, broad dissemination of drug-resistant microbes presents a challenge for the therapy of human bacterial infections (Furuya and Lowy, 2006). At least for some diseases, drug-resistant microbes represent harbingers of a return to the pre-antibiotic era (Neu, 1992). Bacillus anthracis is the causative agent of anthrax, a zoonotic disease transmitted from the cadaver of infected animals to humans by contact, ingestion or inhalation (Mock and Fouet, 2001). Upon entry into phagocytes, spores germinate in the phagosome and vegetative forms first multiply in the cytoplasm (Dixon et al., 2000). Once released into body fluids, B. anthracis resist phagocytosis and replicate in vertebrate tissues (Dixon et al., 1999), forming spores once their host has succumbed to anthrax infection (Koch, 1876). Hallmarks of anthrax are its low infectious dose (fewer than 50 spores cause lethal disease) and explosive replication of vegetative forms, accumulating 1010 colony forming units (CFU) per gram of host tissue (Friedlander et al., 1993). Because of the ease of preparation and the stability of spores, B. anthracis is the most potent and feared biological weapon (Inglesby et al., 1999).
Several key features enable the invasion and replication strategies of B. anthracis. The bacterium secretes two toxins, lethal toxin and edema toxin, that subvert the host immune system to promote pathogen dissemination and implement host killing through cleavage of mitogen-activated protein kinase kinases and inhibition of their signaling pathways (Duesbery et al., 1998; Kim et al., 2008; Rhie et al., 2003). In addition, B. anthracis synthesizes a capsule that protects its vegetative forms from phagocytic killing during infection (Drysdale et al., 2005; Preisz, 1909). The capsule is composed of poly-γ-D-glutamate (PDGA), where glutamyl is polymerized via γ-type amides formed between its α-amino and γ-carboxyl groups (Bruckner et al., 1953). PDGA synthesis, transport and assembly factors are encoded by the capBCADE genes on the pXO2 virulence plasmid (Green et al., 1985; Makino et al., 1988; Okinaka et al., 1999). A model for capsule synthesis can be derived from genetic and biochemical analyses of B. anthracis and B. subtilis, which require pgsBCA and dep (homologs of capBCA and capD, respectively) to synthesize poly-L-γ-glutamic acid (PLGA) (Kimura et al., 2004; Makino et al., 1989) and biochemical anlaysis of B. licheniformis capsule (Troy, 1973a, b) (Fig. 1a). CapB functions as an ATP-dependent ligase and, together with CapC, synthesizes PDGA, which is transported by CapA and CapE across the membrane (Ashiuchi et al., 2001; Ashiuchi et al., 2004; Candela et al., 2005; Candela and Fouet, 2006). In addition, some organisms like B. licheniformis encode a polyglutamyl γ-hydrolase enzyme that appears to modulate the size of PDGA (King et al., 2000).
Fig. 1. Model for capsule synthesis and assembly in the envelope B. anthracis.
A. Poly-γ-D-glutamic acid (PDGA) is synthesized in the cytoplasm by CapBC and capsular polymers are translocated across the bacterial membrane by CapAE. CapD anchors PDGA by catalyzing a transpeptidation reaction, forming an amide bond between capsule and the side chain amino group of meso-diaminopimelic acid (m-DAP) in the peptidoglycan scaffold, composed of the N-acetylglucosamine-(β1–4)-N-acetylmuramic acid (L-alanine-D-iso-glutamine-m-diaminopimelic acid-D-alanine).
B. CapD cleaves PDGA (red and black) to form a γ-glutamyl acyl-intermediate that is resolved by the amino group of m-DAP (green) within muropeptides. Rearranged bonds are shown in a wavy purple line.
CapD (Dep) resides in the envelope of B. anthracis and has two proposed functions, the release of small PDGA fragments into the extracellular milieu and the attachment of capsular filaments to the bacterial envelope (Candela and Fouet, 2005; Uchida et al., 1993) (Fig. 1b). Indeed, B. anthracis strain Sterne expressing capBCAE with and without capD was found to synthesize low- and high-molecular-weight PDGA, respectively; CapD-derived low-molecular-weight PDGA is thought to interfere with host defense mechanisms (Uchida et al., 1993). Unlike wild-type B. anthracis, vegetative forms of capD variants do not co-sediment with capsular material, further supporting the model of CapD-mediated PDGA attachment to the bacterial envelope (Candela and Fouet, 2005).
Results
Virulence properties of B. anthracis Ames capD variants
Previous work studied the contribution of capD towards B. anthracis virulence. When injected into the peritoneal cavity of mice, 106 vegetative forms of B. anthracis Sm, a variant of the Pasteur II strain (Pasteur, 1881), caused disseminated lethal infections, whereas a dep (capD) mutant did not (Makino et al., 2002). A second experiment compared the virulence of wild-type and isogenic capD variant spores derived from the parent strain RPG1, which lacks all anthrax toxin genes (Pezard et al., 1991). Subcutaneous injection of mice with 5×105 RPG1 spores caused lethal infections, however 7.5×108 spores of the capD mutant were required to produce similar mortality (Candela and Fouet, 2005). Neither of these studies analyzed the contribution of capD during anthrax infection, initiated by the inoculation of spores derived from fully virulent B. anthracis isolates to cause lethal disease. To test this, we replaced the capD gene with a spectinomycin resistance cassette into B. anthracis strain Ames, a highly virulent strain that harbors both virulence plasmids (pXO1 and pXO2). Spores of wild-type and capD mutant B. anthracis Ames were examined in a guinea pig model, which recapitulates all aspects of anthrax disease including spore germination, vegetative expansion and dissemination, lethal infection and spore formation (Joyce et al., 2006; Marraffini and Schneewind, 2006). Following subcutaneous injection of 35 B. anthracis Ames wild-type spores, all infected guinea pigs (6/6) developed a lethal infection with a mean time-to-death < 2 days (Fig. 2a). In contrast, injection of 3,000 spores of the isogenic capD mutant caused half of the guinea pigs (3/6) to develop a lethal infection with a mean-time to death > 4 days. Further, sixty hours following challenge with 3×104 spores (10 meanlethal-doses, MLD), the dissemination of capD mutant vegetative forms from the inoculation site to organs of the reticulo-endothelial system was severely impaired, as only 104–106 CFU/g were recovered from splenic tissues. By comparison, 36 and 48 hours following a challenge with 35 spores of the wild-type strain, B. anthracis Ames, produced ~109 CFU/g of spleen tissue. Thus, deletion of capD in B. anthracis Ames caused >100 fold increase in anthrax MLD and >10,000 fold decrease in the dissemination of vegetative forms from the inoculation site. These observations validate CapD as a target for antiinfective anthrax therapy.
Fig. 2. Virulence defects of B. anthracis Ames capD mutants in a guinea pig model of anthrax infection.
A. Kaplan-Meier analysis for guinea pigs infected with B. anthracis Ames wild type or capD spores. Cohorts of six guinea pigs were injected subcutaneously with wild-type or capD mutant spore preparations and monitored for lethal anthrax disease over 7 days (Ivins et al., 1994).
B. Dissemination of B. anthracis from the infection site to the spleen was enumerated as colony forming units in tissue homogenate. Arrows in panel A identify animals used for this study. Tissues from subcutaneous lesions and spleen were removed, homogenized under aseptic conditions with PBS and bacterial loads in tissues displayed as total CFU/g of organ.
B. anthracis CapD functions as a transpeptidase
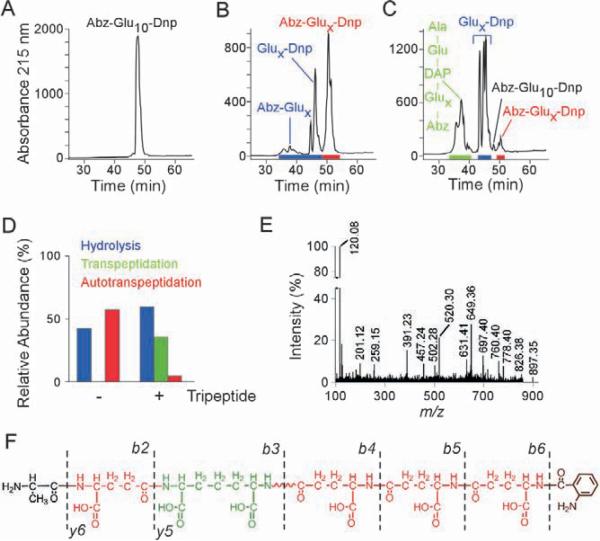
CapD is a member of the γ-glutamyl transferase (GGT) family that is widely conserved in eukaryotes and bacteria (Candela and Fouet, 2005). Although CapD has been proposed to anchor capsular material to the cell wall envelope, the purified enzyme has only been shown to degrade PDGA substrate, which can be used for therapeutic purposes to degrade the capsule of vegetative forms in anthrax infected animals (Candela and Fouet, 2005; Scorpio et al., 2007). Purified CapD is a heterodimer with differentially processed α- and β-subunits, capable of hydrolyzing B. anthracis Ames capsule material (Wu et al., 2008). When incubated with the fluorescence resonance energy transfer (FRET) substrate Abz-[γ-D-Glu]10-Dnp, composed of γ-D-glutamate decamer ([γ-D-Glu]10) with N-terminal ortho-aminobenzoyl (Abz) fluorophore and C-terminal 2,4 dinitrophenyl (Dnp) quencher, the observed increase in fluoresence is a surrogate measure for CapD-catalyzed cleavage (vide infra). Substrate and reaction products were separated by reversed-phase high performance liquid chromatography (rpHPLC) and analyzed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) in positive and negative reflector modes (Fig. 3). These experiments identified not only the expected CapD-cleaved products, Abz-[γ-D-Glu]x-COOH and NH2-[γ-D-Glu]x-Dnp, but also unexpected products, peptides of variable length flanked by fluorophore and quencher (Abz-[γ-D-Glu]2–9-Dnp, Fig. 3ab and Supplemental Table 1). These results can be explained by the following model. CapD forms an acyl-intermediate with its cleaved substrate (Abz-[γ-D-Glu]x-CO), which is released from the enzyme's active site threonine hydroxyl by either hydrolysis, to generate a carboxyl leaving group (Abz-[γ-D-Glu]x-COOH), or by the nucleophilic attack of the amino group, derived from the second cleavage product of CapD (NH2-[γ-D-Glu]x-Dnp). A resultant amide bond restores the linearity of peptides with substrate properties and yields γ-D-Glu polymers of variable lengths.
