Abstract
Background
Accurate computational identification of cis-regulatory motifs is difficult, particularly in eukaryotic promoters, which typically contain multiple short and degenerate DNA sequences bound by several interacting factors. Enrichment in combinations of rare motifs in the promoter sequence of functionally or evolutionarily related genes among several species is an indicator of conserved transcriptional regulatory mechanisms. This provides a basis for the computational identification of cis-regulatory motifs.
Results
We have used a discriminative seeding DNA motif discovery algorithm for an in-depth analysis of 54 seed storage protein (SSP) gene promoters from three plant families, namely Brassicaceae (mustards), Fabaceae (legumes) and Poaceae (grasses) using backgrounds based on complete sets of promoters from a representative species in each family, namely Arabidopsis (Arabidopsis thaliana (L.) Heynh.), soybean (Glycine max (L.) Merr.) and rice (Oryza sativa L.) respectively. We have identified three conserved motifs (two RY-like and one ACGT-like) in Brassicaceae and Fabaceae SSP gene promoters that are similar to experimentally characterized seed-specific cis-regulatory elements. Fabaceae SSP gene promoter sequences are also enriched in a novel, seed-specific E2Fb-like motif. Conserved motifs identified in Poaceae SSP gene promoters include a GCN4-like motif, two prolamin-box-like motifs and an Skn-1-like motif. Evidence of the presence of a variant of the TATA-box is found in the SSP gene promoters from the three plant families. Motifs discovered in SSP gene promoters were used to score whole-genome sets of promoters from Arabidopsis, soybean and rice. The highest-scoring promoters are associated with genes coding for different subunits or precursors of seed storage proteins.
Conclusion
Seed storage protein gene promoter motifs are conserved in diverse species, and different plant families are characterized by a distinct combination of conserved motifs. The majority of discovered motifs match experimentally characterized cis-regulatory elements. These results provide a good starting point for further experimental analysis of plant seed-specific promoters and our methodology can be used to unravel more transcriptional regulatory mechanisms in plants and other eukaryotes.
Background
Designing expression cassettes allowing a precise control of where, when and at which level transcription should occur may ultimately be achieved through synthetic promoter engineering [1]. The basic building blocks for such promoters are regions of cis-regulatory DNA, which in eukaryotes often comprise clusters of cis-regulatory elements (CREs) (called composite motifs, or modules) bound by a combination of transcription factors (TFs). The unraveling of eukaryotic transcriptional regulation is a challenging area of research driving the synergetic development of experimental and computational techniques [2]. Cis-regulatory motifs of plant promoters have commonly been delineated by the experimental manipulation of DNA segments and reporter gene expression assays [3]. Plant cis-regulatory motifs are often reported as consensus sequences, a motif model of limited predictive power [4]. Collections of experimentally characterized plant cis-regulatory elements sequences such as the PLACE database [5] nevertheless remain an invaluable resource e.g. for annotating motifs discovered in sequences that have not been characterized experimentally. The majority of contemporary computational approaches for the discovery of cis-regulatory elements [6] use the position weight matrix (PWM) motif model, based on the frequencies of nucleotides at each position in a collection of regulatory elements. The Seeder DNA motif discovery algorithm, designed for fast and reliable prediction of cis-regulatory elements in eukaryotic promoters, uses a string-based approach to identify motifs that are statistically significant (enriched) in a set of positive sequences as compared to a background set of sequences and it was recently shown to outperform some popular motif discovery tools on biological benchmark data [7].
The maturation of plant seeds, and more specifically protein storage in seeds, is regulated by a combination of hormonal, genetic and metabolic controls [8]. In Arabidopsis, four master regulators of seed maturation have been identified including three TFs of the B3 DNA-binding domain family, namely ABSCISIC ACID INSENSITIVE3 (ABI3), FUSCA3 (FUS3) and LEAFY COTYLEDON2 (LEC2), and a HAP3 subunit of the CCAAT-box binding transcription factor (LEC1) [8-10]. Known dicotyledonous seed maturation regulatory motifs include the RY motif and the ACGT motif, which are targets of B3 and bZIP transcription factors respectively [11]. In rapeseed (Brassica napus L.), a comprehensive analysis of the napA promoter revealed the presence of two regulatory element complexes, the B-box which contains the distB element (GCCACTTGTC) together with the proxB element (TCAAACACC), and the RY/G complex which contains two RY repeats (CATGCA) and one G-box (CACGTG) [12-14]. In bean (Phaseolus vulgaris L.), a comprehensive promoter analysis was performed on the phas promoter by Chandrasekharan et al. [15]. The site-directed substitution mutations analysis within the -295 region of the phas promoter revealed that the G-box, the CCAAAT box, the E-box (CACCGT) and RY elements mediate levels of expression in embryos [15]. Several studies have shown that motifs conferring seed-specific expression reside in the proximal region of the promoter, often within 500 bp upstream of the transcriptional start [e.g. [15-18]]. The analysis of prolamin gene promoters from barley (Hordeum vulgare L.), wheat (Triticum aestivum L.) and maize (Zea mays L.) uncovered a conserved ~30 base pairs (bp) conserved sequence containing two CREs, the GCN4-like (GLM) element (GRTGAGTCAT) (see [19] for the nomenclature of incompletely specified bases), and the prolamin-box (also referred to as the endosperm element) (TGTAAAGT) [20]. An additional element called AACA (AACAAACTCTATC) was further found to be involved in the seed-specific regulation of rice (Oryza sativa L.) glutelin genes [21]. These three CREs (GLM, P-box and AACA) are frequently found in monocotyledonous SSP gene promoters and are bound by TFs of the bZIP, DOF and MYB families, respectively [11].
In this work, we performed de novo motif discovery in 54 SSP gene promoters from Brassicaceae, Fabaceae and Poaceae using discriminative seeding DNA motif discovery, and uncovered the presence of family-specific conserved motifs, the validity of which was corroborated by matching to experimentally characterized plant seed-specific CREs. Furthermore, we show that the discovered motifs constitute signatures of SSP gene promoters in the different species.
Results
Seed storage protein gene promoters contain conserved motifs
Seed storage protein gene promoter sequences (the 500 bp upstream region of the transcriptional start) from Brassicaceae (15 promoters), Fabaceae (17 promoters) and Poaceae (22 promoters) were retrieved from public sequence databases. Discriminative seeding DNA motif discovery [7] was performed separately in each of the three plant families using a background model based on the complete set of promoters from a representative species, namely Arabidopsis (27,234 sequences), soybean (66,155 sequences) and rice (41,019 sequences). Statistically significant conserved cis-regulatory motifs (q-value < 0.05) were identified in SSP gene promoter sequences within each plant family. Discovered motifs were matched to consensus sequences of experimentally characterized plant cis-regulatory elements from the PLACE database [5] using the STAMP suite of tools [22] (Table 1).
