Abstract
Background
Translocations of the Mixed Lineage Leukemia (MLL) gene occur in a subset (5%) of acute myeloid leukemias (AML), and in mixed phenotype acute leukemias in infancy - a disease with extremely poor prognosis. Animal model systems show that MLL gain of function mutations may contribute to leukemogenesis. Wild-type (wt) MLL possesses histone methyltransferase activity and functions at the level of chromatin organization by affecting the expression of specific target genes. While numerous MLL fusion proteins exert a diverse array of functions, they ultimately serve to induce transcription of specific genes. Hence, acute lymphoblastic leukemias (ALL) with MLL mutations (MLLmu) exhibit characteristic gene expression profiles including high-level expression of HOXA cluster genes. Here, we aimed to relate MLL mutational status and tumor suppressor gene (TSG) methylation/expression in acute leukemia cell lines.
Results
Using MS-MLPA (methylation-specific multiplex ligation-dependent probe amplification assay), methylation of 24 different TSG was analyzed in 28 MLLmu and MLLwt acute leukemia cell lines. On average, 1.8/24 TSG were methylated in MLLmu AML cells, while 6.2/24 TSG were methylated in MLLwt AML cells. Hypomethylation and expression of the TSG BEX2, IGSF4 and TIMP3 turned out to be characteristic of MLLmu AML cell lines. MLLwt AML cell lines displayed hypermethylated TSG promoters resulting in transcriptional silencing. Demethylating agents and inhibitors of histone deacetylases restored expression of BEX2, IGSF4 and TIMP3, confirming epigenetic silencing of these genes in MLLwt cells. The positive correlation between MLL translocation, TSG hypomethylation and expression suggested that MLL fusion proteins were responsible for dysregulation of TSG expression in MLLmu cells. This concept was supported by our observation that Bex2 mRNA levels in MLL-ENL transgenic mouse cell lines required expression of the MLL fusion gene.
Conclusion
These results suggest that the conspicuous expression of the TSG BEX2, IGSF4 and TIMP3 in MLLmu AML cell lines is the consequence of altered epigenetic properties of MLL fusion proteins.
Background
Translocations of the Mixed Lineage Leukemia (MLL) gene occur in a subset of acute leukemias. The correlation between MLL translocations and expression of specific gene clusters is so evident that "mixed lineage leukemia", originally applied to biphenotypic acute leukemia cells, is now used to describe the MLL mutant (MLLmu) acute leukemias [1]. High expression levels of a set of HOXA cluster genes are characteristic of MLL mutations in primary acute lymphoblastic leukemia (ALL) cells, and in MLLmu ALL cell lines [1,2]. For acute myeloid leukemia (AML) cell lines, a similar correlation exists between MLL translocations and expression of the gene brain expressed X-linked 2 (BEX2, formerly called BEX1) [3]. In healthy people, BEX2 is expressed in the brain and, more weakly, in pancreas and testis, but not in hematopoetic cells [3,4]. In leukemia cell lines, we found BEX2 expression to be restricted to MLLmu AML. MLL wild-type (MLLwt) AML and ALL cell lines and, notably, also MLLmu ALL cell lines do not transcribe this gene, suggesting that BEX2 expression might be a diagnostic marker for MLLmu AML [3].
Several lines of evidence indicate that epigenetic mechanisms are responsible for the regulation of BEX2 expression: (i) the BEX2 promoter is methylated in MLLwt and unmethylated in MLLmu AML cell lines, thus demonstrating an inverse correlation between gene expression and promoter methylation [5]; (ii) demethylating agents and inhibitors of histone deacetylases (HDAC) induce BEX2 expression in MLLwt cells [5]; (iii) chromatin immunoprecipitation experiments show that histone acetylation plays a role in BEX2 regulation: immunoprecipitation of acetylated histone H3 coprecipitates chromatin from the 5' region of BEX2 in MLLmu, but not in MLLwt cells [5].
BEX1 and BEX2 have recently been described as epigenetically controlled candidate tumor suppressor genes (TSG) in malignant glioma [6]. Promoter hypermethylation of TSG is often seen in malignant diseases and, according to a widely held view, contributes to the rise of malignant cell clones by restraining tumor suppressor gene expression [7]. Moreover, unique profiles of hypermethylated CpG islands have been described which are characteristic of different neoplasias [8,9].
We applied a multiplex methylation detection assay to find out whether the connection between the MLL mutational status and promoter methylation is unique to BEX2 or if this correlation applies to other TSG as well. Results show that MLLwt AML cell lines exhibit a higher propensity for TSG promoter hypermethylation than MLLmu cell lines. This is especially true for Immunoglobulin superfamily member 4 (IGSF4/CADM1), Retinoic acid receptor beta (RARB) and Tissue inhibitor of matrix metalloproteinase 3 (TIMP3), all with MLL-dependent methylation profiles resembling BEX2. According to methylation-specific PCR (MSP), primary AML cells without rearrangement of the MLL gene also show a preference for TSG hypermethylation.
Our experimental results suggest that MLLmu proteins enhance the expression of distinct TSG and that this might be the consequence of altered epigenetic regulatory mechanisms of the fusion proteins.
Results and Discussion
Methylation patterns of TSG differ in MLLmu and MLLwt cell lines
Hypermethylation of CpG islands in the promoter regions of TSG occurs widely in malignancy, resulting in transcriptional inactivation which promotes cancerogenesis [7]. Unique profiles of hypermethylated CpG islands have been described, being characteristic of different neoplasias [8,9]. BEX2 is a newly described TSG, which is silenced by hypermethylation in malignant glioma [6]. With leukemia/lymphoma cell lines as model systems, we have shown that BEX2 hypomethylation and gene expression typifies MLLmu AML [3]. BEX2 silencing in MLLwt cells is the result of epigenetic mechanisms like promoter methylation and histone deacetylation [3,5]. Here, we set out to elucidate whether the described correlation between promoter hypomethylation and genetically rearranged MLL is peculiar to BEX2, or whether other TSG display similar features.