Fig. 3. CapD catalyzes amide bond formation between γ-D-glutamyl peptides and the cell wall crossbridge. Assay mix containing 200 μM Abz-γ-D-Glu10-Dnp and 20 μM CapD was incubated in absence or presence of 1 mM wall peptide (L-Ala-D-iGlu-m-DAP) at 22°C for two hours and reaction products were separated by rpHPLC and analyzed by MALDI-TOF MS.
A. Chromatogram of substrate Abz-γ-D-Glu10-Dnp.
B. Chromatogram of reaction products after incubation of substrate Abz-γ-D-Glu10-Dnp with CapD.
C. Chromatogram of reaction products after incubation of substrates Abz-γ-D-Glu10-Dnp and L-Ala-D-iGlu-m-DAP with CapD. MALDI-TOF analysis of reaction products is provided in Supplemental Tables 1 and 2.
D. Comparison of product yields, expressed as relative abundance (%), for CapD catalysis in panels B and c.
E. Tandem mass spectrometry experiment in the positive reflector mode of m/z 897.35, the CapD transpeptidation product L-Ala-D-iGlu-(Abz-[γ-D-Glu]3)-m-DAP. Daughter ion signals and structural predictions are listed in Table 2.
F. Structural schematic and corresponding b- and y-ions derived from m/z 897.35.
A transpeptidation reaction that reiteratively cuts and mends PDGA substrate, here designated autotranspeptidation, appears futile and cannot explain how capsular material is immobilized in the bacterial envelope. We wondered whether cell wall peptidoglycan functions as a nucleophile for the CapD transpeptidation reaction by attacking the acyl-intermediate. For example, the side chain amino group of m-diaminopimelic acid is involved in multiple transpeptidation reactions that crosslink peptidoglycan or immobilize proteins in the cell wall (Schneewind et al., 1995; Tipper and Strominger, 1965). To test this, CapD was incubated with Abz-[γ-D-Glu]10-Dnp and the peptidoglycan tripeptide, L-Ala-D-iGlu-m-DAP, a component of the B. anthracis envelope (Severin et al., 2004). Reaction products were separated by rpHPLC and analyzed by MALDI-TOF MS, which revealed PDGA linked to peptidoglycan (L-Ala-D-iGlu-(Abz-[γ-D-Glu]x)-m-DAP) and hydrolysis products (Abz-[γ-D-Glu]x, [γ-D-Glu]x-Dnp), but limited autotranspeptidation products (Abz-[γ-D-Glu]2–9-Dnp) (Fig. 3cd, Table 1 and Supplemental Table 2). To reveal the structure of transpeptidation products, the compound with m/z 897.35 was subjected to collisionally-induced dissociation in a tandem mass spectrometry experiment (CID-MS/MS, Fig.3 fg). Observed daughter ion spectra were in agreement with a compound structure in which the γ-carboxyl group of Abz-γ-D-Glu3 is amide linked to the side chain amino group of m-DAP in the peptidoglycan cell wall peptide (L-Ala-D-iGlu-(Abz-[γ-D-Glu]3)-m-DAP) (Table 2). Similar analyses of other transpeptidation products revealed the same capsular anchor structure with variable length PDGA fragments. For example, CID-MS/MS identified the structure of m/z 639.25 as L-Ala-D-iGlu-(Abz-[γ-D-Glu]1)-m-DAP (Supplemental Fig. 1 and Supplemental Table 3). In summary, when offered both PDGA and peptidoglycan substrates, CapD catalyzed a transpeptidation reaction that linked capsular material to cell wall crossbridges (Fig. 1). To ascertain whether the transpeptidation reaction occurs in vivo, B. anthracis murein sacculi with attached capsule were isolated and treated with purified CapD to cut long PDGA strands (Supplemental Fig. 2). Murein sacculi were washed and peptidoglycan cleaved with muramidase. Released fragments were separated by rpHPLC and analyzed by MALDI-TOF MS (Supplemental Fig. 2), which revealed three cell wall fragments (m/z 1051.53, 1009.47 and 645.34) with glutamate amide linked to the side chain amino group of m-DAP (see Supplemental Tables 4–6 for CID-MS/MS), in agreement with the model that PDGA is anchored to peptidoglycan crossbridges in the envelope of B. anthracis (Fig. 1).
Table 1.
CapD catalyzes the transfer of γ-D-glutamyl peptides to the cell wall tripeptide L-Ala-γ-D-Glu-m-DAP in vitro. Transpeptidase reaction products were recovered from rpHPLC (see Fig. 3c) and analyzed by MALDI-TOF MS in positive reflectron mode. Program ProteinProspector was used for m/z calculations (http://prospector.ucsf.edu).
| Ion | Observed m/z | Calculated m/z | Predicted structure |
|---|---|---|---|
| [M+H] | 639.25 | 639.26 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu-Abz) |
| [M+H] −H2O | 621.24 | 621.26 | |
| [M+Na] | 661.24 | 661.26 | |
| [M+H] | 768.30 | 768.30 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu2-Abz) |
| [M+H] −H2O | 750.29 | 750.30 | |
| [M+H] | 897.35 | 897.35 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu3-Abz) |
| [M+H] −H2O | 879.34 | 879.35 | |
| [M+H] | 1026.40 | 1026.39 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu4-Abz) |
| [M+H] −H2O | 1008.40 | 1008.39 | |
Table 2.
CapD covalently links γ-D-glutamyl peptides to side chain amino group of m-DAP. m/z 897.35 (Table 1) was subjected to tandem MS/MS analysis. CID daughter ions and predicted structure assignments are listed. Program ProteinProspector was used for m/z calculations (http://prospector.ucsf.edu)
| Ion | Observed m/z | Calculated m/z | Predicted structure |
|---|---|---|---|
| [M+H] | 897.35 | 897.35 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu3-Abz) |
| b6 | 760.40 | 760.30 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu3) |
| b6 +H2O | 778.40 | 778.31 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu3) |
| b5 | 631.41 | 631.26 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu2) |
| b5 +H2O | 649.36 | 649.27 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu2) |
| b4 | 502.28 | 502.21 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu) |
| b4 +H2O | 520.30 | 520.22 | L-Ala-γ-D-Glu-m-DAP(-γ-D-Glu) |
| b3 +H2O | 391.23 | 391.18 | L-Ala-γ-D-Glu-m-DAP |
| b2 | 201.12 | 201.09 | L-Ala-γ-D-Glu |
| y6 | 826.38 | 826.31 | γ-D-Glu-m-DAP(-γ-D-Glu3-Abz) |
| y5 | 697.40 | 697.27 | m-DAP(-γ-D-Glu3-Abz) |
| Internal [M+H] | 259.15 | 259.09 | γ-D-Glu-γ-D-Glu |
High throughput screening for CapD inhibitors
To monitor CapD activity under high-throughput screening (HTS) conditions, we measured FRET of CapD-mediated γ-type peptide bond hydrolysis with the substrate Abz-[γ-D-Glu]5-Dpn (Fig. 4a). Reaction conditions were optimized for 384-well format HTS of the NSRB library (National Screening Laboratory for the Regional Centers of Excellence in Biodefense and Emerging Infecious Diseases, Harvard Medical School) for inhibitors and assay conditions were standardized for 90–120 min incubation with 75–90% fluorescence at saturation point (Fig. 4b). The quality of pilot screens was assessed with the Z'-factor, a measure for data variability within the dynamic range of the assay (Zhang et al., 1999). As this assay achieved a Z'=0.9, 146,205 compounds were scored in duplicate for CapD inhibition. Raw data were filtered to exclude highly fluorescent compounds and a Z-score was derived for each measurement, which allowed classification of 345 hits (0.24% of all compounds) into weak, medium and strong hits (61 compounds exhibiting greater than 50% inhibition of CapD).
Fig. 4. High-throughput screening for CapD inhibitors.
A. CapD-mediated cleavage of FRET-substrate Abz-γ-D-Glu5-Dnp (red arrows) is monitored as fluorescence emission at 415 nm (saturation kinetics). The order of addition of reactants and incubation times are displayed on the flow chart and were optimized by performing a pilot screen with 9216 compounds.