Table 1.
DNA motifs discovered in the promoters of plant seed-storage protein genes
| Plant family | Motif ID | q-value | PLACE ID | STAMP alignment | E value |
| Brassicaceae | B1 | 1.60e-07 | RYREPEATBNNAPA |
MKCCATGCAAAN ---CATGCA--- |
5.02-08 |
| B2 | 5.45e-04 | GADOWNAT |
AYKTGTCACYCY ACGTGTC----- |
6.86e-08 | |
| B3 | 1.84e-02 | RYREPEATBNNAPA |
NYWCATGCANNY ---CATGCA--- |
9.68e-08 | |
| Fabaceae | F1 | 1.43e-07 | LEGUMINBOXLEGA5 |
NNRCCATGCATR TAGCCATGCAWR |
4.73e-12 |
| F2 | 2.06-03 | RYREPEATLEGUMINBOX |
RNNCATGCANNN ---CATGCAY-- |
1.05e-09 | |
| F3 | 7.24e-03 | TATABOX1 |
TMNCTATAAATA ---CTATAAATA |
1.58e-12 | |
| F4 | 9.05e-03 | E2FBNTRNR |
KAMGCGGCNAMN ---GCGGCAAA- |
9.03e-05 | |
| F5 | 4.53e-02 | ACGTSEED2 |
NSACWCNTCMWY ACACACGTCAA- |
1.32e-08 | |
| Poaceae | P1 | 6.84e-08 | GLMHVCHORD |
KRTGAGTCATNN -RTGASTCAT-- |
1.52e-13 |
| P2 | 2.17e-05 | PROLAMINBOXOSGLUB1 |
ANNTTGCAAAMN ----TGCAAAG- |
4.41e-06 | |
| P3 | 1.85e-04 | EMHVCHORD |
NYRTAAAGTNNW -TGTAAAGT--- |
6.45e-11 | |
| P4 | 2.94e-03 | TATABOX1 |
NANCTATAAAWR ---CTATAAATA |
6.12e-10 | |
| P5 | 9.17e-03 | BIHD1OS |
KNTTGTCATNTW ---TGTCA---- |
6.65e-06 | |
| P6 | 1.14e-02 | GCAACREPEATZMZEIN |
NMWAAAGCAANN -GCAACGCAAC- |
5.47e-03 | |
| P7 | 2.82e-02 | O2F3BE2S1 |
WNNACATRCWWR TCCACGTACT-- |
1.55e-05 |
q-value, statistical significance of motif
PLACE ID, identifier of PLACE consensus sequence matching motif
STAMP alignment, alignment of motif consensus sequence (top) with PLACE consensus sequence (bottom)
E value, expectation value of the STAMP alignment
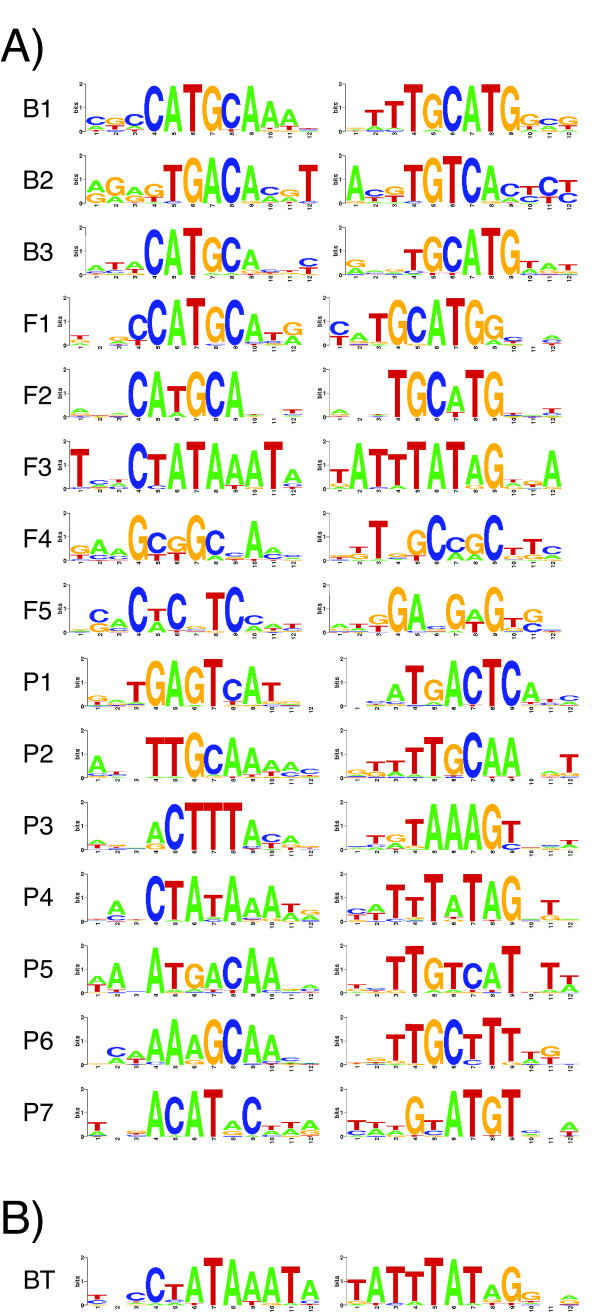
Figure 1A shows sequence logos of the significant motifs enriched in SSP gene promoters from Brassicaceae (B1-B3), Fabaceae (F1-F5), and Poaceae (P1-P7). Three motifs were statistically significant (q-value ≤ 0.05) in the Brassicaceae SSP gene promoters, corresponding to two RY-like motifs and one ACGT-like motif (motifs B1-B3).
Figure 1.
Sequence logos of motifs enriched in seed storage protein gene promoter sequences. A) Sequence logos of significant DNA motifs discovered in SSP gene promoter sequence from Brassicaceae (B1-3), Fabaceae (F1-5) and Poaceae (P1-P7). B) Sequence logos of the TATA-box motif identified in Brassicaceae SSP gene promoter sequences. Left, forward motif, right, reverse complement of motif.
Five significant motifs were found in the Fabaceae SSP gene promoters, including two RY-like motifs and one ACGT-like motif (motifs F1, F2, F5). Motif F3 is a TATA-box motif and is discussed below. The fourth motif discovered (motif F4) is possibly related to the E2Fb motif (GCGGCAAA) found in the tobacco (Nicotiana tabacum L.) ribonucleotide reductase 2 (RNR2) gene promoter [23]. The Fabaceae E2Fb-like motif (motif F4) does not have similarity to any known plant seed-specific cis-regulatory elements; it is thus a novel putative SSP gene promoter cis-regulatory motif.