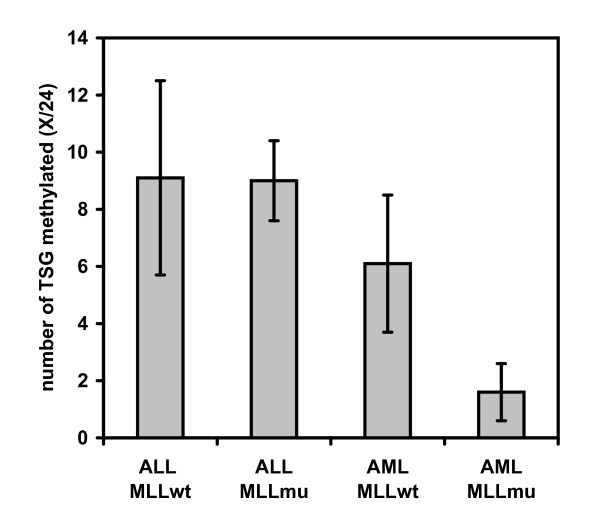
Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) was performed to assess the methylation status of 24 TSG in 28 acute leukemia cell lines, 50% with MLL translocations (see Additional file 1). ALL-derived cell lines showed a higher percentage of methylated TSG than AML cell lines (Fig. 1). The mean number of methylated TSG in ALL cell lines was independent of MLL mutational status (Fig. 1). In contrast, 25% of all TSG analyzed were methylated in MLLwt, but only 7% in MLLmu AML cell lines (Fig. 1). Provided that the TSG tested are representative, MLLmu AML cell lines display a significantly lower tendency to TSG hypermethylation than MLLwt cell lines.
Figure 1.
Low tendency for TSG methylation in MLL mu AML cell lines. Methylation status of 24 TSG was determined by MS-MLPA (see also: Additional file 1). Mean numbers of methylated TSG and standard deviations in the different groups of leukemia cell lines are shown. Note that MLLmu AML, but not MLLmu ALL cell lines show reduced TSG methylation.
Methylation status of BEX2, IGSF4, RARB and TIMP3 is indicative of MLLmu AML
Expression array analysis allows classification of tumors, including the discrimination of acute leukemias with/without MLL translocations [1]. Likewise, profiles of promoter hypermethylation are unique for different types of cancer and have also been used to identify lymphomatous entities [8-12]. We evaluated our MS-MLPA data to find out whether MLLmu and MLLwt cell lines could be distinguished via their respective TSG methylation patterns (see Additional file 1). In addition to the 24 TSG included in this assay, we tested methylation of BEX2 by a methylation-sensitive DNA restriction- and quantitative real-time PCR-assay.
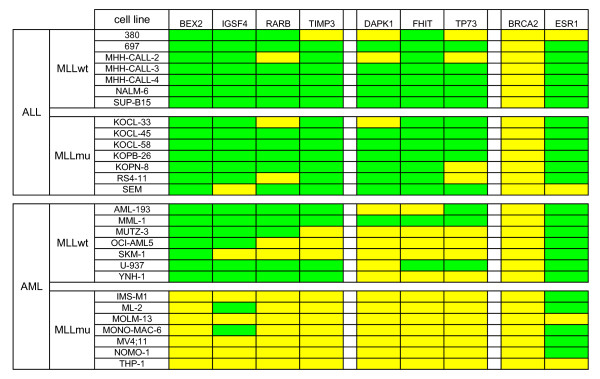
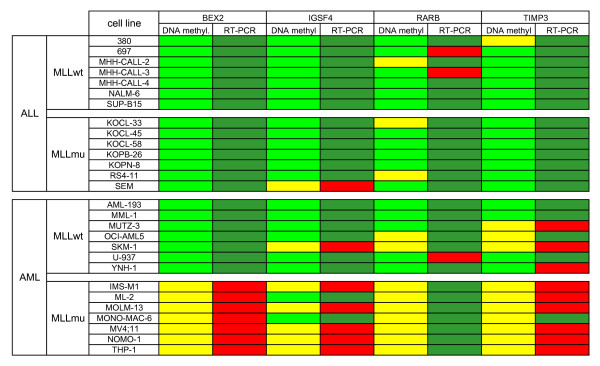
Promoter methylation of TSG BEX2, IGSF4, RARB and TIMP3 distinguished MLLmu and MLLwt AML: 0/7 MLLmu and 6/7 MLLwt AML cell lines exhibited methylation of at least two of these four TSG (p-value = 0.002, Fisher's exact test) (Fig. 2). Neither these four genes nor any other set of TSG analyzed distinguished between MLLmu and MLLwt ALL cell lines, suggesting that a myeloid gene expression background was obligatory for the MLL-specific methylation pattern (see Fig. 2, Additional file 1).
Figure 2.
TSG methylation predictive for classification of acute leukemia cell lines. Methylation was assessed by MS-MLPA (see also Additional file 1) or by DNA digestion with methylation-sensitive HhaI plus promoter-specific quantitative real-time PCR (for analysis of BEX2). Green: methylation level ≥ 10%; yellow: methylation level < 10%. BEX2, IGSF4, RARB and TIMP3 are methylated in ALL and in MLLwt AML, but not in MLLmu AML. This correlation is statistically significant with p-values < 0.05 for BEX2, RARB and TIMP3 and with a p-value of 0.051 for IGSF4 (Fisher's exact test). Methylation analysis of DAPK1, FHIT and TP73 discriminates between ALL and AML cell lines (p-values ≤ 0.01, Fisher's exact test). Other TSG (e.g. BRCA2 and ESR1) are methylated or unmethylated in the majority of cell lines with no preference for subtype.
Most of the 24 TSG analyzed did not allow the histological origin of the cells to be inferred, being either methylated (e.g. ESR1) or unmethylated (e.g. BRCA2) in the majority of cell lines (see Fig. 2, Additional file 1). Furthermore, deletions, notably those of the TSG CDKN2A and CDKN2B, occurred in ALL as well as in AML cell lines (see Additional file 1). However, the methylation pattern of three genes (DAPK1, FHIT, TP73) was highly characteristic (p-values = 0.01, Fisher's exact test) of either ALL or AML (Fig. 2). These results show that analysis of TSG methylation may also be used to recognize distinct leukemic entities. Noteworthy in the context of our study, methylation patterns of BEX2, IGSF4, RARB and TIMP3 allowed discrimination between MLLmu and wt AML cell lines (Fig. 2).