B. Flow chart of assays used for hit validation. The primary screen used the FRET-based assay performed in a 384-well format followed by single sample measurement (Hit validation of FRET-based assay). The secondary screen used CapD-mediated cleavage of PDGA analyzed by agarose gel electrophoresis and methylene blue staining of capsular material.
The 61 compounds in the strong hit category were tested for their CapD inhibitory attributes, by measuring both hydrolysis of FRET substrate and capsule degradation visualized by agarose gel electrophoresis and methylene blue staining (Fig. 4b). A total of 52 hits were confirmed as strong inhibitors by the FRET assay (Supplemental Fig. 3). Only 14 out of 54 compounds inhibited CapD-mediated degradation of isolated capsular filaments, suggesting that most hit compounds inhibited CapD-mediated cleavage of peptide substrates but not recognition of the physiological capsule substrate. Hits were subjected to computational analysis (Molinspiration: www.molinspiration.com) of molecular properties (i.e. molecular weight, polar surface area, number of hydrogen bond donors and acceptors, and bond rotation) to determine violations of Lipinski's rule-of-five (Lipinski et al., 2001; Rishton, 2003; Seidler et al., 2003). Nine compounds displayed drug-like properties and were submitted to the NSRB informatics group for analysis of promiscuous inhibitory activity. Five compounds displayed no inhibitory activities in 15 unrelated assays and were therefore considered for further analysis as potential CapD inhibitors.
Inhibition of capsule attachment in vivo
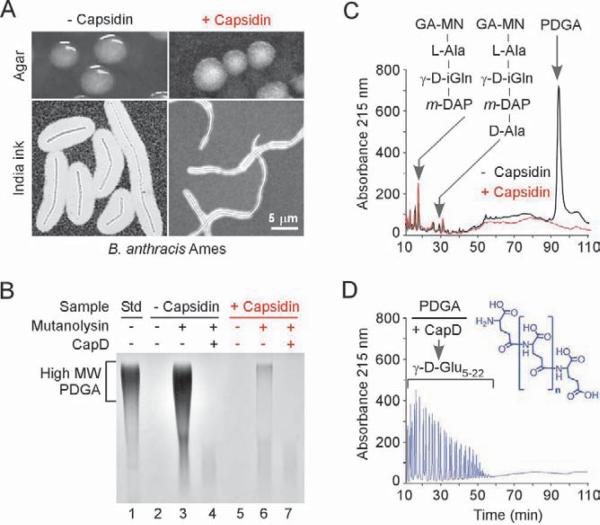
The five hit compounds were tested for their ability to block CapD-mediated capsule formation by vegetative cells. B. anthracis Ames was grown on agar plates supplemented with 1 mM compound in the presence of 5% CO2 and serum to induce capsule synthesis (Green et al., 1985). B. anthracis formed colonies in the presence of all five compounds, indicating that these molecules cannot function as antibiotics. One compound caused B. anthracis Ames to form colonies with the morphology of non-encapsuled cells: rough-edged colonies with ground-glass appearance, distinct from the smooth-slimy surface of colonies formed by encapsulated cells (Green et al., 1985) (Fig. 5a, top panels). The inhibitory compound, #11, with the chemical designation 4-[(4-bromophenyl)thio]-3-(diacetylamino)benzoic acid (ChemBankID 1985080) and a molecular weight of 408.27, was named capsidin. Microscopic visualization of capsule with India ink-staining further documented the inhibitory properties of capsidin: thin, irregularly shaped capsule layers surrounded bacterial cells that had been incubated with the inhibitor (Fig. 5a, bottom panels).
Fig. 5. Inhibition of CapD-mediated deposition of capsule in the envelope of B. anthracis.
A. B. anthracis strain Ames was grown on agar in 5% CO2 atmosphere with or without capsidin (#11). Top panels show photographs of colonies grown on agar plates. Bottom panels show light microscopy images of formalin-fixed bacteria stained with India ink.
B. CapD mediated deposition of capsule in murein sacculi (lanes 2 and 5) in the presence or absence of capsidin (#11) was measured by mutanolysin cleavage of peptidoglycan and separation of solubilized PDGA by agarose gel electrophoresis (lanes 3 and 6). Solubilized capsule material was treated with purified CapD to verify the chemical nature of PDGA (lanes 4 and 7). Purified soluble PDGA served as a standard (std) (lane 1).
C. rpHPLC chromatography of mutanolysin-digested cell wall envelope material from B. anthracis strain Ames grown with (red) or without (black) capsidin (#11). Retention times for eluted peptidoglycan disaccharide-tripeptide and -tetrapeptide as well as PDGA capsule are indicated with arrows (see Table 3 and Supplemental Table 4 for structural analysis of wall peptides).
D. Capsular material in the PDGA peak that eluted at 93–96 min (panel C) was treated with CapD and subjected to rpHPLC using a methanol gradient in sodium phosphate buffer. Eluted PDGA compounds were desalted and analyzed by mass spectrometry (Supplemental Table 5). The structure of PDGA polymer is shown in blue (n = number of glutamate residues 5–22).
To further investigate the inhibitory effect of capsidin, a cell wall fraction containing peptidoglycan and anchored capsular material (PDGA) was extracted and purified from B. anthracis that had been grown in the presence or absence of 1 mM inhibitor. Suspensions of murein sacculi were subjected to agarose gel electrophoresis and methylene blue staining; as expected, the integrity of the cell wall envelope prevented PDGA capsule to enter the gel matrix (compare lanes 1 and 2 Fig. 5b and lanes 1 and 2 Supplemental Fig. 4). However, treatment of murein sacculi with mutanolysin, an N-acetylmuramidase that cleaves the β(1–4) glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine (Yokogawa et al., 1974), permitted the release of high-molecular weight capsular material (lane 3, Fig. 5b). Incubation of the mutanolysin-released capsular material with CapD caused its complete degradation (lane 4, Fig. 5b). Murein sacculi of B. anthracis that had been grown in the presence of capsidin harbored little capsule, as the mutanolysin treatment released insufficient material for visualization by methylene blue staining (lanes 5–7, Fig. 5b). Thus, capsidin treatment inhibited the CapD-mediated deposition of PDGA capsule into the cell wall envelope of B. anthracis.
For structural identification of peptidoglycan and capsule components, mutanolysin-treated material was separated by rpHPLC (Fig. 5c) and individual peaks were analyzed by MALDI-TOF MS. Fractions containing monomeric peptidoglycan subunits eluted early (18–36 min), whereas cross-linked subunits were retained longer (39–60 min) (Supplemental Table 7). The structures of two major peptidoglycan subunits, disaccharide-tripeptide [N-acetylglucosamine (GA)-(β1–4)-N-acetylmuramic acid (MN)-L-Ala-D-iGln-m-DAP], and tetrapeptide [GA-MN-L-Ala-D-iGln-m-DAP-D-Ala], were confirmed by amino acid composition analysis and CID MS/MS (Table 3). A striking difference between the two envelope preparations is the absorbance peak at 93–96 min, which was present only in the envelope of B. anthracis that had not been treated with inhibitor. MALDI-TOF MS of the desalted peak fraction failed to discern discrete ion signals, as would be expected of linear capsule fragments of highly variable lengths. Amino acid composition of the purified material revealed Ala:Glu/Gln:m-DAP at a molar ratio of 2:106:1, indicating that the 93–96 min peak indeed contains polyglutamate linked to peptidoglycan (Table 3). To probe for the presence of γ-peptide bonds, cell wall anchored polyglutamate was incubated with CapD and reaction products were separated by rpHPLC (Fig. 5d). Mass spectrometry of eluted reaction products identified a series of compounds comprised of γ-D-Glu5–22 (Supplemental Table 8), but also wall peptides with glutamate attached to the side chain amino group of m-DAP (m/z 645.3, 1009.47 and 1051.53, Supplemental Tables 4–6). Thus, mutanolysin treatment of murein sacculi isolated from B. anthracis strain Ames released peptidoglycan fragments without and with capsule, as PDGA is anchored by CapD to the side chain amino group of m-DAP. As control, murein sacculi of the capD mutant do not harbor cell wall anchored capsule (Supplemental Fig. 4). Finally, capsidin treatment blocked CapD-mediated anchoring of the PDGA capsule to the cell wall envelope of B. anthracis, but did not interfere with synthesis or secretion of capsular material (Supplemental Fig. 5).
Table 3.
Identification of major cell wall compounds found in encapsulated B. anthracis Ames (− Capsidin) and in cells treated with inhibitor (+ Capsidin). Compounds were separated by rpHPLC using a methanol gradient in sodium phosphate buffer and desalted before MALDI-TOF MS analysis in positive and negative reflectron mode. Amino acid analysis was carried out by ion-exchange HPLC following acidic hydrolysis.
| Sample | Retention (min) | [M+Na]+ |
[M-H]− |
Amino acid composition1) |
Predicted structure2) | ||||
|---|---|---|---|---|---|---|---|---|---|
| observed | calculated | observed | calculated | Ala | Glx | DAP | |||
| ± Capsidin | 18 | 848.42 | 848.36 | 824.19 | 824.36 | 1 | 1 | 1 | GA-MN-L-Ala-γ-D-iGln-m-DAP3) |
| ± Capsidin | 29 | 919.44 | 919.40 | 895.20 | 895.40 | 2 | 1 | 1 | GA-MN-L-Ala-γ-D-iGln-m-DAP-D-Ala3) |
| − Capsidin | 93–96 | n. a. | n. a. | n. a. | n. a. | 2 | 106 | 1 | L-Ala-γ-D-iGln-m-DAP(-γ-D-Glu)x-D-Ala |
n. a. Not applicable
Molar ratios of amino acids are derived from experimental data rounded to the nearest integral number. The analysis does not discriminate between Glu and Gln (Glx).