Motifs enriched in the promoters of Poaceae SSP genes (seven significant motifs) are distinct from those observed in the two other plant families. The first motif discovered (motif P1) is most similar to the GCN4-like motif (GLM). The second motif (motif P2) is similar to a variant of the prolamin-box motif (TGCAAAG) found in a rice glutelin promoter [18]. This sequence has also been suggested to act as a prolamin-box variant in a wheat glutenin promoter [24]. The third motif (motif P3) is a strong match to the typical prolamin-box (TGTAAAGT). Motif P4 is a TATA-box motif and is discussed below. The fifth motif (motif P5) has some core similarity with a rice BELL homeodomain transcription factor binding site [25]. It is also similar to an Skn-1-like motif identified in a rice glutelin gene promoter [26]. Motif P6 is related to the GCAA motif found in a maize zein promoter [27]. Motif P7 does not have similarity to any known monocotyledonous seed promoter motif but is weakly related to an opaque-2 recognition site [28].
Seed storage protein gene promoters contain TATA-box motifs
The third motif discovered in Fabaceae (motif F3), and the fourth motif discovered in Poaceae SSP gene promoters (motif P4), are highly similar to a TATA-box motif (CTATAAATA). In Fabaceae SSP gene promoters, the best matching subsequences to the TATA-box motif (motif F3) are localized between positions -20 to -30 upstream of the transcription start site (interquartile range of 7.0 bp). No TATA-box motif was initially discovered in Brassicacea SSP gene promoters. To investigate whether Brassicaceae SSP gene promoters also contain a TATA-box motif, we searched the Brassicaceae promoter sequences with the TATA-box motif found in Fabaceae (motif F4). Scoring promoter sequences with the F4 motif's PWM returned a highly similar TATA-box motif (Figure 1B, motif BT). In both Brassicaceae and Fabaceae, most best matching subsequences to the TATA-box motif are also localized approximately -20 to -30 upstream of the transcriptional start (Figure 2).
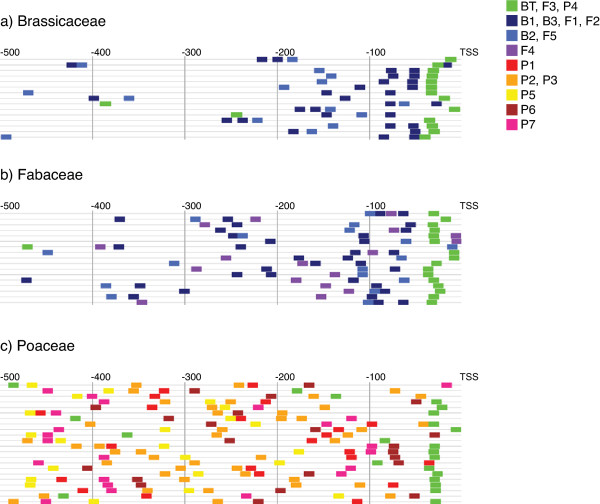
Figure 2.
Position of cis-regulatory motifs on seed storage protein gene promoter sequences. The positions of the best matching subsequence to motifs discovered in SSP gene promoters from (a) Brassicaceae, (b) Fabaceae and (c) Poaceae are mapped onto promoter sequences. (Motifs in Brassicaceae (B1-3), Fabaceae (F1-5) and Poaceae (P1-P7), respectively).
Some seed storage regulatory motifs are highly localized
The position of the best matching subsequences to discovered motifs (putative CREs) in promoter sequences, identified by the Seeder algorithm [7], is illustrated in Figure 2. The distribution of best matching subsequence positions (deciles) is represented in Additional file 1. Several patterns emerge from this map: (i) the TATA-box motif is highly localized to positions approx. between -20 to -30 upstream of the transcriptional start in Brassicaceae, Fabaceae and Poaceae SSP promoters; (ii) Brassicaceae and Fabaceae SSP promoters have one RY motif localized in close proximity upstream of the TATA-box, and one additional RY motif and one ACGT motif at variable position upstream of the TATA-box; (iii) Poaceae SSP promoters are characterized by one GLM, two P-box, one Skn-1 and one GCAA motifs scattered at variable positions upstream of the transcriptional start.
The combination of Fabaceae seed storage motifs is a signature of seed storage protein gene promoters in the soybean genome
The recently sequenced soybean genome is predicted to contain over 65,000 protein-coding genes (Soybean Genome Project, DoE Joint Genome Institute http://www.phytozome.net/soybean). This publicly available genome sequence set was used to retrieve 66,155 promoter sequences. We used the Fabaceae PWMs (F1-5) to identify the best matching promoter sequences from the soybean genome by a PWM scoring and sequence matching strategy. In order to assign a function to the genes whose promoters were enriched in these five motifs, we manually annotated the top-ten matching gene sequences from the genome. The translated gene sequences corresponding to the top-ten scoring promoters were aligned with the Swiss-Prot database (plant sequences) using the Smith-Waterman algorithm. All of the top-scoring promoters are associated with soybean genes coding for different subunits of glycinin, β-conglycinin or 7S globulin (Table 2). Similar results were obtained in Arabidopsis and rice (Additional file 2), where eight out of the top-ten scoring Arabidopsis promoters are associated with SSP genes and the top-ten scoring rice promoters are all associated with SSP genes.
Table 2.
Top-ten scoring soybean promoters for the presence of Fabaceae seed-storage protein gene promoter motifs
| Gene ID | PWM rank | Hit id | Hit description | E value |
| Glyma03 g32030.1 | 1 | P04776 | Glycinin Gy1 | 0.0 |
| Glyma19 g34780.1 | 2 | P11828 | Glycinin Gy3 | 0.0 |
| Glyma20 g28650.2 | 3 | P13916 | β-conglycinin, alpha chain | 0.0 |
| Glyma03 g32020.2 | 4 | P04405 | Glycinin Gy2 | 2.0e-251 |
| Glyma10 g04280.1 | 5 | P02858 | Glycinin Gy4 | 0.0 |
| Glyma13 g18450.1 | 6 | P04347 | Glycinin Gy5 | 3.0e-285 |
| Glyma10 g39150.1 | 7 | P11827 | Beta-conglycinin, alpha' chain | 3.0e-105 |
| Glyma20 g28640.1 | 8 | P25974 | β-conglycinin, beta chain | 7.0e-298 |
| Glyma20 g28460.2 | 9 | P25974 | β-conglycinin, beta chain | 1.0e-269 |
| Glyma03 g39940.1 | 10 | P13917 | Basic 7S globulin 1 | 7.0e-302 |
Gene ID, gene identifier (soybean genome assembly and annotation Glyma1)
PWM rank, total PWM matching score rank
C-B rank, total Cluster-Buster score rank
Hit ID, hit identifier (Uniprot/Swiss-Prot)
Description, hit description
E value, hit alignment expectation value.