MS-MLPA is a screening technique and obtained results rely on the analysis of one single CpG-site in the promoter region. To verify the validity of the methylation status as determined by MS-MLPA or HhaI-restriction-sensitive PCR assay (for BEX2), we performed MSP after bisulfite conversion of DNA for BEX2, IGSF4 and TIMP3 (see Additional file 2). The respective CpGs tested by MSP and MS-MLPA were located at different promoter sites. The accuracy of MS-MLPA/HhaI-restriction-sensitive PCR to predict the methylation status of these three TSG as determined by MSP was reassuringly high (83%), supporting the validity of MS-MLPA for use in our screening.
In conclusion, most MLLwt cell lines show hypermethylation of BEX2, IGSF4 and TIMP3, while MLLmu cell lines are preferentially unmethylated.
Methylation of tumor suppressor genes in primary MLLmu and MLLwt AML cells
CpG island hypermethylation occurs more often in cell lines than in primary tumor cells [11,13]. Controversial opinions exist on the question whether or not cell lines, including leukemia cell lines, show the same methylation profiles as the analogous primary cells [13-15]. To investigate the possible concordance of TSG methylation in cell lines and primary cells, we tested samples from MLLmu and MLLwt AML patients for TSG methylation. MS-MLPA assay detected low numbers of methylated TSG (mean 0.5/24 +/- 1.0 TSG) in primary AML cells, which is in accordance with earlier findings applying the same technique [16]. MSP has been shown to be a suitable and more sensitive technique for primary AML samples than MS-MLPA [17].
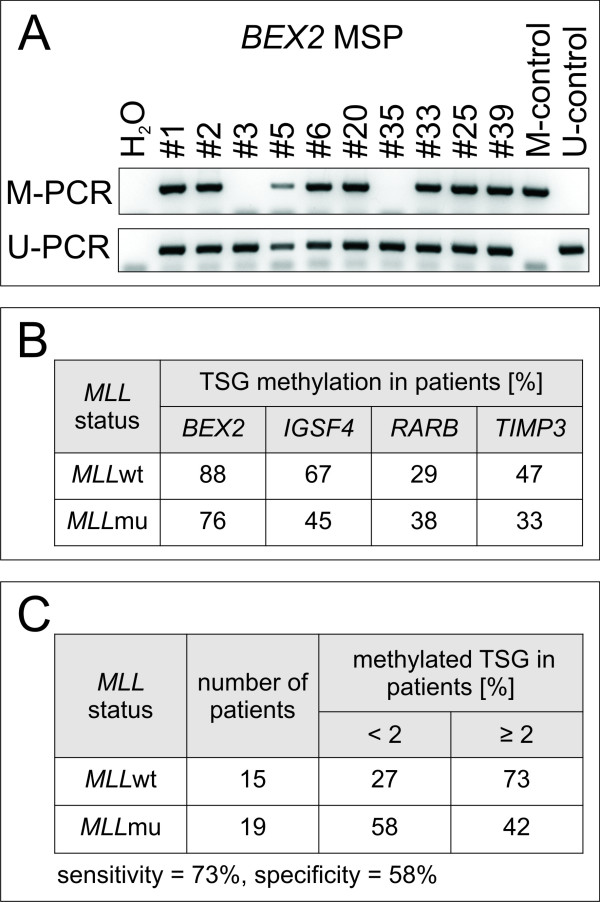
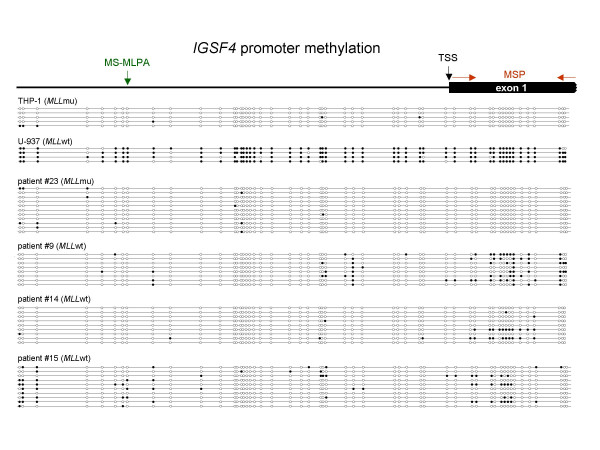
Therefore, we performed MSP after bisulfite conversion of DNA and detected methylated and unmethylated TSG promoter sequences using primers specific for BEX2, IGSF4, RARB and TIMP3, the most informative genes in our system (Fig. 3). For BEX2, IGSF4 and TIMP3, MSP indicated a trend towards an association between promoter methylation and MLLwt status (see Fig. 3B and Additional file 3). This correlation could not be observed for RARB, rendering this gene less interesting for our study. Bisulfite sequencing of the IGSF4 promoter region confirmed MSP results: MLLwt AML patients (#9, #14 and #15) and MLLwt cell line U-937 harbored clones with CpG methylation adjacent to the transcriptional start site of IGSF4, unlike MLLmu patient #23 and cell line THP-1 (see Fig. 4, Additional file 3).
Figure 3.
TSG methylation in primary AML samples. Methylation status was assessed by MSP for BEX2, IGSF4, TIMP3 and RARB in a panel of MLLwt and MLLmu AML patients. A) Representative results for BEX2 MSP are shown. Patients #1 - #17 carry MLLwt, patients #18 - #40 harbor MLL rearrangements (see Additional file 3). B) Summary of methylation data obtained by MSP in patients. Lower percentages of MLLmu patients than MLLwt patients show promoter hypermethylation of BEX2, IGSF4 and TIMP3 (see also Additional file 3). C) Note that methylation of two out of three TSG (BEX2, IGSF4, TIMP3) is more frequent in MLLwt patients than in MLLmu patients.
Figure 4.