Abbreviations: GA, glucosamine; iGln, iso-glutamine; m-DAP, meso-diaminopimelic acid; MN, N-acetylmuramic acid
Structures for m/z 848.42 and 919.44 were confirmed by tandem MS/MS spectrometry (not shown).
Inhibition of CapD by capsidin
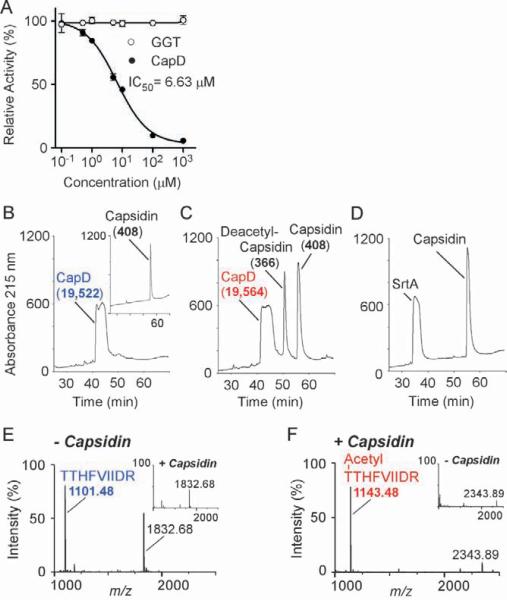
GGTs cleave glutathione and form a γ-glutamyl acyl-intermediate, which, for some enzymes, is resolved by water to generate glutamate (Keillor et al., 2005; Tate and Meister, 1981). We asked whether capsidin inhibits this reaction, measuring cleavage of γ-L-glutamyl-p-nitroanilide (γ-L-Glu-p-NA) by mammalian GGT as an increase in absorbance at 410 nm (p-NA). Capsidin had no effect on mammalian GGT (Fig. 6a). The dose-response of CapD inhibition by capsidin, measured as FRET of cleaved substrate Abz-[γ-D-Glu]5-Dnp, fit a sigmoidal function with an IC50 value of 6.63 μM. CapD did not cleave the GGT substrate γ-L-Glu-p-NA (Supplemental Table 9), nor was CapD activity affected by the GGT inhibitors 6-diazo-5-oxo-L-norleucine and acivicin (data not shown).
Fig. 6. Characterization of CapD inhibition by capsidin.
A. Capsidin inhibits CapD, but not GGT. Purified CapD (closed circles) and mammalian GGT (open circles) were incubated with increasing concentrations of capsidin (#11) followed by addition of substrate, Abz-γ-D-Glu5-Dnp (CapD) and γ-L-glutamyl-p-nitroanilide (γ-L-Glu-pNA)(GGT). CapD activity was monitored as fluorescence emission at 415 nm, whereas GGT activity was measured as absorbance at 410 nm. Relative activity is displayed as the mean with standard deviations of three independent reactions. IC50 was calculated by fitting data with variable slope sigmoidal dose-response function using software package GraphPad Prism 5.
B-F. Capsidin acetylates CapD. rpHPLC elution profile of CapD not treated (B) or treated (C) with inhibitor was recorded at 215 nm; the inset in panel b shows the rpHPLC elution profile of Capsidin alone. Peak fractions were subjected to electrospray ionization mass spectrometry (ESMS) for mass determination and the corresponding masses are indicated in parenthesis next to each peak (see Supplemental Table 10). As a control, panel D shows the rpHPLC elution profile recorded at 215 nm for Sortase A incubated with capsidin. Untreated and capsidin-treated CapD were further subjected to tryptic digestion and reaction products separated by rpHPLC and analyzed by MALDI-TOF. Panel E shows the mass spectrogram of eluate collected at 39% acetonitrile from sample without capsidin revealing an ion with m/z of 1101.48. This ion was not detected in the corresponding fraction of capsidin-treated CapD (inset). Instead, data in panel F shows that tryptic digestion of capsidin-treated CapD yielded a new product eluting at 44% acetonitrile with m/z 1143.48. This species was absent from the CapD sample not treated with Capsidin (inset). The identity of both ions, m/z 1101.48 and m/z 1143.48, was confirmed by tandem mass spectrometry (Supplemental Tables 11 and 12). Both ions correspond to the first 9 amino acids of the small subunit of CapD with the first residue, Thr-352 being acylated upon Capsidin incubation.
We asked whether treatment with capsidin causes a chemical modification of CapD and incubated the enzyme in the presence or absence of inhibitor for two hours. Separation of inhibitor and enzyme by rpHPLC and electrospray ionization mass spectrometry ESI-MS measurements of eluted peak fractions revealed a mass gain of 42 for CapD that had been treated with the inhibitor (Fig. 6bc). Only the small subunit of the enzyme, which encompasses the active site, displayed a mass difference of 42 (19,522.2 and 19,564.2, respectively; Supplemental Table 10). Peaks eluting at 57% and 51% acetonitrile contained intact capsidin (mass 408.0) and a smaller compound with a mass of 366.0, respectively. The observed mass difference (42.0) suggests that one of the acetyl groups was transferred from the diacetyl imide moiety of capsidin to CapD. As a control, purified sortase A from Staphylococcus aureus, another transpeptidase that immobilizes polymers to the peptidoglycan crossbridges (Mazmanian et al., 1999), was incubated with capsidin. No mass differences were observed for sortase A and inhibitor (Fig. 6d) and capsidin failed to inhibit sortase A activity (Ton-That et al., 2000) (data not shown).
To identify the amino acid residue modified by capsidin, CapD preparations were subjected to trypsin digestion. Tryptic fragments were separated by rpHPLC and analyzed by MALDI-TOF mass spectrometry. The 39% acetonitrile fraction of the sample prepared from untreated CapD contained a compound with m/z 1101.48, which is in agreement with a calculated m/z of 1101.61 for the tryptic peptide TTHFVIIDR (Fig. 6e). The first Thr of this peptide corresponds to the catalytic active Thr-352 at the N-terminus of the small subunit (Supplemental Table 11). This peptide was not detected in the capsidin-treated sample. Instead, a new tryptic fragment of m/z 1143.48 eluted at 44% acetonitrile. The mass difference of 42.0 indicates the addition of one acetyl group to the active site fragment of capsidin-treated CapD. Tandem mass spectrometry confirmed the sequence acetyl-TTHFVIIDR for m/z 1143.48 (Supplemental Table 12). In conclusion, capsidin acetylates the catalytically active residue Thr-352 to inhibit the enzymatic activity of CapD.
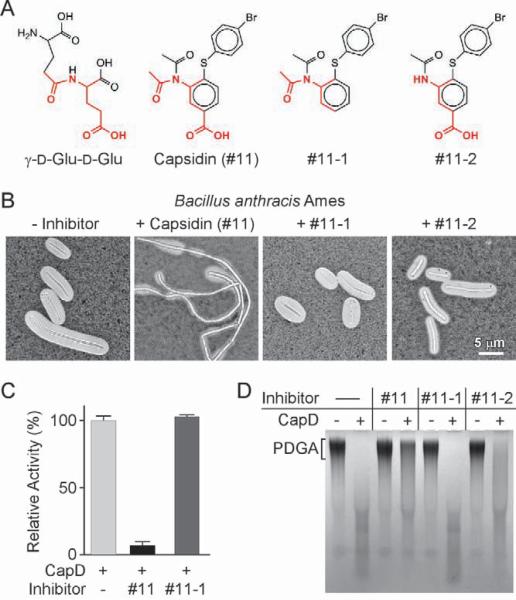
Inhibitory features of Capsidin
Capsidin contains a diacetyl imide moiety that appears to be important for its inhibitory activity. This moiety together with the benzoic acid ring creates a structure that resembles γ-D-glutamyl-γ-D-glutamate (Fig. 7a). Two variants were synthesized that lacked either the carboxyl or the acetyl group of capsidin (Fig. 7a and Supplemental Information; #11-1 and 11-2, respectively). Both compounds failed to prevent encapsulation of B. anthracisin vivo when offered at equal or higher concentrations as capsidin (Fig. 7b, and data not shown). The intrinsic fluorescence of compound #11-2 interfered with FRET, and compound #11-1 failed to inhibit CapD hydrolysis in this assay (Fig. 7c). When analyzed for CapD hydrolysis of PDGA, neither compound 11-1 nor 11-2 displayed inhibitory activity (Fig. 7d). Together, these findings suggest that structural attributes of capsidin that bear compelling resemblance with PDGA are indeed absolutely required for its inhibitory activity. Further, these structural features likely lock one of the two acetyl groups in a donor position to modify the active site of the enzyme. Two possible models for inhibition by capsidin are presented in Supplemental Fig. 6.
Fig. 7. Inhibitory features of capsidin.
A. Chemical structures of γ-D-Glu-D-Glu, compound #11 (capsidin: 4-[(4-bromophenyl)thio]-3-(diacetylamino)benzoic acid) and two synthesized derivatives (#11-1 and #11-2).