The promoters of soybean genes coding for different seed storage protein subunits vary in motif composition
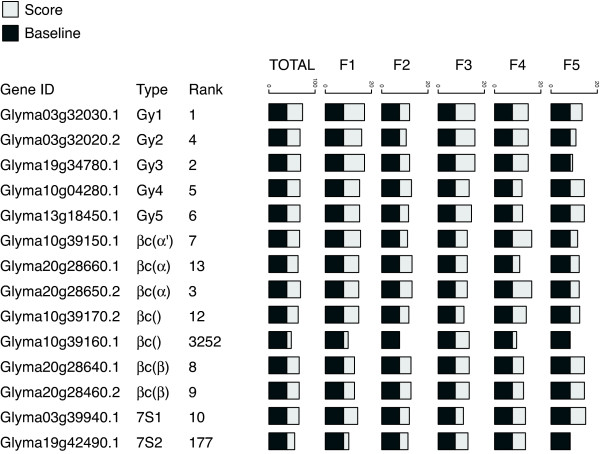
Although genes coding for different soybean SSP subunits have been shown to be expressed specifically in seeds during maturation, some subunits are differentially expressed (in cotyledons vs. embryonic axes; at different time points) [29]. We investigated whether there were also differences in promoter motif composition. Soybean major SSP sequences from the Swiss-Prot database, namely glycinin subunits (Gy1-Gy5) [30], β-conglycinin subunits (α,α', β) [31] and basic 7S globulins [32], were aligned against all soybean predicted peptides (from the genome sequence). We identified 12 soybean peptide sequences with high similarity (percent identity over the alignment > 0.90, expected value < 1.0e-250), and an additional two sequences with moderate similarity (percent identity over the alignment > 0.50, expected value < 1.0e-50). Figure 3 shows the PWM scores for each Fabaceae SSP promoter motif in soybean SSP gene promoters compared with a baseline (the mean score of all 66,155 soybean promoters). The promoters of genes coding respectively for glycinins Gy1 (Glyma03 g32030.1), Gy2 (Glyma03 g32020.2), Gy3 (Glyma19 g34780.1), Gy4 (Glyma10 g04280.1) and Gy5 (Glyma13 g18450.1) scored relatively high (ranks 1, 4, 2, 5 and 6) for the presence of Fabaceae SSP gene promoter motifs. The promoters of all genes coding for the β-conglycinin subunits, namely α' (Glyma10 g39150.1), α (Glyma20 g28660.1, Glyma20 g28650.2) and β (Glyma20 g28640.1, Glyma20 g28460.2) were among the top-15 scoring promoters (ranks 7, 13, 3, 8, 9) out of the 66,155 soybean promoters. The promoters of the gene coding for the basic 7S globulin 1 (Glyma03 g39940.1) was also among the top-ten promoters (rank 10), while that of the gene coding for the basic 7S globulin 2 (Glyma19 g42490.1) scored lower (rank 177). The products of two genes flanking gene Glyma10 g39150.1 on chromosome 10 (Glyma10 g39160.1, Glyma10 g39170.2) are equivalently good matches to the three β-conglycinin subunits (α, α' and β) (percent identity > 0.50, expected value < 1.0e-50), making a precise annotation difficult for those two genes. Interestingly, the promoter of Glyma10 g39160.1 scored very low (rank 3,252) while that of Glyma10 g39170.2 was among the top-15 scoring promoters (rank 12).
Figure 3.
PWM score and rank of Fabaceae SSP gene promoter motifs in 14 soybean SSP gene promoters. The PWM matrix score associated with Fabaceae SSP gene promoter motifs in 14 soybean SSP gene promoters is compared to the average score obtained in 66,155 soybean promoters (baseline). Gy (1-5), glycinin subunit (1-5); βc (α', α, β), β-conglycinin subunit (α', α, β).
Discussion
We have applied the Seeder discriminative DNA motif discovery algorithm to an in-depth analysis of SSP gene promoters from Brassicaceae, Fabaceae and Poaceae. Most discovered motifs match experimentally characterized cis-regulatory element consensus sequences, which strongly supports the validity of the discovered motifs.
The analysis of Brassicaceae SSP gene promoters highlighted the presence of three significant motifs corresponding to two RY motifs and one ACGT motif. It is interesting to contrast this result with that obtained from the analysis of promoters of Arabidopsis seed-specific marker genes where one RY motif and one ACGT motif were significantly enriched [7]. The three motifs match components of the RY/G complex experimentally characterized in the rapeseed napA promoter [12]. The analysis of Brassicaceae SSP gene promoter sequences using the Seeder algorithm did not initially reveal enrichment in a TATA-box motif. This could be explained by the proportion of promoters containing a TATA-box in the background set of sequences, or by the relatively low complexity of TATA-box motifs which makes them hard to discriminate from background, particularly if we take into account the fact that promoter sequences are generally A/T rich [33]. We used a PWM corresponding to a putative Fabaceae TATA-box motif to retrieve, in Brassicaceae SSP gene promoter sequences, a motif highly localized around position -20 to -30 relative to the transcriptional start site. The localization, the information content of the motif and the fact that it is very similar to TATA-box motifs found in Fabaceae and Poaceae SSP gene promoters suggest that this motif indeed corresponds to a Brassicaceae SSP gene promoter TATA-box motif, in accordance with reported occurrences of TATA-box motifs in the promoters of e.g. napA and napB [34,35].
Fabaceae SSP gene promoters have also revealed enrichment in two RY motifs. The RY motif has long been known to be conserved in legume seed-protein gene promoters [36] and RY CREs have been proven to be functional e.g. in soybean [37,38] and broad bean (Vicia faba L.) [39]. A novel, E2Fb-like motif was discovered in Fabaceae SSP gene promoters. E2F transcription factors are involved in the control of cell cycle [40]. The role of this E2Fb-like motif in seed-specific gene expression will require further experimental verification.
Position weight matrices corresponding to motifs discovered in Fabaceae, Arabidopsis and rice SSP gene promoters were used to score the respective whole genome sets of promoter sequences. The top-ten scoring promoters are associated with SSP-coding genes in soybean and rice, as are eight out of the top-ten scoring promoters in Arabidopsis. This combination of a few motifs is thus sufficient to constitute a signature of SSP gene promoters. The fact that the promoter of some soybean genes coding for SSP protein subunits did score relatively low to the combination of Fabaceae SSP gene promoter motifs may indicate alternative regulatory mechanisms for those genes. Furthermore, the promoters of other soybean SSP protein genes such as those coding for albumin-1 (Glyma13 g26330.1, Glyma13 g26340.1) and 2S albumin (Glyma12 g34160.1, Glyma13 g36400.1) did also score relatively low (data not shown) and could be regulated by a different set of TFs.