Bisulfite sequencing of the IGSF4 promoter region. The IGSF4 promoter region (559 bp, 52 CpG sites) was sequenced after bisulfite conversion of DNA in two cell lines and selected AML patients. CpGs are represented as open dots (unmethylated) or filled dots (methylated). In MLLwt AML patients and in the MLLwt cell line U-937 clones with CpG methylation next to the transcriptional start site (TSS) were detected whereas no methylation was detectable in the MLLmu patient analyzed and in the MLLmu cell line THP-1. Note that CpG sites analyzed by the MS-MLPA probe (green) and MSP primers (red) are not identical.
Using methylation of at least two out of the three relevant genes (BEX2, IGSF4 and TIMP3) as classifier for the MLL mutational status, the true positive identification rate (sensitivity) for MLLmu samples was 73%, with a specificity of 58%, supporting the conclusion that methylation of BEX2, IGSF4 and TIMP3 is more frequent in MLLwt than in MLLmu patients (Fig. 3C). Testing a larger cohort of patients will be necessary to confirm a statistically significant positive correlation between MLL mutational status and TSG methylation.
Inverse correlation of TSG methylation and expression
Hypermethylation of CpG islands in the promoter regions of TSG generally leads to the silencing of the respective genes [18,19]. To test whether BEX2, IGSF4, RARB and TIMP3 were epigenetically regulated, we analyzed mRNA expression of these genes in cell lines by quantitative real-time PCR. Figure 5 shows that three of the four genes analyzed (BEX2, IGSF4 and TIMP3) demonstrated an inverse correlation between TSG promoter methylation and gene expression consistent with epigenetic gene regulation. Predictive value for true methylation of these genes was 0.93, and accuracy 0.94. In contrast, RARB remained silent in most cell lines, independent of methylation status (Fig. 5). A similar result has been described for RARA: primary AML cells show low RARA2 expression although the promoter is not methylated confirming that regulation of gene transcription is a multi-factor process, not solely reliant on promoter methylation/demethylation [20]. Thus, (i) MLLmu AML cell lines express higher levels of BEX2, IGSF4 and TIMP3 than MLLwt cell lines, and (ii) these three TSG appear to be regulated by epigenetic mechanisms.
Figure 5.
Inverse correlation between TSG methylation and expression. Hypomethylation (yellow) parallels RNA expression (red) of BEX2, IGSF4 and TIMP3 in MLLmu AML. MLLwt AML cell lines show hypermethylation (light green) and transcriptional silencing (green) of these. In contrast, RARB expression is low (green) even in cell lines that carry the unmethylated promoter (yellow). Excluding RARB, TSG hypermethylation inversely correlated with gene transcription in 94% of the cases. TSG methylation status is shown as determined by MS-MLPA (see Additional file 1). Gene expression was assessed in triplicates by quantitative real-time PCR (RT-PCR) analysis. For BEX2, IGSF4 and TIMP3, cell line IMS-M1 was applied as calibrator; for RARB, expression levels in cell line 697 were set as 1. Cell lines that reached relative expression levels ≥ 0.05 were considered positive (red).
RARB was excluded from further studies, as we neither observed an inverse correlation between RARB promoter methylation and gene expression in cell lines nor did we see a positive correlation between MLLmu and RARB hypomethylation in primary AML cells.
According to our findings, TSG hypomethylation and mRNA expression are characteristic features of MLLmu AML cells. This is in apparent conflict with the notion that MLL fusion proteins act as strong oncogenes, a view that is supported by the fact that MLLmu confers a dismal prognosis in AML. We hypothesize that BEX2, IGSF4 and TIMP3 are sentinel MLL fusion gene targets, their function being overridden by the strong oncogene MLLmu. A better understanding of how MLL fusion proteins impede TSG promoter methylation might help to illuminate a novel epigenetic role for MLLmu in leukemic cells.
DNA demethylation and histone acetylation induce expression of TSG
To verify epigenetic regulation of BEX2, IGSF4 and TIMP3, we applied the DNA-demethylating agent 5-Aza-2'-deoxycytidine (Aza) and the HDAC inhibitor trichostatin A (TSA). Our previous results had shown that these agents induced expression of BEX2 in MLLwt AML cell lines [5]. We show here that Aza and TSA also triggered mRNA expression of IGSF4 and TIMP3 (Table 1). To confirm activity of TSA, histone H4K12 acetylation, a modification associated with active transcription, was analyzed (see Additional file 4). These results confirmed that epigenetic mechanisms are responsible for TSG silencing in MLLwt cell lines and suggest that TSG hypomethylation in MLLmu cell lines might be caused by alterations of epigenetic MLL functions as result of the various translocations.
Table 1.
Effects of Aza and TSA on TSG expression in MLLwt AML cell lines.
| cell line | treatment | TSG | ||
| BEX2 | IGSF4 | TIMP3 | ||
| AML-193 | TSA | - | - | + |
| Aza | ++ | ++ | +++ | |
| Aza + TSA | ++ | ++ | +++ | |
| SKM-1 | TSA | (+) | + | + |
| Aza | - | (+) | + | |
| Aza + TSA | + | + | ++ | |
| U-937 | TSA | - | + | ++ |
| Aza | - | +++ | +++ | |
| Aza + TSA | - | +++ | +++ | |
Induction of TSG expression by Aza (5 μM, 4 d) and TSA (2 μM, 1 d) when compared to untreated control cells: - < 2-fold, + ≥ 2-fold; ++ ≥ 10-fold, +++ ≥ 100-fold gene expression. Expression levels were determined by quantitative real-time PCR. Note that cell line SKM-1 shows hypomethylation and expression of IGSF4 and TIMP3 (Fig. 5). Experiments were performed at least twice. Brackets: effect variable.