B. B. anthracis strain Ames was grown on agar with 5% CO2 atmosphere in the presence or absence of compounds #11, #11-1and #11-2. Light microscopy images of formalin-fixed bacteria stained with India ink.
C. CapD mediated hydrolysis of 400 pmol Abz-γ-D-Glu5-Dnp in the presence of 20 pmol enzyme and 2 nmol compounds #11 and #11-1. The mean of 3 control reactions (no inhibitor) was defined as 100%.
D. CapD-mediated degradation of purified PDGA (20 μg) in the presence or absence of compounds #11, #11-1 and #11-2 was measured by agarose gel electrophoresis and methylene blue staining.
Phagocytic killing of B. anthracis Ames in whole blood
We sought to examine the therapeutic impact of capsidin treatment on B. anthracis. Because the pharmacological and pharmacokinetic attributes of capsidin are still unknown, we employed an in vitro assay for anthrax pathogenesis. One ml of human blood, freshly isolated by vein puncture and treated with lepirudin to prevent coagulation, was unable to promote phagocytosis and killing of 105 B. anthracis Ames vegetative forms; instead, the fully encapsulated vegetative forms engaged in explosive replication as occurs during anthrax infection (Fig. 8ab). In contrast, incubation of human blood with the capD variant of B. anthracis strain Ames led to phagocytic killing of the non-encapsulated vegetative forms, causing a three log reduction in viable bacteria over a two hour observation period (Fig. 8ab). B. anthracis strain Ames vegetative forms were treated with capsidin for two hours and washed prior to incubation with human blood. The large reduction in capsule assembly by capsidin pre-treatment enabled human phagocytes to kill 99% of the vegetative forms (Fig. 8ab).
Fig. 8. Capsidin treatment enables phagocytic clearance of B. anthracis Ames.
A. B. anthracis Ames spores were germinated in the presence of 5% CO2 with or without capsidin (#11). Formalin-fixed bacteria were stained with India ink and capsule visualized by light microscopy.
B. Approximately 105 spores of B. anthracis Ames or an isogenic capD mutant were germinated with or without capsidin and incubated with one ml of heparinized human blood. Aliquots were removed at times 0, 60 and 120 min, blood cells lysed with saponin and bacteria enumerated as CFU/ml by inoculation on agar. At the time of infection (0 min), 105 vegetative forms were added to 1 ml human blood. Mean CFU/ml are used to calculate fold changes, fold killing (<100) or growth (>100) at 60 and 120 min intervals over time 0.
Although capsidin specifically inhibits CapD, large amounts of this compound were needed to block capsule attachment in vivo (Figs. 5a and 8). As both of these in vivo assays employ serum or blood products, we asked whether capsidin is altered and separated the inhibitor incubated in control buffer, serum or blood by rpHPLC. The results demonstrated that upon incubation with serum or blood, but not buffer, capsidin is converted slowly to a 366.0 Da species, suggesting that the inhibitor suffered the loss of one of its acetyl groups (Supplemental Fig. 7). In contrast, incubation of inhibitor with bacterial extracts did not precipitate the inactivation of capsidin (data not shown). These findings, together with the data in Fig. 2, validate CapD as a target for the development of antiinfective therapy of anthrax infections with antibiotic-resistant strains. Inhibitory properties of capsidin may be exploited by chemical modification of this compound in an effort to develop structurally related variants with prolonged stability in animal tissue.
Discussion
The cell wall envelope of Gram-positive bacteria represents a surface organelle that functions not only as an exoskeletal element for the physical integrity of the microbe but also promotes interactions between each bacterium and its environment (Navarre and Schneewind, 1999). Enzymatic reactions that promote assembly of the peptidoglycan scaffold represent valuable targets for antibiotic therapy (Strominger, 1968), however many pathogens have evolved resistance mechanisms that cause therapy failures (Walsh, 1993). During assembly, peptidoglycan precursors form a single large macromolecule, the murein sacculus, enveloping each microbe in a 50–100 nm thick wall structure (Salton, 1952; Strominger et al., 1967). Cell wall peptidoglycan is covalently and non-covalently decorated with teichoic acids, capsule, polysaccharides and proteins (Archibald et al., 1961; Ghuysen, 1968; Schneewind et al., 1992). The sum of these molecular decorations provide the bacterial envelope with species- and strain-specific properties that enable microbial virulence, interactions with the host immune system and the development of disease (Navarre and Schneewind, 1999).
Bacterial transpeptidases offer unique targets for the development of inhibitory compounds with therapeutic values. Penicillin-sensitive transpeptidases catalyze the formation of crosslinks between the amino group of crossbridges and their neighboring wall peptides (Tipper and Strominger, 1965). These enzymes cleave peptidoglycan precursor at D-Ala-D-Ala, forming ester-linked intermediates with an active site serine (Yocum et al., 1979). Another class of transpeptidase employs a similar mechanism to cleave m-DAP-D-Ala, forming amide bonds between the carboxyl group of m-DAP and the ε-amino group of lysine at the C-terminal end of Braun's murein lipoprotein (Braun and Sieglin, 1970; Braun and Wolff, 1970; Magnet et al., 2007). Sortases cleave LPXTG motifs and related sequences in sorting signals of surface proteins; the thioester enzyme intermediate is resolved by nucleophilic attack of the amino group in the peptidoglycan crossbridge (Navarre and Schneewind, 1994; Schneewind et al., 1995; Ton-That et al., 1999). Finally, we show here that CapD acts as a fourth class of transpeptidase by utilizing the amino group of cell wall crossbridges to immobilize capsular polymer into the bacterial envelope. Thus, the amino groups of crossbridges are not only essential for peptidoglycan assembly they are also the site of covalent modification with protein and capsule.
CapD is a member of the γ-glutamyl transferase (GGT) family. Mammalian GGT enzymes transfer γ-glutamate from glutathione to amino acids or peptides (Tate and Meister, 1981). This transferase activity could not be demonstrated for bacterial GGT enzymes, and bacteria are thought to specifically degrade glutathione (Okada et al., 2006). Results presented here expand the spectrum of nucleophile acceptors for GGT enzymes to the amino group of cell wall muropeptides. The products of the CapD catalyzed reaction, linear PDGA filaments tethered by amide bond to the side chain of m-DAP, prevent opsono-phagocytic killing of vegetative forms. The CapD catalytic mechanism of B. anthracis is likely shared with other bacteria that also harbor CapD enzymes, e.g. Bacillus subtilis, Bacillus licheniformis, or Staphylococcus epidermidis (Candela and Fouet, 2006; Kocianova et al., 2005). Although stereochemical attributes of poly-γ-glutamates or chemical composition of peptidoglycan differ between microbes, availability of capsular γ-carboxyl groups and crossbridge amino groups represent conserved features (Schleifer and Kandler, 1972).
Considering the essential role of CapD for bacterial escape from phagocytic clearance, its location in the envelope of bacilli and staphylococci and its specificity for poly-γ-glutamate and peptidoglycan substrates, CapD satisfies key criteria for targets of antiinfective therapy (Ehrlich and Bertheim, 1907). We therefore sought to identify inhibitors that prevent CapD-mediated anchoring of capsule to the bacterial cell wall envelope, thereby allowing immune cells to phagocytose and kill the invading pathogen. To achieve this goal, we exploited a FRET assay for chemical library screening towards the identification of an inhibitor, capsidin, which blocks CapD activity in vitro and in vivo and fulfills the predictions for an antiinfective in blocking an important virulence strategy of B. anthracis. Capsidin functions as a non-competitive inhibitor that specifically acetylates the active site threonine of CapD transpeptidase, but not the active site of other transpeptidases. Thus, inhibition of CapD in vitro and in bacterial cultures in vivo appears specific. However the compound is slowly deacetylated in vivo and chemical modifications must be introduced to capsidin in order to develop lead compounds with therapeutic applications in infectious disease. Future work in this area must therefore elucidate the chemical, pharmacological and pharmacokinetic attributes of capsidin derivatives to further explore the therapeutic value of these compounds for the treatment of anthrax infections with drug-resistant B. anthracis strains.
Experimental Procedures
Bacterial strains and growth conditions
B. anthracis strain Ames and its variants were grown in brain heart infusion (BHI) or Luria Bertani (LB) broth at 37°C. Growth media were supplemented with kanamycin (20 μg ml−1) or spectinomycin (200 μg ml−1) as needed. B. anthracis chromosomal DNA was extracted with the Wizard Genomic DNA Purification Kit (Promega). Sporulation was induced by diluting overnight BHI cultures 1:100 into 2×SG medium (Leighton and Doi, 1971) and further incubation at 37°C. To induce capsule production, bacteria were grown on NBY agar supplemented with 0.8% sodium bicarbonate and 10% heat inactivated horse serum (Gibco) at 37°C in 5% CO2. To block capsule formation in 5% CO2, 1 mM capsidin was added to the agar. For extraction of murein sacculi, sterile nitrocellulose membrane round filters (82 mm diameter; Millipore) were placed on top of NBY/bicarbonate/horse serum agar plates and inoculated with B. anthracis Ames grown overnight on LB agar. Plates were incubated for 20 hours at 37°C in 5% CO2.