In soybean, experimental SSP gene promoter analyses have focused on the α' and β subunits of β-conglycinin [17,41-45]. Experimental analyses have revealed the importance of the proximal region (~250 bp upstream of the transcription start site) and the presence of several factors binding the promoters (soybean embryo factors SEF) and the presence of a RY cis-regulatory element. The study by Fujiwara and Beachy [38] disproved a cis-regulatory role for the binding sites of SEF3 and SEF4 located within the proximal promoter and confirmed the role of the RY element in seed-specific gene regulation. The work by Yoshino et al. [46,47] on the promoter of the α subunit of β-conglycinin also suggests a role for RY elements in seed-specific gene regulation. The promoters of genes coding for glycinin subunits Gy2 and Gy3 have also been analyzed experimentally [37,48,49] yet although an A/T-rich SEF-binding sequence has been identified, the only clearly confirmed cis-regulatory element therein is a RY element. Our results suggest that soybean SSP promoters may be characterized by four cis-regulatory motifs, in addition to a TATA-box motif.
Motifs enriched in the promoters of Poaceae SSP genes were all good matches to experimentally characterized plant seed-specific cis-regulatory elements including a GLM motif, two prolamin-box-like motifs, a Skn-1-like motif and a TATA-box motif. A recent study [50] has identified a barley protein homologous to the Arabidopsis FUSCA3 that regulates SSP genes and binds RY boxes; this was the first report of a possible implication of the RY motif in seed-specific gene regulation in a monocotyledonous plant species. Our computational analysis did not reveal significant enrichment in RY motifs among Poaceae SSP gene promoters. This however does not necessarily refute a possible role for B3-type transcription factors and RY-like elements in the transcriptional regulation of some Poaceae SSP genes, which could be an attribute of a limited number of genes only, and not a general feature of Poaceae SSP gene promoters.
On the other hand, motifs containing the AAAG core of Dof transcription factor binding sites [51] were found only in Poaceae SSP gene promoters. Soybean Dof-type transcription factor have been reported to be involved in the regulation of the lipid content in soybean seeds [52], and a prolamin-box motif has been reported in pea (Pisum sativum L.) [53]. However, prolamin-box motifs have been reported mostly in Poaceae promoters [e.g. [18,20,24,52,54-56]]. Indeed, our results suggest that prolamin-box-like motifs are conserved in Poaceae SSP gene promoters, but are not featured in Brassicaceae or Fabaceae SSP gene promoters.
Conclusion
Presented results highlight motifs that are conserved in SSP gene promoters within three plant families. Promoter/motif combinations generated in this analysis can be further validated experimentally, e.g. in a framework such as that used by [15]. Most motifs conserved in SSP gene promoters have a high degree of similarity with experimentally characterized cis-regulatory elements; this is an indicator that they are indeed functional in seed-specific gene regulation. The same methodology can be applied to analyze various data sets and decipher transcriptional regulation mechanisms in plants and other eukaryotes.
Methods
Sequence data collection
The Uniprot database [57] release 14.6 was parsed using Bioperl [58] and a total of 233 plant SSP were retrieved (annotated as seed storage protein in description or keywords). Those records were matched to 230 UniRef100 entries [59]. Database references (EMBL) were used to retrieve a maximum of one promoter (500 bp upstream of the transcriptional start) per UniRef100 cluster using the BioPerl toolkit [58]. Transcriptional start positions were retrieved from The Arabidopsis Information Resource website http://www.arabidopsis.org and the Rice Genome Annotation Project http://rice.plantbiology.msu.edu website for Arabidopsis and rice respectively. In other species, the transcriptional start positions were retrieved in the literature [20,35,60-77]. The transcription start sites were predicted in 13 promoters for which transcriptional start data was unavailable in GenBank or literature, using the TSSP software from Softberry Inc. http://www.softberry.ru. One representative sequence among sequences with percentage identity > 0.90 over clustalw alignment [78] was selected for further analysis. This process returned 15 Brassicaceae SSP gene promoter sequences, 17 Fabaceae SSP gene promoter sequences and 22 Poaceae SSP gene promoter sequences (listed in Additional file 3). Background sets of promoter sequences (500 bp upstream of annotated mRNAs) from Arabidopsis, soybean and rice sequences were retrieved using BioPerl and genome annotation data available for each species in generic feature format (GFF). A set of 27,234 promoters Arabidopsis protein-coding gene promoters were retrieved using The Arabidopsis Information Resource release 8 (TAIR8) http://www.arabidopsis.org. A set of 66,155 predicted soybean promoters were retrieved using the Glyma1.0 chromosome-scale assembly and genome annotation (Soybean Genome Project, DoE Joint Genome Institute) http://www.phytozome.net/soybean. A set of 41,019 rice (Oryza sativa) promoters was retrieved using the rice genome assembly and annotation release 5.0 http://rice.plantbiology.msu.edu.
Computation of background distributions and motifs
For all sequence species, background SMD distributions were computed using a seed length of six and matches on both strands [7]. For motif discovery in Brassicaceae, we used a background model based on Arabidopsis promoters, for Fabaceae we used a background model based on soybean promoters, and for motif discovery in Poaceae we used a background model based on rice promoters. Background models were computed using the Seeder::Background perl module [7]. The Seeder algorithm was used to perform motif discovery in SSP gene promoters using a seed-length of six and a motif length of 12. The top-five motifs were compared to known plant motifs in the PLACE database (Higo, et al., 1998) using the STAMP web server (Mahony and Benos, 2007). For each group of promoters, quartiles and deciles for the motif positions were computed using a custom perl script implementing the median-unbiased estimator algorithm [79].
Scoring of soybean promoter sequences
Scoring of the three promoter sets from soybean, Arabidopsis and rice was performed using PWMs as follow: for each given promoter, for a given PWM (in descending order of significance), each (unmasked) position is scored [80], and the position at which the score is maximum is masked; the process is repeated for each motif. Individual scores (for each motif) and the total score (for all motifs) are reported for each promoter sequence.
Annotation of soybean genes
Smith-Waterman alignments of the soybean predicted peptides corresponding to the top-ten scoring promoters was performed against the Uniprot release 14.6 (plant sequences) using a TimeLogic DeCypher system (Active Motif, Inc., 1914 Palomar Oaks Way, Suite 150, Carlsbad, CA. 92008) with BLOSUM62 scoring matrix, gap opening penalty -12, gap extension penalty -2 and an E value threshold of 1e-5. The top-scoring protein from Uniprot was reported for each soybean predicted peptide. For retrieving soybean genes corresponding to a reference set of soybean SSP [Swiss-Prot:P04776, Swiss-Prot:P04405, Swiss-Prot:P11828, Swiss-Prot:P02858, Swiss-Prot:P04347, Swiss-Prot:P11827, Swiss-Prot:P13916, Swiss-Prot:P13916, Swiss-Prot:P25974, Swiss-Prot:P25974, Swiss-Prot:P13917, Swiss-Prot:P13917, Swiss-Prot:Q8RVH5, Swiss-Prot:Q8RVH5], alignment against all soybean predicted peptides (66,210 sequences) was performed. For each reference sequence, the soybean predicted peptide among hits with significance < 1e-100 and percent identities > 90% over the alignment maximizing the alignment score was attributed as best match.