Bex2 transcription depends on expression of MLL-ENL
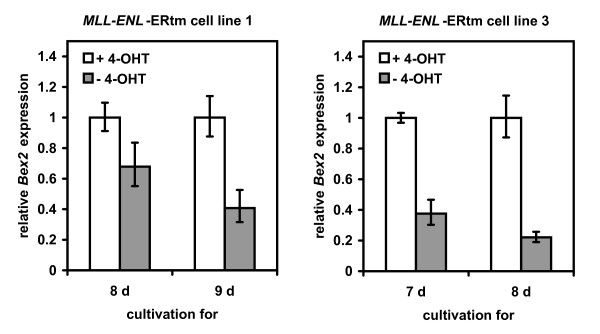
To verify whether MLL fusion proteins were responsible for altered expression of TSG, we applied a mouse bone marrow cell line system that expresses MLL-ENL fused to the ligand-binding domain of the estrogen receptor. Removal of 4-hydroxytamoxifen (4-OHT) from the medium tethers MLL-ENL to a heat shock protein complex, thereby blocking the activation of the fusion protein [21]. Withdrawal of 4-OHT led to a marked decrease of Bex2 expression in this mouse cell system confirming that MLL fusion proteins serve to upregulate this gene (Fig. 6).
Figure 6.
MLL fusion proteins regulate Bex2 expression. In presence of 4-OHT (100 nM) MLL-ENL-ERtm mouse cell lines expressed MLL-ENL and Bex2. Withdrawal of 4-OHT for seven to nine days induced downregulation of MLL-ENL and suppression of Bex2 transcription. Bex2 expression levels with standard deviations were determined in two different MLL-ENL-ERtm cell line clones by quantitative real-time PCR analysis.
These results suggest that expression of TSG in MLLmu cell lines is attributable to the presence of MLL fusion proteins, raising the question whether the MLL fusion proteins directly target TSG promoters by blocking DNA methylation processes, or whether these effects are secondary.
It is difficult to attribute a specific oncogenic function to any single region of MLL fusion genes. Characteristic of the complexity of the situation is that many MLL translocations are reciprocal, and that both fusion genes are transcribed and may function oncogenically. However, MLL-ENL alone was sufficient to induce Bex2 in the transgene system indicating that the reciprocal fusion product was not required for regulation of this TSG. Furthermore, we observed TSG hypomethylation in various MLLmu cell lines with a variety of fusion partners. Together, these observations suggested that the inhibitory effect of MLL fusion genes on TSG promoter methylation was mediated by the N-terminal part of MLL and not by the fusion partner.
Both, the N-terminal and the C-terminal part of MLL have been implicated in transcriptional and/or epigenetic regulation. Located 3', the Su(var)3-9, enhancer-of-zeste, trithorax (SET) domain confers histone H3K4 methyltransferase activity to the protein [22]. However, histone H3K4 methylation is a marker for epigenetic activation and it appears unlikely that the loss of an activating function should lead to expression of TSG. Located upstream of the MLL breakpoint cluster region and retained in MLL fusion genes is the CXXC DNA methyltransferase homology domain, which binds CpG rich regions particularly when these are unmethylated [23]. This domain is essential for target gene recognition and is required for transformation by MLL fusion proteins [24]. Therefore, the CXXC domain might direct MLL to transcribed genes or possibly protect CpG islands against methylation. The N-terminal part of MLL has also been reported to bind menin [25]. MLL and menin are required for cellular transformation and both proteins cooperatively regulate expression of the genes p27Kip1 and p18Ink4c [25,26].
Thus, regulation of TSG expression and alterations of epigenetic functions of MLL fusion proteins are feasible consequences of MLL translocations, but whether the TSG hypomethylation in MLLmu cell lines described by us is the direct effect of MLL fusion proteins or whether the hypomethylation is rather an indirect consequence, remains to be resolved. For two reasons we favor the latter explanation: MLLmu ALL cell lines do not show the TSG methylation profile that is typical for MLLmu AML suggesting that additional, tissue-specific factors play a role in this context. Furthermore, ChIP-DNA mircroarrays did not identify TIMP3 as target of wild-type MLL, the other two genes not being tested [27]. Future studies will have to provide evidence whether the MLL fusion proteins show altered DNA binding specificities than the wild-type protein allowing access to the promoters of the named TSG.
Conclusion
We describe the conspicuous expression of TSG BEX2, IGSF4 and TIMP3 in MLLmu AML cells. In MLLwt cell lines these genes are silenced by promoter methylation. Transcription could be reactivated by treatment with demethylating agents and HDAC inhibitors. In MLLmu AML cell lines, constitutive expression of BEX2, IGSF4 and TIMP3 was accompanied by promoter hypomethylation. Ectopic expression of MLL-ENL drove upregulation of Bex2 in a tamoxifen-inducible mouse model indicating that the MLL fusion genes were responsible for upregulation of TSG mRNA. Taken together, our results suggest that hypomethylation of TSG characteristic of MLLmu AML cells may be the consequence of MLL gene alterations, including elements responsible for epigenetic regulation.
Methods
Human cell lines
The continuous cell lines were either taken from the stock of the cell bank (DSMZ - German Collection of Microorganisms and Cell Cultures) or were generously provided by the original investigators. Detailed references and cultivation protocols have been described previously [28,29]. The following cell lines were tested for TSG methylation: (i) B-cell precursor (BCP) ALL-derived cell lines expressing MLLwt: 380, 697, MHH-CALL-2, MHH-CALL-3, MHH-CALL-4, NALM-6, SUP-B15; (ii) BCP ALL-derived cell lines with MLLmu: KOCL-33 t(11;19), KOCL-45 t(4;11), KOCL-58 t(4;11), KOPB-26 t(9;11), KOPN-8 t(11;19), RS4;11 t(4;11), SEM t(4;11); (iii) AML-derived cell lines with MLLwt: AML-193 (M5), MML-1 (M1), MUTZ-3 (M4), OCI-AML5 (M4), SKM-1 (M5), U-937 (M5), YNH-1 (M1); (iv) AML-derived cell lines with MLLmu: IMS-M1 (M5, t(9;11)), ML-2 (M4, t(6;11)), MOLM-13 (M5, t(9;11)), MONO-MAC-6 (M5, t(9;11)), MV4;11 (M5, t(4;11)), NOMO-1 (M5, t(9;11)), THP-1 (M5, t(9;11)).
Human tissue samples
After informed consent was given, bone marrow or peripheral blood specimens were obtained during routine clinical assessment of 40 AML patients. The collection of patient samples for analysis of genetic changes was approved by the local ethics committee. DNA extraction was performed from unselected cells from bone marrow or peripheral blood. The MLLwt/mu status of primary samples was assessed with long-distance PCR analysis, as described previously [30].