Allelic replacement
The B. anthraciscapD variant was generated by allelic replacement using the pTS1 derivative pLM4, containing a thermosensitive origin of replication (Marraffini and Schneewind, 2006). The plasmid carried two DNA fragments with 1 kbp DNA sequence each flanking the capD gene albeit that the first 59 and last 81 nucleotides of capD were left intact. A spectinomycin resistance marker was obtained following BamHI digestion of plasmid pJRS312 (a pUC18-aad9; GenBank M69221) and inserted between both DNA fragments using blunt-ended cloning (pJRS312 was a gift from Dr. T. Koehler). Sequences of primers used for cloning of the upstream and downstream DNA fragments were as follows: (AAGAGCTCTATTTAAAAAACTCAGATTACGT) and (AAGGTACCATTAAGCTGACTATCAAACAGAA), upstream DNA fragment; (AAGGTACCTTAATTAAAGACGAGAGAG) and (AACCCGGGAATACATTTCATTCTACGATAAT), downstream DNA fragment. Plasmid DNA purified from the E. coli K1077 strain was electroporated into B. anthracis Ames and transformants grown at 30°C (permissive temperature) on LB agar (20 μg ml−1 kanamycin; 200 μg ml−1 spectinomycin). Isolated transformants were grown in cultures and incubated at 43°C overnight (restrictive temperature). To ensure loss of pLM4-based plasmid, cultures were diluted four times into fresh media with no antibiotic selection and propagated at 30°C on LB agar containing spectinomycin. Colonies were examined for kanamycin sensitivity and their DNA analyzed by PCR for the presence or absence of the mutant allele. Nucleic acid sequences of wild-type and mutant alleles were verified by DNA sequencing.
Purification of murein sacculi and capsule
Bacteria were grown on NBY agar supplemented with 0.8% sodium bicarbonate and 10% heat inactivated horse serum (Gibco) at 37°C in 5% CO2, with or without 1 mM Capsidin. Bacteria were scraped off filters and suspended in 250 mM Tris-HCl, pH 7.5. Cells in this suspension were lyzed in the presence of 4% SDS following incubation at 100°C for 30 min. Cell wall envelope was sedimented by ultracentrifugation at 120,000 ×g for 2 hours at 22°C. Capsular material in the supernatant was precipitated with 4 volumes ethanol and used as PDGA substrate to assess CapD hydrolytic activity. The sediment was treated with 200 μg ml−1 proteinase K (Sigma) for 12 hours at 37°C in 20 mM HEPES-KOH, pH 7.5 and the enzyme was heat-inactivated for 30 min at 95°C. Murein sacculi were washed three times with 20 mM HEPES-KOH, pH 7.5, extracted twice with 8 M LiCl, followed by two washes with distilled water. Treatment of murein sacculi with mutanolysin (200 μg ml−1 in 20 mM HEPES-KOH, pH 7.5) was performed for 12 hours at 37°C and the enzyme was inactivated by heating samples for 5 min at 95°C. Following ultracentrifugation, soluble peptidoglycan fragments with capsule material were recovered in the supernatant.
Sample preparation and chromatography of muropeptides was performed as described (Glauner et al., 1988) with the following modifications. Muramic acid of solubilized muropeptides was reduced to muramitol prior to rpHPLC; 600 μl of sample were mixed with 200 μl sodium borate buffer, pH 9.0 into which 6 mg ml−1 solid sodium borate was added. Samples were incubated for 20 min at room temperature and the reaction was stopped by lowering the pH to 2.5 with 80 μl 20% ortho-phosphoric acid. Reduced muropeptides were separated on a Hypersil octadecylsilane column (4.6 by 250 mm; particle size: 5 μm; Thermo Electron Co.) and eluted over a period of 80 min with a linear gradient of 5% (vol/vol) methanol in 40 mM NaH2PO4, pH 2.7, to 15% (vol/vol) methanol in 40 mM NaH2PO4, pH 3.2 (50% of elution buffer). A second gradient was applied consisting of 15% to 30% methanol (100% elution buffer) using the same NaH2PO4 buffers. Flow rate (0.5 ml min−1) and column temperature (52°C) were maintained constant during the course of the experiments. Peaks were detected by recording absorbance at 215 nm, and eluate fractions collected and desalted by a second rpHPLC run. A linear gradient of 0% to 30% (vol/vol) acetonitrile in 0.1% (vol/vol) trifluoroacetic acid was applied over 30 min, followed by a steeper gradient of 30% to 100% acetonitrile in 0.1% (vol/vol) trifluoroacetic acid over 10 min with constant flow rate (0.5 ml min−1). Eluate fractions absorbing at 215 nm were collected and dried under vacuum.
Desalted muropeptide samples were dissolved in 5 μl 30% acetonitrile, 0.1% trifluoroacetic acid. Aliquots (0.5 μl) were mixed with an equal volume of saturated matrix stock solution (10 mg ml−1 α-cyano-4-hydroxy-cinnamic acid in 50% acetonitrile, 0.1% trifluoroacetic acid) and spotted for mass spectrometry analysis on a Reflectron MALDI-TOF instrument (ABI Biosystems).
Amino acid composition
Muropeptide hydrolysis and amino acid analysis on a Beckman 7300 Analyzer were performed at the W.M. Keck Foundation Biotechnology Resource Laboratory, Yale University.
PDGA hydrolysis by CapD
Peptidoglycan samples containing PDGA (10 μl aliquots ~ 60 μg) were treated with 5 μM recombinant CapD in 20 mM HEPES-KOH, pH 7.5 for 2 hours at 37°C. PDGA was separated by gel electrophoresis in 2.5% agarose, followed by staining with methylene blue. For CapD treatment of rpHPLC fractions, dried samples were dissolved in100 μl H2O, neutralized with 10 μl 1N NaOH (pH 6–8) and dialyzed against water for 24 hours at 4°C. Samples were treated with 1 μM recombinant CapD for 4 hours at 37°C. Enzyme inactivation was achieved by heating samples for 5 min at 96°C.
In vitro transpeptidation reaction
200 μM Abz-(γ-D-Glu10)-Dnp (Biopeptide Co., Inc) was incubated with 20 μM CapD in the presence or absence of chemically synthesized L-Ala-γ-D-Glu-m-DAP (AnaSpec, Inc.) for 2 hours at 22°C in reaction buffer (20 mM HEPES-KOH, pH 7.5). Reaction products resulting from hydrolysis or transpeptidation reactions were analyzed by rpHPLC.
HTS assay and data analysis
The CapD assay measuring hydrolysis of FRET-substrate was adapted and validated for the HTS experiment as follows. The FRET-substrate Abz-γ-D-Glu5-Dnp (Biopeptide Co., Inc) was used instead of Abz-(γ-D-Glu10)-Dnp because of its lower intrinsic fluroescence. The quality of the CapD assay was assessed as follows. 192 wells of a black polystyrene 384-well plate were loaded with 30 μl of a solution containing 20 pmol CapD in 25 mM HEPES-KOH, 0.1% Tween-20, pH 7.5. 30 μl aliquots of the same HEPES buffer alone were dispensed into the remaining half of the wells. The reaction was started by adding 400 pmol substrate in a 10 μl volume to each well. This experiment was conducted in triplicate using three 384-well plates prepared in the same manner. A time course of the reaction was carried out at 22°C and fluorescence increase was recorded with an Envision plate reader (excitation 315 nm, emission 415 nm). Saturation was reached after 240 min (endpoint in relative fluorescence units: 2,382,081 ± 44,138). This time course experiment was repeated twice and the quality of the CapD screening assay was assessed using the Z'-factor:
where SD+/− and Ave+/− represent standard deviation and average of positive and negative controls (Zhang et al., 1999). To screen for fast reacting inhibitors during HTS, the reaction was allowed to proceed for 90–120 min only corresponding to 75–90% of the fluorescence value at saturation point. For the HTS experiment, a Seiko pin-transfer robot equipped with a Caliper Twister II robotic arm was used to add 1 nmol test compound solubilized in 100 nL DMSO (100%) to each well containing the assay mix and CapD as described above. Pre-incubation with compound proceeded for 60 min at 22°C prior addition of substrate. Fluorescence was recorded 90–120 min later using the Envision plate reader. Each plate included 16 no-compound controls and 16 wells no-compound, no-CapD controls. The entire HTS was carried out in duplicate. Raw data were filtered to exclude highly fluorescent compounds. The cutoff was defined as fluorescence intensity at saturation point plus 3-fold standard deviation from validation experiment. The experimental mean and standard deviation were calculated for each plate to derive a standard Z score for every measurement using the equation Z = (X-Mean)/(SD). Positive hits with negative Z scores were used to classify screen hits as strong (Z<−8), medium (−8<Z<−5) and weak (−5<Z<−4) inhibitor candidates.
IC50 measurement
CapD (0.1 μM) was pre-incubated in 50 μl reaction buffer (10 mM HEPES-KOH, 5% glycerol, 0.1% Tween-20, pH 7.5) with increasing amounts of capsidin (0.1–1,000 μM) for 15 min at 22°C. The reaction was started by adding 10 μM Abz-γ-D-Glu5-Dnp, incubated 120 min at 37°C and quenched by heat treatment at 95°C for 5 min. Product formation was monitored as fluorescence emission at 415 nm. Mammalian GGT (1 μM GGT from equine kidney; Sigma) was pre-incubated in 1 ml reaction buffer with increasing amounts of capsidin (0.1–1,000 μM) for 15 min at 22°C. Reaction was carried out with 100 μM (γ-L-Glu-pNA for 120 min at 37°C. Substrate cleavage was measured as absorbance at 410 nm.