Authors' contributions
FF and MVS designed the study. FF performed programming and data analysis. MVS supervised the project. Both authors have participated in writing the manuscript and have read and approved the final version.
Supplementary Material
Minimum, maximum and sample deciles for the position of SSP gene promoter motifs. The minimum, maximum and deciles of positions of best matching subsequences to motifs discovered in Brassicaceae (B1-3, BT), Fabaceae (F1-5, FT), and Poaceae (P1-5) are plotted on an axis corresponding to a promoter sequence of 500 bp.
List of top-scoring Arabidopsis and rice promoters for the presence of seed storage protein gene promoter motifs. Species, binomial name of species; GeneID, accession number; Description, functional gene annotation.
List of seed-storage protein gene promoters included in the analysis. Uniprot ID, UniProt/SwissProt sequence identifier; GenPept ID, GenPept accession number; GenBank ID, GenBank accession number; Start, start coordinate for the coding sequence; Stop, stop coordinate for the coding sequence; Strand, strand (+/-) of the coding sequence; Species, binomial name of species.
Acknowledgments
Acknowledgements
The authors thank Mathieu Blanchette for critical reading of the manuscript, and acknowledge the Natural Sciences and Engineering Research Council of Canada (NSERC) for a Discovery grant to M.V.S. and an NSERC Postgraduate Scholarship (PGS D) to F.F We also acknowledge le Fonds de recherche sur la nature et les technologies (FQRNT) and the Centre Sève for financial support.
Contributor Information
François Fauteux, Email: francois.fauteux2@mail.mcgill.ca.
Martina V Strömvik, Email: martina.stromvik@mcgill.ca.
References
- Venter M. Synthetic promoters: genetic control through cis engineering. Trends Plant Sci. 2007;12:118–124. doi: 10.1016/j.tplants.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Elnitski L, Jin VX, Farnham PJ, Jones SJ. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006;16:1455–1464. doi: 10.1101/gr.4140006. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T. The structure of plant gene promoters. In: Setlow JK, editor. Genetic Engineering: Principles and Methods. Vol. 19. New-York: Plenum Press; 1997. pp. 15–47. [Google Scholar]
- Schneider TD. Consensus sequence Zen. Applied bioinformatics. 2002;1:111–119. [PMC free article] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Higo H. PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998;26:358–359. doi: 10.1093/nar/26.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GuhaThakurta D. Computational identification of transcriptional regulatory elements in DNA sequence. Nucleic Acids Res. 2006;34:3585–3598. doi: 10.1093/nar/gkl372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauteux F, Blanchette M, Stromvik MV. Seeder: Discriminative Seeding DNA Motif Discovery. Bioinformatics. 2008;24:2303–2307. doi: 10.1093/bioinformatics/btn444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. Combined networks regulating seed maturation. Trends Plant Sci. 2007;12:294–300. doi: 10.1016/j.tplants.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008;54:608–620. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L. The Arabidopsis Book. The American Society of Plant Biologists, Rockville, MD; 2008. Storage Reserve Accumulation in Arabidopsis: Metabolic and Developmental Control of Seed Filling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Carbonero P. Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol. 2005;49:645–651. doi: 10.1387/ijdb.052046jc. [DOI] [PubMed] [Google Scholar]
- Ellerstrom M, Stalberg K, Ezcurra I, Rask L. Functional dissection of a napin gene promoter: identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol Biol. 1996;32:1019–1027. doi: 10.1007/BF00041385. [DOI] [PubMed] [Google Scholar]
- Stalberg K, Ellerstom M, Ezcurra I, Ablov S, Rask L. Disruption of an overlapping E-box/ABRE motif abolished high transcription of the napA storage-protein promoter in transgenic Brassica napus seeds. Planta. 1996;199:515–519. doi: 10.1007/BF00195181. [DOI] [PubMed] [Google Scholar]
- Ezcurra I, Ellerstrom M, Wycliffe P, Stalberg K, Rask L. Interaction between composite elements in the napA promoter: both the B-box ABA-responsive complex and the RY/G complex are necessary for seed-specific expression. Plant Mol Biol. 1999;40:699–709. doi: 10.1023/A:1006206124512. [DOI] [PubMed] [Google Scholar]
- Chandrasekharan MB, Bishop KJ, Hall TC. Module-specific regulation of the beta-phaseolin promoter during embryogenesis. Plant J. 2003;33:853–866. doi: 10.1046/j.1365-313X.2003.01678.x. [DOI] [PubMed] [Google Scholar]
- Lindstrom JT, Vodkin LO, Harding RW, Goeken RM. Expression of soybean lectin gene deletions in tobacco. Dev Genet. 1990;11:160–167. doi: 10.1002/dvg.1020110206. [DOI] [PubMed] [Google Scholar]
- Chamberland S, Daigle N, Bernier F. The legumin boxes and the 3' part of a soybean beta-conglycinin promoter are involved in seed gene expression in transgenic tobacco plants. Plant Mol Biol. 1992;19:937–949. doi: 10.1007/BF00040526. [DOI] [PubMed] [Google Scholar]
- Wu C, Washida H, Onodera Y, Harada K, Takaiwa F. Quantitative nature of the Prolamin-box, ACGT and AACA motifs in a rice glutelin gene promoter: minimal cis-element requirements for endosperm-specific gene expression. Plant J. 2000;23:415–421. doi: 10.1046/j.1365-313x.2000.00797.x. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. 1985;13:3021–3030. doi: 10.1093/nar/13.9.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Heyworth A, Pywell J, Kreis M. Nucleotide sequence of a B1 hordein gene and the identification of possible upstream regulatory elements in endosperm storage protein genes from barley, wheat and maize. Nucleic Acids Res. 1985;13:7327–7339. doi: 10.1093/nar/13.20.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaiwa F, Yamanouchi U, Yoshihara T, Washida H, Tanabe F, Kato A, Yamada K. Characterization of common cis-regulatory elements responsible for the endosperm-specific expression of members of the rice glutelin multigene family. Plant Mol Biol. 1996;30:1207–1221. doi: 10.1007/BF00019553. [DOI] [PubMed] [Google Scholar]
- Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboute ME, Clement B, Sekine M, Philipps G, Chaubet-Gigot N. Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell. 2000;12:1987–2000. doi: 10.1105/tpc.12.10.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MS, Flavell RB. Identification of an enhancer element for the endosperm-specific expression of high molecular weight glutenin. Plant Cell. 1990;2:1171–1180. doi: 10.1105/tpc.2.12.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Song F, Goodman RM, Zheng Z. Up-regulation of OsBIHD1, a rice gene encoding BELL homeodomain transcriptional factor, in disease resistance responses. Plant Biol (Stuttg) 2005;7:459–468. doi: 10.1055/s-2005-865851. [DOI] [PubMed] [Google Scholar]
- Washida H, Wu CY, Suzuki A, Yamanouchi U, Akihama T, Harada K, Takaiwa F. Identification of cis-regulatory elements required for endosperm expression of the rice storage protein glutelin gene GluB-1. Plant Mol Biol. 1999;40:1–12. doi: 10.1023/A:1026459229671. [DOI] [PubMed] [Google Scholar]
- So JS, Larkins BA. Binding of an endosperm-specific nuclear protein to a maize beta-zein gene correlates with zein transcriptional activity. Plant Mol Biol. 1991;17:309–319. doi: 10.1007/BF00040627. [DOI] [PubMed] [Google Scholar]
- Vincentz M, Leite A, Neshich G, Vriend G, Mattar C, Barros L, Weinberg D, de Almeida ER, de Carvalho MP, Aragao F, et al. ACGT and vicilin core sequences in a promoter domain required for seed-specific expression of a 2S storage protein gene are recognized by the opaque-2 regulatory protein. Plant Mol Biol. 1997;34:879–889. doi: 10.1023/A:1005874404706. [DOI] [PubMed] [Google Scholar]
- Meinke DW, Chen J, Beachy RN. Expression of Storage-Protein Genes during Soybean Seed Development. Planta. 1981;153:130–139. doi: 10.1007/BF00384094. [DOI] [PubMed] [Google Scholar]
- Nielsen NC, Dickinson CD, Cho TJ, Thanh VH, Scallon BJ, Fischer RL, Sims TL, Drews GN, Goldberg RB. Characterization of the glycinin gene family in soybean. Plant Cell. 1989;1:313–328. doi: 10.1105/tpc.1.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada JJ, Barker SJ, Goldberg RB. Soybean beta-conglycinin genes are clustered in several DNA regions and are regulated by transcriptional and posttranscriptional processes. Plant Cell. 1989;1:415–425. doi: 10.1105/tpc.1.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Hirano H. Nucleotide sequence of the basic 7S globulin gene from soybean. Plant Physiol. 1994;105:1019–1020. doi: 10.1104/pp.105.3.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Krishnamachari A. Computational analysis of plant RNA Pol-II promoters. Biosystems. 2006;83:38–50. doi: 10.1016/j.biosystems.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Ericson ML, Muren E, Gustavsson HO, Josefsson LG, Rask L. Analysis of the promoter region of napin genes from Brassica napus demonstrates binding of nuclear protein in vitro to a conserved sequence motif. Eur J Biochem. 1991;197:741–746. doi: 10.1111/j.1432-1033.1991.tb15966.x. [DOI] [PubMed] [Google Scholar]
- Josefsson LG, Lenman M, Ericson ML, Rask L. Structure of a gene encoding the 1.7 S storage protein, napin, from Brassica napus. J Biol Chem. 1987;262:12196–12201. [PubMed] [Google Scholar]
- Dickinson CD, Evans RP, Nielsen NC. RY repeats are conserved in the 5'-flanking regions of legume seed-protein genes. Nucleic Acids Res. 1988;16:371. doi: 10.1093/nar/16.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelievre JM, Oliveira LO, Nielsen NC. 5'CATGCAT-3' Elements Modulate the Expression of Glycinin Genes. Plant Physiol. 1992;98:387–391. doi: 10.1104/pp.98.1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Beachy RN. Tissue-specific and temporal regulation of a beta-conglycinin gene: roles of the RY repeat and other cis-acting elements. Plant Mol Biol. 1994;24:261–272. doi: 10.1007/BF00020166. [DOI] [PubMed] [Google Scholar]
- Baumlein H, Nagy I, Villarroel R, Inze D, Wobus U. Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 1992;2:233–239. [PubMed] [Google Scholar]
- Inze D, Veylder LD. Cell Cycle Regulation in Plant Development. Annu Rev Genet. 2006;40:77–105. doi: 10.1146/annurev.genet.40.110405.090431. [DOI] [PubMed] [Google Scholar]
- Chen ZL, Schuler MA, Beachy RN. Functional analysis of regulatory elements in a plant embryo-specific gene. Proc Natl Acad Sci USA. 1986;83:8560–8564. doi: 10.1073/pnas.83.22.8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZL, Pan NS, Beachy RN. A DNA sequence element that confers seed-specific enhancement to a constitutive promoter. EMBO J. 1988;7:297–302. doi: 10.1002/j.1460-2075.1988.tb02812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD, Bernier F, Lessard PA, Beachy RN. Nuclear factors interact with a soybean beta-conglycinin enhancer. Plant Cell. 1989;1:623–631. doi: 10.1105/tpc.1.6.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard PA, Allen RD, Bernier F, Crispino JD, Fujiwara T, Beachy RN. Multiple nuclear factors interact with upstream sequences of differentially regulated beta-conglycinin genes. Plant Mol Biol. 1991;16:397–413. doi: 10.1007/BF00023991. [DOI] [PubMed] [Google Scholar]
- Lessard PA, Allen RD, Fujiwara T, Beachy RN. Upstream regulatory sequences from two beta-conglycinin genes. Plant Mol Biol. 1993;22:873–885. doi: 10.1007/BF00027372. [DOI] [PubMed] [Google Scholar]
- Yoshino M, Kanazawa A, Tsutsumi KI, Nakamura I, Shimamoto Y. Structure and characterization of the gene encoding alpha subunit of soybean beta-conglycinin. Genes Genet Syst. 2001;76:99–105. doi: 10.1266/ggs.76.99. [DOI] [PubMed] [Google Scholar]
- Yoshino M, Nagamatsu A, Tsutsumi K, Kanazawa A. The regulatory function of the upstream sequence of the beta-conglycinin alpha subunit gene in seed-specific transcription is associated with the presence of the RY sequence. Genes Genet Syst. 2006;81:135–141. doi: 10.1266/ggs.81.135. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kitamura Y, Fukazawa C. The glycinin box: a soybean embryo factor binding motif within the quantitative regulatory region of the 11S seed storage globulin promoter. Mol Gen Genet. 1994;243:353–357. doi: 10.1007/BF00301071. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kitamura Y, Arahira M, Fukazawa C. cis-acting regulatory regions of the soybean seed storage 11S globulin gene and their interactions with seed embryo factors. Plant Mol Biol. 1993;21:973–984. doi: 10.1007/BF00023596. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Gonzalez N, Diaz I, Parcy F, Carbonero P, Vicente-Carbajosa J. FUSCA3 from barley unveils a common transcriptional regulation of seed-specific genes between cereals and Arabidopsis. Plant J. 2008;53:882–894. doi: 10.1111/j.1365-313X.2007.03382.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999;17:209–214. doi: 10.1046/j.1365-313X.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 2007;52:716–729. doi: 10.1111/j.1365-313X.2007.03268.x. [DOI] [PubMed] [Google Scholar]