Methylation-specific multiplex ligation-dependent probe amplification assay
The MS-MLPA assay (ME001B; MRC-Holland, Amsterdam, Netherlands) simultaneously detects copy number changes and CpG methylation of the promoter regions of 24 different TSG. This semi-quantitative technique is based on digestion of DNA with the methylation-sensitive restriction enzyme HhaI (Fermentas, St. Leon-Rot, Germany) and a subsequent multiplex PCR followed by fragment analysis via capillary electrophoresis [31]. MS-MLPA data were analysed using a Microsoft Excel spreadsheet designed specifically for the ME001B assay. Levels of methylation were calculated by comparing the relative peak area of the HhaI digested ligation product with the corresponding ligation product from the undigested sample. Peak areas were normalised relative to neighbouring control ligation products prior to comparison as recommended by MRC-Holland. The spreadsheet for the analysis of the ME001B kit is freely available for download and use on the National Genetics Reference Laboratory website at http://www.ngrl.org.uk/Manchester/mlpapubs.html. For assessment of BEX2 promoter methylation, HhaI-digested and undigested DNA was used as template for subsequent quantitative real-time PCR using SYBR GREEN PCR Master Mix (Applied Biosystems, Foster City, CA, USA) in a 7500 Applied Biosystems real-time PCR system. The sequence flanked by primer pair A lacked HhaI sites, and was used as endogenous control. Sequence amplified by primer pair B contained two HhaI sites. The percentage of non-cleaved and thus methylated template in comparison to the undigested sample was calculated using the ΔΔCt-method. BEX2 A forward: 5'-GGT TGG TGA GAA GGA GGG TG-3'; BEX2 A reverse: 5'-GAG ACA CGA GTG ACG ACT GCA-3'; BEX2 B forward: 5'-TGG AGA GGA CGG AGA TGA GTG-3'; BEX2 B reverse: 5'-CAC CCT CCT TCT CAC CAA CC-3'. Methylation was scored positive when the calculated methylation percentage was ≥ 10%.
Methylation-specific polymerase chain reaction (MSP)
Bisulfite conversion of DNA was performed as described by the supplier (Active Motif, Rixensart, Belgium). For detecting TSG promoter methylation, we performed nested PCR with first round primers amplifying converted DNA independently of the methylation status (bisulfite-specific PCR), while second round primers for M- and U-PCR specifically recognized the methylated or unmethylated versions of the promoter. PCR products of the initial bisulfite-specific PCR were diluted 1:100 for subsequent M- and U-PCR. PCR conditions and primer sequences are listed in Additional file 5. Epitect PCR Control DNA (Qiagen, Hilden, Germany) was used as control for methylated and unmethylated templates.
Bisulfite sequencing
To confirm methylation status of the IGSF4 promoter, DNA of cell lines and AML patients was bisulfite converted according to the manufacturer's instructions (Active Motif). Subsequently, amplification of the IGSF4 promoter region (559 bp) was performed using primers IGSF4 BSP fwd and IGSF4 BSP rev, specifically binding bisulfite converted DNA (for primer sequence and PCR conditions see Additional file 5). Resulting IGSF4 fragments were purified, cloned into pGEM-T Easy vector (Promega, Madison, WI, USA) and sequenced. Sequences were evaluated using BiQ Analyzer http://biq-analyzer.bioinf.mpi-sb.mpg.de and had to conform to at least 90% bisulfite conversion rate. In addition, identical clones were excluded from the analysis.
Expression of tumor suppressor genes
Quantitative PCR was performed on a 7500 Applied Biosystems (Darmstadt, Germany) real-time PCR system using the manufacturer's protocol. RNA was prepared using the Trizol reagent (Invitrogen, Karlsruhe, Germany). For mRNA quantification, reverse transcription was performed using the SuperScript II reverse transcriptase kit (Invitrogen). TaqMan probes (Applied Biosystems) were used to quantify human BEX2 (Hs 00218464m1), IGSF4 (Hs 00204937m1), RARB (Hs 00233407m1) and TIMP3 (Hs 00927216m1) expression levels with TBP as endogenous control. Expression of mouse Bex2 was assessed using the SYBR GREEN PCR Master Mix (Applied Biosystems) with Tbp as internal control. Bex2 forward: 5'-GCG AGC GGG ACA GAT TGA C-3'; Bex2 reverse: 5'-TCC ATT TCT CCT GGG CCT ATC-3'. Tbp forward: 5'-ACC AGA ACA ACA GCC TTC CAC-3'; Tbp reverse: 5'-TGC CGT AAG GCA TCA TTG GAC-3'. Relative expression levels were calculated using the ΔΔCt-method.
Induction of MLL-ENL
Mouse bone marrow cell lines MLL-ENL-ERtm 1 and 3 were maintained in RPMI 1640 medium supplemented with 10% FBS (Sigma, Taufkirchen, Germany), IL-3 (5 ng/ml), GM-CSF (5 ng/ml), IL-6 (5 ng/ml), SCF (50 ng/ml) and 4-OHT (100 nM). Murine cytokines were obtained from Richter-Helm (BioLogics, Hamburg, Germany) and 4-OHT was purchased from Sigma. 4-OHT releases MLL-ENL from a heat-shock protein complex thereby activating the fusion protein [21]. Note that MLL-ENL is essentially required for the continuous proliferation of this cell line.
Treatment with demethylating agents and inhibitors of histone deacetylases
Aza (Sigma) was used to verify the effect of methylation on expression of TSG. The HDAC inhibitor TSA (Sigma) was applied for testing the role of histone acetylation for TSG expression. Cells were seeded at a cell density of 5 × 105 cells/ml, Aza was added at a final concentration of 5 μM. Control cells were treated with 0.05% DMSO. After 2 d, half of the medium was replenished with medium with/without Aza (5 μM). TSA (2 μM final concentration) was added for the last 24 h of cultivation. After 4 d, cells were harvested to prepare RNA and protein.