MS analysis of capsidin treated CapD
Recombinant CapD (100 μM) was incubated in 10 mM HEPES-KOH, 10% glycerol, pH 7.5, with or without 1mM capsidin in a 500 μl volume for 2 hours at room temperature. A control reaction was performed under identical conditions using recombinant sortase A from Staphylococcus aureus. For molecular mass determination, samples were immediately separated by rpHPLC on a PLRP-S column (particle size 5μm; Thermo Electron Co.) using a linear gradient of 5–60% acetonitrile (vol/vol) in 0.1% formic acid (vol/vol) over 60 min. Absorbance was monitored at 215 and 280 nm and peak fractions were subjected to electrospray ionization mass spectrometry (ESMS).
For protein fragmentation, samples were digested with trypsin (100 μg ml2212;1) for 12 hours at 37°C. Tryptic fragments were separated by rpHPLC on a Betasil C18 column (particle size 5μm; Thermo Electron Co.) using a linear gradient of 0–100% acetonitrile (vol/vol) in 0.1% formic acid over 100 min (flow rate: 0.5 ml × ml−1). Absorbance was monitored at 215 nm and peak fractions were analyzed by mass spectrometry on a Reflectron MALDI-TOF instrument (ABI Biosystems).
Virulence studies
Overnight cultures of bacteria in BHI were diluted 1:10 into 2×SG medium and incubated at 37°C for 72 h. B. anthracis spores (wild type and capD) were washed three times with sterile distilled water and examined for the presence of vegetative cells by microscopy. Contaminating vegetative cells, typically <1% of spore preparations, were heat-killed by incubating the spore preparation at 65°C for 30 min prior to additional washes. Colony-forming units were enumerated after serial dilution and plating on LB agar. Six female Hartley guinea pigs (350 to 400 g) were injected subcutaneously with 100 μl of spore suspension in PBS into the right leg with a 0.5-ml insulin syringe. Animals were monitored for anthrax disease over 7 days following infection. All animals that developed severe symptoms of disease were killed immediately. Lethal dose values were calculated as previously described (Reed and Muench, 1938).
To assess killing of B. anthracis in whole blood, spores (105 ml−1) were germinated with or without capsidin in BHI supplemented with 10% fetal bovine serum, 0.8 % NaHCO3 and incubated under 5% CO2 atmosphere at 37°C without aeration for 2 hours. Germination and capsule production were verified by India ink staining of formalin-fixed bacteria on an Olympus Provis axial microscope (1,000 × magnification) equipped with CCD camera. Newly germinated bacterial cells were washed and incubated with 1 ml lepirudin (50 μg/ml) treated human whole blood.
Supplementary Material
Acknowledgements
We acknowledge Ruiying Wu for help with CapD purification, Sue Chiang and members of the NSRB for technical assistance during HTS, Derek Elli, Christina Procacci and Kristy Skurauskis for technical assistance with BSL-3 procedures, and Justin Kern, Anthony Maresso, Luciano Marraffini and Vilasak Thammavongsa for discussion. The authors acknowledge membership within and support from the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (GLRCE, National Institute of Allergy and Infectious Diseases Award 1-U54-AI-057153).
Footnotes
Supporting information Additional supporting information may be found in the online version of this article.
References
- Archibald AR, Armstrong JJ, Baddiley J, Hay JB. Teichoic acids and the structure of bacterial cell walls. Nature. 1961;191:570–572. doi: 10.1038/191570a0. [DOI] [PubMed] [Google Scholar]
- Ashiuchi M, Nawa C, Kamei T, Song JJ, Hong SP, Sung MH, Soda K, Misono H. Physiological and biochemical characteristics of poly gamma-glutamate synthetase complex of Bacillus subtilis. Eur J Biochem. 2001;268:5321–5328. doi: 10.1046/j.0014-2956.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- Ashiuchi M, Shimanouchi K, Nakamura H, Kamei T, Soda K, Park C, Sung MH, Misono H. Enzymatic synthesis of high-molecular-mass poly-gamma-glutamate and regulation of its stereochemistry. Appl Environ Microbiol. 2004;70:4249–4255. doi: 10.1128/AEM.70.7.4249-4255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Sieglin U. The covalent murein-lipoprotein structure of the Escherichia coli cell wall. Eur. J. Biochem. 1970;13:336–346. doi: 10.1111/j.1432-1033.1970.tb00936.x. [DOI] [PubMed] [Google Scholar]
- Braun V, Wolff H. The murein-lipoprotein linkage in the cell wall of Escherichia coli. Eur. J. Biochem. 1970;14:387–391. doi: 10.1111/j.1432-1033.1970.tb00301.x. [DOI] [PubMed] [Google Scholar]
- Bruckner V, Kovacs J, Denes G. Structure of poly-D-glutamic acid isolated from capsulated strains of B. anthracis. Nature. 1953;172:508. doi: 10.1038/172508a0. [DOI] [PubMed] [Google Scholar]
- Candela T, Fouet A. Bacillus anthracis CapD, belonging to the gamma-glutamyltranspeptidase family, is required for the covalent anchoring of capsule to peptidoglycan. Mol Microbiol. 2005;57:717–726. doi: 10.1111/j.1365-2958.2005.04718.x. [DOI] [PubMed] [Google Scholar]
- Candela T, Mock M, Fouet A. CapE, a 47-amino-acid peptide, is necessary for Bacillus anthracis polyglutamate capsule synthesis. J Bacteriol. 2005;187:7765–7772. doi: 10.1128/JB.187.22.7765-7772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela T, Fouet A. Poly-gamma-glutamate in bacteria. Mol. Microbiol. 2006;60:1091–1098. doi: 10.1111/j.1365-2958.2006.05179.x. [DOI] [PubMed] [Google Scholar]
- Chain E, Florey HW, Gardner AD, Heatley NG, Jennings MA, Orr-Ewing J, Sanders AG. Penicillin as a chemotherapeutic agent. Lancet. 1940;ii:226–228. [Google Scholar]
- Chain EB. Thiry years of penicillin therapy. Proc. R. Soc. London. 1971;179:293–319. doi: 10.1098/rspb.1971.0098. [DOI] [PubMed] [Google Scholar]
- Dixon TC, Meselson M, Guillemin J, Hanna PC. Anthrax. N. Engl. J. Med. 1999;341:815–826. doi: 10.1056/NEJM199909093411107. [DOI] [PubMed] [Google Scholar]
- Dixon TC, Fadl AA, Koehler TM, Swanson JA, Hanna PC. Early Bacillus anthracis-macrophage interactions: intracellular survival survival and escape. Cell. Microbiol. 2000;2:453–463. doi: 10.1046/j.1462-5822.2000.00067.x. [DOI] [PubMed] [Google Scholar]
- Domagk G. Ein Beitrag zur Chemotherapie der bakteriellen Infektionen. Deutsche Medizinische Wochenschrift. 1935;61:250–253. [Google Scholar]
- Drysdale M, Heninger S, Hutt J, Chen Y, Lyons CR, Koehler TM. Capsule synthesis by Bacillus anthracis is required for dissemination in murine inhalation anthrax. Embo J. 2005;24:221–227. doi: 10.1038/sj.emboj.7600495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Cleveland DW, Vande Woude GF. Proteolytic inactivation of Map-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- Ehrlich P, Bertheim A. Über p-Aminophenyl-Arsinsäure. Berichte der Deutschen Chemischen Gesellschaft. 1907;40:3292–3297. [Google Scholar]
- Friedlander AM, Welkos SL, Pitt MLM, Ezzell JW, Worsham PL, Rose KJ, Ivins BE, Lowe JR, Howe GB, Mikesell P, Lawrence WB. Postexposure prophylaxis against experimental inhalation anthrax. J. Infect. Dis. 1993;167:1239–1242. doi: 10.1093/infdis/167.5.1239. [DOI] [PubMed] [Google Scholar]
- Furuya EY, Lowy FD. Antimicrobial-resistant bacteria in the community setting. Nat Rev Microbiol. 2006;4:36–45. doi: 10.1038/nrmicro1325. [DOI] [PubMed] [Google Scholar]
- Ghuysen J-M. Use of bacteriolytic enzymes in determination of wall structure and their role in cell metabolism. Bacteriol. Rev. 1968;32:425–464. [PMC free article] [PubMed] [Google Scholar]
- Glauner B, Holtje J-V, Schwarz U. The composition of the murein of Escherichia coli. J. Biol. Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 1985;49:291–297. doi: 10.1128/iai.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, Jahrling PB, Hauer J, Layton M, McDade J, Osterholm MT, O'Toole T, Parker G, Perl T, Russel PK, Tonat K. Anthrax as a biological weapon: medical and public health management. Working group on civilian defense. JAMA. 1999;281:2127–2137. doi: 10.1001/jama.281.22.2127. [DOI] [PubMed] [Google Scholar]
- Ivins BE, Fellows PF, Nelson GO. Efficacy of a standard human anthrax vaccine against Bacillus anthracis spore challenge in guinea-pigs. Vaccine. 1994;12:872–874. doi: 10.1016/0264-410x(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Joyce J, Cook J, Chabot D, Hepler R, Shoop W, Xu Q, Stambaugh T, Aste-Amezaga M, Wang S, Indrawati L, Bruner M, Friedlander A, Keller P, Caulfield M. Immunogenicity and protective efficacy of Bacillus anthracis poly-gamma-D-glutamic acid capsule covalently coupled to a protein carrier using a novel triazine-based conjugation strategy. J. Biol. Chem. 2006;281:4831–4843. doi: 10.1074/jbc.M509432200. [DOI] [PubMed] [Google Scholar]