- Shirsat A, Wilford N, Croy R, Boulter D. Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol Gen Genet. 1989;215:326–331. doi: 10.1007/BF00339737. [DOI] [PubMed] [Google Scholar]
- VicenteCarbajosa J, Moose SP, Parsons RL, Schmidt RJ. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P. An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. Plant J. 1998;16:53–62. doi: 10.1046/j.1365-313x.1998.00275.x. [DOI] [PubMed] [Google Scholar]
- Muller M, Knudsen S. The nitrogen response of a barley C-hordein promoter is controlled by positive and negative regulation of the GCN4 and endosperm box. Plant J. 1993;4:343–355. doi: 10.1046/j.1365-313X.1993.04020343.x. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- Rodin J, Sjodahl S, Josefsson LG, Rask L. Characterization of a Brassica napus gene encoding a cruciferin subunit: estimation of sizes of cruciferin gene families. Plant Mol Biol. 1992;20:559–563. doi: 10.1007/BF00040615. [DOI] [PubMed] [Google Scholar]
- Ryan AJ, Royal CL, Hutchinson J, Shaw CH. Genomic sequence of a 12S seed storage protein from oilseed rape (Brassica napus c.v. jet neuf) Nucleic Acids Res. 1989;17:3584. doi: 10.1093/nar/17.9.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depigny-This D, Raynal M, Aspart L, Delseny M, Grellet F. The cruciferin gene family in radish. Plant Mol Biol. 1992;20:467–479. doi: 10.1007/BF00040606. [DOI] [PubMed] [Google Scholar]
- Takei Y, Yamauchi D, Minamikawa T. Nucleotide sequence of the canavalin gene from Canavalia gladiata seeds. Nucleic Acids Res. 1989;17:4381. doi: 10.1093/nar/17.11.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Schuler MA, Godette WD, Zenger V, Beachy RN, Slightom JL. The glycosylated seed storage proteins of Glycine max and Phaseolus vulgaris. Structural homologies of genes and proteins. J Biol Chem. 1986;261:9228–9238. [PubMed] [Google Scholar]
- Sims TL, Goldberg RB. The glycinin Gy1 gene from soybean. Nucleic Acids Res. 1989;17:4386. doi: 10.1093/nar/17.11.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura Y, Arahira M, Itoh Y, Fukazawa C. The complete nucleotide sequence of soybean glycinin A2B1a gene spanning to another glycinin gene A1aB1b. Nucleic Acids Res. 1990;18:4245. doi: 10.1093/nar/18.14.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown D, Ellis TH, Gatehouse JA. The sequence of a gene encoding convicilin from pea (Pisum sativum L.) shows that convicilin differs from vicilin by an insertion near the N-terminus. Biochem J. 1988;251:717–726. doi: 10.1042/bj2510717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbigin EJ, Delumen BO, Chandler PM, Gould A, Blagrove RJ, March JF, Kortt AA, Higgins TJV. Pea Convicilin - Structure and Primary Sequence of the Protein and Expression of a Gene in the Seeds of Transgenic Tobacco. Planta. 1990;180:461–470. doi: 10.1007/BF02411442. [DOI] [PubMed] [Google Scholar]
- Rerie WG, Whitecross MI, Higgins TJV. Nucleotide-Sequence of an a-Type Legumin Gene from Pea. Nucleic Acids Research. 1990;18:655–655. doi: 10.1093/nar/18.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatehouse JA, Bown D, Gilroy J, Levasseur M, Castleton J, Ellis TH. Two genes encoding 'minor' legumin polypeptides in pea (Pisum sativum L.). Characterization and complete sequence of the LegJ gene. Biochem J. 1988;250:15–24. doi: 10.1042/bj2500015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumlein H, Wobus U, Pustell J, Kafatos FC. The legumin gene family: structure of a B type gene of Vicia faba and a possible legumin gene specific regulatory element. Nucleic Acids Res. 1986;14:2707–2720. doi: 10.1093/nar/14.6.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weschke W, Baumlein H, Wobus U. Nucleotide sequence of a field bean (Vicia faba L.var.minor) vicilin gene. Nucleic Acids Res. 1987;15:10065. doi: 10.1093/nar/15.23.10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derose RT, Ma DP, Kwon IS, Hasnain SE, Klassy RC, Hall TC. Characterization of the Kafirin Gene Family from Sorghum Reveals Extensive Homology with Zein from Maize. Plant Molecular Biology. 1989;12:245–256. doi: 10.1007/BF00043202. [DOI] [PubMed] [Google Scholar]
- Sumner-Smith M, Rafalski JA, Sugiyama T, Stoll M, Soll D. Conservation and variability of wheat alpha/beta-gliadin genes. Nucleic Acids Res. 1985;13:3905–3916. doi: 10.1093/nar/13.11.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski JA. Structure of wheat gamma-gliadin genes. Gene. 1986;43:221–229. doi: 10.1016/0378-1119(86)90210-6. [DOI] [PubMed] [Google Scholar]
- Scheets K, Hedgcoth C. Nucleotide-Sequence of a Gamma-Gliadin Gene - Comparisons with Other Gamma-Gliadin Sequences Show the Structure of Gamma-Gliadin Genes and the General Primary Structure of Gamma-Gliadins. Plant Science. 1988;57:141–150. doi: 10.1016/0168-9452(88)90080-5. [DOI] [Google Scholar]
- Pedersen K, Devereux J, Wilson DR, Sheldon E, Larkins BA. Cloning and Sequence-Analysis Reveal Structural Variation among Related Zein Genes in Maize. Cell. 1982;29:1015–1026. doi: 10.1016/0092-8674(82)90465-2. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Hyndman R, Fan Y. Sample quantiles in statistical packages. The American Statistician. 1996;v50:361(365). [Google Scholar]
- Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Minimum, maximum and sample deciles for the position of SSP gene promoter motifs. The minimum, maximum and deciles of positions of best matching subsequences to motifs discovered in Brassicaceae (B1-3, BT), Fabaceae (F1-5, FT), and Poaceae (P1-5) are plotted on an axis corresponding to a promoter sequence of 500 bp.
List of top-scoring Arabidopsis and rice promoters for the presence of seed storage protein gene promoter motifs. Species, binomial name of species; GeneID, accession number; Description, functional gene annotation.
List of seed-storage protein gene promoters included in the analysis. Uniprot ID, UniProt/SwissProt sequence identifier; GenPept ID, GenPept accession number; GenBank ID, GenBank accession number; Start, start coordinate for the coding sequence; Stop, stop coordinate for the coding sequence; Strand, strand (+/-) of the coding sequence; Species, binomial name of species.