Histone purification and Western blot analysis
Histones were purified according to the protocol of the supplier (Active Motif). Then, 3.3 μg or 10 μg protein were separated by electrophoresis (15% SDS gels) to detect histones and acetylated histones, respectively. Anti acetyl histone H4K12 antiserum was obtained from Biomol/Upstate (Hamburg, Germany), anti histone H4 monoclonal Ab was purchased from Abcam (Cambridge, United Kingdom). Specific bands on nitrocellulose membranes were visualized with the biotin/streptavidin-horseradish peroxidase system (Amersham, Freiburg, Germany) in combination with the "Renaissance Western Blot Chemoluminescence Reagent" protocol (DuPont, Bad Homburg, Germany).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
SR designed parts of the study and performed MS-MLPA, MSP analysis, sequencing of bisulfite-converted DNA and co-wrote the manuscript, WGD established MLPA, CM performed MLLwt and MLLmu analysis for primary AML samples, RM provided patient samples and gave good advice, MS performed knock-down experiments, RS provided MLL-ENL-ERtm mouse cell lines and gave good advice, AW established the spreadsheets for the evaluation of MS-MLPA experiments, HGD provided cell lines and critically read the manuscript, HQ designed the study and wrote the manuscript.
Supplementary Material
Methylation status of 24 TSG in 28 acute leukemia cell lines according to MS-MLPA. Shown is the promoter methylation status of TSG. Green: methylation ≥ 10%; yellow: methylation < 10%. TSG are arranged from left to right according to gene locus. For TSG CDKN2A and CDKN2B, ploidy status is indicated by numbers. Data shown are from one experiment per cell line. Reproducibility of MS-MLPA results was confirmed by up to four repetitions of analyses with selected cell lines.
MSP analyses of BEX2, IGSF4 and TIMP3 in AML cell lines. Methylation status of BEX2, IGSF4 and TIMP3 were determined in MLLwt and MLLmu AML cell lines by MSP to control methylation status as determined by MS-MLPA. Results for M- and U-PCR are shown as well as the methylation status according to MS-MLPA. Performance of MS-MLPA as a classification system for methylated or unmethylated TSG was evaluated using a confusion matrix. Overall, the results of the techniques were in good concordance with an accuracy of 0.83. In detail, accuracy was 0.86 for BEX2, 0.93 for IGSF4 and 0.71 for TIMP3.
Methylation analyses in primary AML samples. Results of promoter methylation analysis according to MSP of BEX2, IGSF4, RARB and TIMP3 are shown for MLLmu and MLLwt AML patients
Activating histone modifications induced by TSA. TSA effected acetylation of histone H4K12, as assessed by Western blot analysis. Aza treatment was not alone sufficient to induce histone H4K12 acetylation. MLLwt cell lines AML-193, SKM-1 and U-937 were treated with TSA (2 μM, 1 d), Aza (5 μM, 4 d) or a combination of both reagents.
Primers for methylation-specific PCR. Primers and reaction conditions for BSP and MSP are listed.
Acknowledgments
Acknowledgements
Acknowledgements: We thank Rod AF MacLeod for critically reading the manuscript. This study was in part supported by a grant from the Deutsche Krebshilfe (107819) to RM.
Contributor Information
Sonja Röhrs, Email: sro07@dsmz.de.
Wilhelm G Dirks, Email: wdi@dsmz.de.
Claus Meyer, Email: claus.meyer@em.uni-frankfurt.de.
Rolf Marschalek, Email: rolf.marschalek@em.uni-frankfurt.de.
Michaela Scherr, Email: M.Scherr@t-online.de.
Robert Slany, Email: rslany@biologie.uni-erlangen.de.
Andrew Wallace, Email: AndrewW@CMMC.nhs.uk.
Hans G Drexler, Email: hdr@dsmz.de.
Hilmar Quentmeier, Email: hqu@dsmz.de.
References
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Quentmeier H, Dirks WG, MacLeod RAF, Reinhardt J, Zaborski M, Drexler HG. Expression of HOX genes in acute leukemia cell lines with and without MLL translocations. Leukemia Lymphoma. 2004;45:567–574. doi: 10.1080/10428190310001609942. [DOI] [PubMed] [Google Scholar]
- Quentmeier H, Tonelli R, Geffers R, Pession A, Uphoff CC, Drexler HG. Expression of BEX1 in acute myeloid leukemia with MLL rearrangements. Leukemia. 2005;19:1488–1489. doi: 10.1038/sj.leu.2403820. [DOI] [PubMed] [Google Scholar]
- Yang QS, Xia F, Gu SH, Yuan HL, Chen JZ, Yang QS, Ying K, Xie Y, Mao YM. Cloning and expression pattern of a spermatogenesis-related gene, BEX1, mapped to chromosome Xq22. Biochem Genet. 2002;40:1–12. doi: 10.1023/A:1014565320998. [DOI] [PubMed] [Google Scholar]
- Fischer C, Drexler HG, Reinhardt J, Zaborski M, Quentmeier H. Epigenetic regulation of brain expressed X-linked-2, a marker for acute myeloid leukemia with mixed lineage leukemia rearrangements. Leukemia. 2007;21:374–377. doi: 10.1038/sj.leu.2404493. [DOI] [PubMed] [Google Scholar]
- Foltz G, Ryu GY, Yoon JG, Nelson T, Fahey J, Frakes A, Lee H, Field L, Zander K, Sibenaller Z, Ryken TC, Vibhakar R, Hood L, Madan A. Genome-wide analysis of epigenetic silencing identifies BEX1 and BEX2 as candidate tumor suppressor genes in malignant glioma. Cancer Res. 2006;66:6665–6674. doi: 10.1158/0008-5472.CAN-05-4453. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Huang HJS, Petrelli NJ, Zhang X, O'Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;25:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- Esteller M. Profiling aberrant DNA methylation in hematologic neoplasms: a view from the tip of the iceberg. Clin Immunol. 2003;109:80–88. doi: 10.1016/S1521-6616(03)00208-0. [DOI] [PubMed] [Google Scholar]
- Rahmatpanah FB, Carstens S, Guo J, Sjahutera O, Taylor KH, Duff D, Shi H, Davis JW, Hooshmand SI, Chitma-Matsiga R, Caldwell CW. Differential DNA methylation patterns of small B-cell lymphoma subclasses with different clinical behavior. Leukemia. 2006;10:1855–1862. doi: 10.1038/sj.leu.2404345. [DOI] [PubMed] [Google Scholar]
- Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, Taylor KH, Sjahputera O, Andresi M, Wooldridge JE, Caldwell CW. Discovery of novel epigenetic markers in non-Hodgkin's lymphoma. Carcinogenesis. 2007;28:60–70. doi: 10.1093/carcin/bgl092. [DOI] [PubMed] [Google Scholar]
- Smiraglia DJ, Rush LJ, Frühwald MC, Dai Z, Held WA, Costello JF, Lang JC, Eng C, Li B, Wright FA, Caligiuri MA, Plass C. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Human Molecular Genetics. 2001;10:1413–1419. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- Ueki T, Walter KM, Skinner H, Jaffee E, Hruban RH, Goggins M. Aberrant CpG island methylation in cancer cell lines arises in the primary cancers from which they were derived. Oncogene. 2002;21:2114–2117. doi: 10.1038/sj.onc.1205275. [DOI] [PubMed] [Google Scholar]
- Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, Esteller M. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- Hess CJ, Errami A, Berkhof J, Denkers F, Ossenkoppele GJ, Nygren AO, Schuurhuis GJ, Waisfisz Q. Concurrent methylation of promoters from tumor associated genes predicts outcome in acute myeloid leukemia. Leukemia Lymphoma. 2008;49:1132–1141. doi: 10.1080/10428190802035990. [DOI] [PubMed] [Google Scholar]
- Ekmekci CG, Gutierrez MI, Siraj AK, Ozbek U, Bhatia K. Aberrant methylation of multiple tumor suppressor genes in acute myeloid leukemia. Am J Hematol. 2004;77:233–240. doi: 10.1002/ajh.20186. [DOI] [PubMed] [Google Scholar]
- Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437X(95)80044-1. [DOI] [PubMed] [Google Scholar]
- Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- Glasow A, Barrett A, Petrie K, Guptka R, Boix-Chornet M, Zhou DC, Grimwade D, Gallagher R, von Lindern M, Waxman S, Enver T, Hildebrandt G, Zelent A. DNA methylation-independent loss of RARA gene expression in acute myeloid leukemia. Blood. 2008;111:2374–2377. doi: 10.1182/blood-2007-05-088344. [DOI] [PubMed] [Google Scholar]
- Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, Borkhardt A, Chanda SK, Walker J, Soden R, Hess JL, Slany RK. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24:617–628. doi: 10.1128/MCB.24.2.617-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/S1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Birke M, Schreiner S, Garcia-Cuellar MP, Mahr K, Titgemeyer F, Slany RK. The MT domain of the proto-oncoprotein MLL binds to CpG-containing DNA and discriminates against methylation. Nucleic Acids Research. 2002;30:958–965. doi: 10.1093/nar/30.4.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caslini C, Yang Z, El-Osta M, Milne TA, Slany R, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67:7275–7283. doi: 10.1158/0008-5472.CAN-06-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Hughes CM, Lloyd R, Yang Z, Rozenblatt-Rosen O, Dou Y, Schnepp RW, Krankel C, Livolsi VA, Gibbs D, Hua X, Roeder RG, Meyerson M, Hess JL. Menin and MLL cooperatively regulated expression of cyclin-dependent kinase inhibitors. Proc Natl Acad Sci USA. 2005;102:749–754. doi: 10.1073/pnas.0408836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chavalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci USA. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HG. Guide to Leukemia-Lymphoma Cell Lines Braunschweig. 2005.
- Drexler HG, Quentmeier H, MacLeod RAF. Malignant hematopoietic cell lines: in vitro models for the study of MLL gene alterations. Leukemia. 2004;18:227–232. doi: 10.1038/sj.leu.2403236. [DOI] [PubMed] [Google Scholar]
- Meyer C, Schneider B, Reichel M, Angermüller S, Strehl S, Schnittger S, Schoch C, Jansen MW, van Dongen JJ, Pieters R. Diagnostic tool for the identification of MLL rearrangements including unknown partner genes. Proc Natl Acad Sci USA. 2005;102:449–454. doi: 10.1073/pnas.0406994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz O, Hess CJ, Schouten JP, Errami A. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;33:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methylation status of 24 TSG in 28 acute leukemia cell lines according to MS-MLPA. Shown is the promoter methylation status of TSG. Green: methylation ≥ 10%; yellow: methylation < 10%. TSG are arranged from left to right according to gene locus. For TSG CDKN2A and CDKN2B, ploidy status is indicated by numbers. Data shown are from one experiment per cell line. Reproducibility of MS-MLPA results was confirmed by up to four repetitions of analyses with selected cell lines.
MSP analyses of BEX2, IGSF4 and TIMP3 in AML cell lines. Methylation status of BEX2, IGSF4 and TIMP3 were determined in MLLwt and MLLmu AML cell lines by MSP to control methylation status as determined by MS-MLPA. Results for M- and U-PCR are shown as well as the methylation status according to MS-MLPA. Performance of MS-MLPA as a classification system for methylated or unmethylated TSG was evaluated using a confusion matrix. Overall, the results of the techniques were in good concordance with an accuracy of 0.83. In detail, accuracy was 0.86 for BEX2, 0.93 for IGSF4 and 0.71 for TIMP3.
Methylation analyses in primary AML samples. Results of promoter methylation analysis according to MSP of BEX2, IGSF4, RARB and TIMP3 are shown for MLLmu and MLLwt AML patients
Activating histone modifications induced by TSA. TSA effected acetylation of histone H4K12, as assessed by Western blot analysis. Aza treatment was not alone sufficient to induce histone H4K12 acetylation. MLLwt cell lines AML-193, SKM-1 and U-937 were treated with TSA (2 μM, 1 d), Aza (5 μM, 4 d) or a combination of both reagents.
Primers for methylation-specific PCR. Primers and reaction conditions for BSP and MSP are listed.