- Keillor JW, Castonguay R, Lherbet C. Gamma-glutamyl transpeptidase substrate specificity and catalytic mechanism. Methods Enzymol. 2005;401:449–467. doi: 10.1016/S0076-6879(05)01027-X. [DOI] [PubMed] [Google Scholar]
- Kim C, Wilcox-Adelman S, Sano Y, Tang WJ, Collier RJ, Park JM. Antiinflammatory cAMP signaling and cell migration genes co-opted by the anthrax bacillus. Proc Natl Acad Sci U S A. 2008;105:6150–6155. doi: 10.1073/pnas.0800105105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Tran LS, Uchida I, Itoh Y. Characterization of Bacillus subtilis gamma-glutamyltransferase and its involvement in the degradation of capsule poly-gamma-glutamate. Microbiology. 2004;150:4115–4123. doi: 10.1099/mic.0.27467-0. [DOI] [PubMed] [Google Scholar]
- King EC, Blacker AJ, Bugg TD. Enzymatic breakdown of poly-gamma-D-glutamic acid in Bacillus licheniformis: identification of a polyglutamyl gamma-hydrolase enzyme. Biomacromolecules. 2000;1:75–83. doi: 10.1021/bm990001n. [DOI] [PubMed] [Google Scholar]
- Koch R. Die Aetiologie der Milzbrand-Krankheit, begruendet auf die Entwicklungsgeschichte des Bacillus Anthracis. Beitraege zur Biologie der Pflanzen. 1876;2:277–310. [Google Scholar]
- Kocianova S, Vuong C, Yao Y, Voyich JM, Fischer ER, DeLeo FR, Otto M. Key role of poly-gamma-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J Clin Invest. 2005;115:688–694. doi: 10.1172/JCI23523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton TJ, Doi RH. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971;246:3189–3195. [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Magnet S, Bellais S, Dubost L, Fourgeaud M, Mainardi JL, Petit-Frere S, Marie A, Mengin-Lecreulx D, Arthur M, Gutmann L. Identification of the L,D-transpeptidases responsible for attachment of the Braun lipoprotein to Escherichia coli peptidoglycan. J Bacteriol. 2007;189:3927–3931. doi: 10.1128/JB.00084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Sasakawa C, Uchida I, Terakado N, Yoshikawa M. Cloning and CO2-dependent expression of the genetic region for encapsulation from Bacillus anthracis. Mol Microbiol. 1988;2:371–376. doi: 10.1111/j.1365-2958.1988.tb00041.x. [DOI] [PubMed] [Google Scholar]
- Makino S, Uchida I, Terakado N, Sasakawa C, Yoshikawa M. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 1989;171:722–730. doi: 10.1128/jb.171.2.722-730.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Watarai M, Cheun HI, Shirahata T, Uchida I. Effect of the lower molecular capsule released from the cell surface of Bacillus anthracis on the pathogenesis of anthrax. J Infect Dis. 2002;186:227–233. doi: 10.1086/341299. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Schneewind O. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol. Microbiol. 2006;62:1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Mock M, Fouet A. Anthrax. Annu. Rev. Microbiol. 2001;55:647–671. doi: 10.1146/annurev.micro.55.1.647. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu HC. The crisis in antibiotic resistance. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- Okada T, Suzuki H, Wada K, Kumagai H, Fukuyama K. Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate. Proc Natl Acad Sci U S A. 2006;103:6471–6476. doi: 10.1073/pnas.0511020103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okinaka R, Cloud K, Hampton O, Hoffmaster A, Hill K, Keim P, Koehler T, Lamke G, Kumano S, Manter D, Martinez Y, Ricke D, Svensson R, Jackson P. Sequence, assembly and analysis of pX01 and pX02. J. Appl. Microbiol. 1999;87:261–262. doi: 10.1046/j.1365-2672.1999.00883.x. [DOI] [PubMed] [Google Scholar]
- Pasteur L. Le vaccin du charbon. Comptes Rendus Hebdomadaires des Scéances de l'Académie des Sciences. 1881;92:666–668. [Google Scholar]
- Pezard C, Berche P, Mock M. Contribution of individual toxin components to virulence of Bacillus anthracis. Infect. Immun. 1991;59:3472–3477. doi: 10.1128/iai.59.10.3472-3477.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisz H. Experimentelle studien ueber virulenz, empfaenglichkeit und immunitaet beim milzbrand. Zeitschr. Immunitaet.-Forsch. 1909;5:341–452. [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. American Journal of Hygiene. 1938;27:493–497. [Google Scholar]
- Rhie GE, Roehrl MH, Mourez M, Collier RJ, Mekalanos JJ, Wang JY. A dually active anthrax vaccine that confers protection against both bacilli and toxins. Proc. Natl. Acad. Sci. USA. 2003;100:10925–10930. doi: 10.1073/pnas.1834478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishton GM. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov Today. 2003;8:86–96. doi: 10.1016/s1359644602025722. [DOI] [PubMed] [Google Scholar]
- Salton MRJ. Cell wall of Micrococcus lysodeikticus as the substrate of lysozyme. Nature. 1952;170:746–747. doi: 10.1038/170746a0. [DOI] [PubMed] [Google Scholar]
- Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- Schneewind O, Fowler A, Faull KF. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- Scorpio A, Chabot DJ, Day WA, O'Brien DK, Vietri NJ, Itoh Y, Mohamadzadeh M, Friedlander A. Poly-γ-glutamate capsuledegrading enzyme treatment enhances phagocytosis and killing of encapsulated Bacillus anthracis. Antimicrob Agents Chemother. 2007;51:215–222. doi: 10.1128/AAC.00706-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidler J, McGovern SL, Doman TN, Shoichet BK. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J Med Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- Severin A, Tabei K, Tomasz A. The structure of the cell wall peptidoglycan of Bacillus cereus RSVF1, a strain closely related to Bacillus anthracis. Microb. Drug Resist. 2004;10:77–82. doi: 10.1089/1076629041310082. [DOI] [PubMed] [Google Scholar]
- Strominger JL, Izaki K, Matsuhashi M, Tipper DJ. Peptidoglycan transpeptidase and D-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Fed. Proc. 1967;26:9–18. [PubMed] [Google Scholar]
- Strominger JL. Penicillin-sensitive enzymatic reactions in bacterial cell wall synthesis. Harvey Lectures. 1968;64:179–213. [PubMed] [Google Scholar]
- Tate SS, Meister A. Gamma-glutamyl transpeptidase: catalytic, structural and functional aspects. Mol. Cell. Biochem. 1981;39:357–368. doi: 10.1007/BF00232585. [DOI] [PubMed] [Google Scholar]
- Tipper DJ, Strominger JL. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-alanine. Proc. Natl. Acad. Sci. USA. 1965;54:1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Mazmanian SK, Faull KF, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH(2)-Gly(3) substrates. J Biol Chem. 2000;275:9876–9881. doi: 10.1074/jbc.275.13.9876. [DOI] [PubMed] [Google Scholar]
- Troy FA. Chemistry and biosynthesis of the poly(-D-glutamyl) capsule in Bacillus licheniformis. II. Characterization and structural properties of the enzymatically synthesized polymer. J Biol Chem. 1973a;248:316–324. [PubMed] [Google Scholar]
- Troy FA. Chemistry and biosynthesis of the poly(-D-glutamyl) capsule in Bacillus licheniformis. I. Properties of the membrane-mediated biosynthetic reaction. J Biol Chem. 1973b;248:305–315. [PubMed] [Google Scholar]
- Uchida I, Makino S, Sasakawa C, Yoshikawa M, Sugimoto C, Terakado N. Identification of a novel gene, dep, associated with depolymerization of the capsular polymer in Bacillus anthracis. Mol. Microbiol. 1993;9:487–496. doi: 10.1111/j.1365-2958.1993.tb01710.x. [DOI] [PubMed] [Google Scholar]
- Walsh CT. Vancomycin resistance: decoding the molecular logic. Science. 1993;261:308–309. doi: 10.1126/science.8392747. [DOI] [PubMed] [Google Scholar]
- Walsh CT. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406:775–781. doi: 10.1038/35021219. [DOI] [PubMed] [Google Scholar]
- Wu R, Richter S, Zhang R, Elmer A, Missikas D, Joachimiak A. Submitted for publication. 2008. Crystal structure of Bacillus anthracis CapD, a γ-glutamyl transpeptidase involved in capsule biogenesis. [Google Scholar]
- Yocum RR, Waxman DJ, Rasmussen JR, Strominger JL. Mechanism of penicillin action: penicillin and substrate bind covalently to the same active site serine in two bacterial D-alanine carboxypeptidases. Proc. Natl. Acad. Sci. USA. 1979;76:2730–2734. doi: 10.1073/pnas.76.6.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa K, Kawata S, Nishimura S, Ikeda Y, Yoshimura Y. Mutanolysin, bacteriolytic agent for cariogenic streptococci: partial purification and properties. Antimicrob. Agents Chemother. 1974;6:156–165. doi: 10.1128/aac.6.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.