Abstract
Background
It has been previously demonstrated that a single nucleotide polymorphism (SNP) in the IL13 promoter region, IL13 -1055T>C (rs1800925), was associated with susceptibility to severe malaria in Thais. In the present study, fine association mapping for a cytokine gene cluster including IL4, IL5, and IL13 on chromosome 5q31 was conducted using the same malaria subjects to refine the region containing a primary variant or a haplotype susceptible to severe malaria.
Methods
A total of 82 SNPs spanning 522 kb of the 5q31 region were analysed in 368 patients with Plasmodium falciparum malaria (203 mild malaria and 165 severe malaria patients).
Results
Only rs1881457 located in the promoter region of IL13, which is in linkage disequilibrium with rs1800925 (r2 = 0.73), showed a significant association with severe malaria after adjusting for multiple testing (P = 0.046 by permutation test). This SNP was in a haplotype block spanning 97 kb (from rs2069812 to rs2240032). The detected haplotype block contained the RAD50 gene and the promoter of IL13, but not the other genes.
Conclusion
A haplotype block in which a primary polymorphism associated with severe malaria is likely to be encoded was identified in Thai malaria patients.
Background
Over the course of the last decade a number of studies have provided evidence for a linkage between the blood infection level of Plasmodium falciparum and the human chromosome 5q31 region in African populations [1-4]. In addition to malaria, the 5q31 region shows a linkage to the response against other infectious diseases such as schistosomiasis [5] and leishmaniasis [6]. The 5q31-33 region contains genes encoding the T helper 2-type cytokines (the interleukin genes IL3, IL4, IL5, IL9, and IL13) and other immunologically active genes such as interferon regulatory factor-1 (IRF1). These genes are strong candidates for controlling the outcome of malaria infection.
In a previous study, three single nucleotide polymorphisms (SNPs) in the promoter regions of IL3, IL4, and IL13 were investigated. Of which, a SNP in the IL13 promoter region, IL13 -1055T>C (rs1800925), was found to be associated with susceptibility to severe malaria in Thais [7]. However, a number of candidate genes or polymorphisms still remain to be analyzed. In addition, no other polymorphisms surrounding rs1800925 were analyzed and thus the possibility that the association of rs1800925 with severe malaria may have resulted from linkage disequilibrium (LD) from other polymorphisms could not be excluded. The aim of this study is to better define the genomic region showing the association with severe malaria on the 5q31 region.
Methods
Subjects
A total of 368 adult patients with P. falciparum malaria (165 patients with severe P. falciparum malaria and 203 patients with mild malaria) living in northwest Thailand were enrolled in this study. All patients underwent treatment at the Hospital for Tropical Diseases, Faculty of Tropical Medicine, Mahidol University. Malarial infection by P. falciparum was confirmed by a positive blood smear for the asexual form of P. falciparum. Clinical manifestations of severe and mild malaria were classified according to the following definitions and criteria. A patient was classified as severe malaria when he/she has one or more of the following signs: high parasitaemia (>100,000 parasite/ml), hypoglycaemia (glucose <22 nmol/l), severe anaemia (haematocrit <20% or haemoglobin <7.0 g/dl), and a serum creatinine level of more than 3.0 mg/dl. In the present study, patients with cerebral malaria were not analyzed. Mild malaria was characterized by fever without other any underlying causes of infections and no manifestations of severe malaria as described above. All individuals were 13 years of age or older, and the mean ages of patients with mild malaria and those with severe malaria were 25.3 and 23.8, respectively. This study was approved by the institutional review board of the Faculty of Tropical Medicine, Mahidol University, and the Research Ethics Committees of the Faculty of Medicine, The University of Tokyo, and the Graduate School of Comprehensive Human Sciences, University of Tsukuba. Informed consent was obtained from all participants.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using a QIAamp Blood Kit (Qiagen, Hilden, Germany). A total of 82 SNPs within a 522 kb region on human chromosome 5q31 were genotyped by using the DigiTag2 assay [8] or TaqMan assay (Table 1). These SNPs were selected to capture the LD structure on 5q31 in Asian populations [9].
Table 1.
Allele frequencies and association tests for SNPs in 5q31 cytokine cluster
| SNP rs# | Gene | Allelic state | Frequency of derived allele | Association P value | |||
| Ancestral | Derived | Severe malaria | Mild malaria | Raw | Permutation | ||
| rs162887 | SLC22A4 | C | T | 0.369 | 0.402 | 0.381 | 1.000 |
| rs3792876 | SLC22A4 | C | T | 0.234 | 0.236 | 0.944 | 1.000 |
| rs3792878 | SLC22A4 | G | A | 0.954 | 0.932 | 0.235 | 0.996 |
| rs3805665 | SLC22A4 | G | A | 0.23 | 0.237 | 0.823 | 1.000 |
| rs3805668 | SLC22A4 | G | A | 0.229 | 0.236 | 0.818 | 1.000 |
| rs270608 | SLC22A4 | A | G | 0.338 | 0.379 | 0.328 | 1.000 |
| rs270607 | SLC22A4 | C | T | 0.372 | 0.397 | 0.515 | 1.000 |
| rs2073839 | SLC22A4 | C | T | 0.234 | 0.236 | 0.950 | 1.000 |
| rs3828673 | SLC22A4 | G | A | 0.234 | 0.236 | 0.950 | 1.000 |
| rs3792885 | SLC22A4 | A | T | 0.229 | 0.236 | 0.818 | 1.000 |
| rs272842 | SLC22A4 | T | C | 0.348 | 0.299 | 0.181 | 0.985 |
| rs3761659 | SLC22A4 | C | G | 0.234 | 0.236 | 0.950 | 1.000 |
| rs3805673 | SLC22A4 | G | A | 0.228 | 0.237 | 0.771 | 1.000 |
| rs273915 | SLC22A4 | G | C | 0.375 | 0.397 | 0.563 | 1.000 |
| rs272887 | SLC22A4 | C | T | 0.372 | 0.397 | 0.515 | 1.000 |
| rs273909 | SLC22A4 | T | C | 0.071 | 0.093 | 0.307 | 1.000 |
| rs272879 | SLC22A4 | G | C | 0.344 | 0.297 | 0.201 | 0.992 |
| rs272873 | SLC22A4 | C | T | 0.167 | 0.175 | 0.773 | 1.000 |
| rs2306772 | SLC22A4 | G | A | 0.234 | 0.236 | 0.950 | 1.000 |
| rs272867 | C | T | 0.344 | 0.298 | 0.205 | 0.992 | |
| rs3788987 | SLC22A5 | G | A | 0.232 | 0.238 | 0.843 | 1.000 |
| rs2631362 | SLC22A5 | T | C | 0.365 | 0.376 | 0.769 | 1.000 |
| rs2631359 | SLC22A5 | G | A | 0.363 | 0.379 | 0.671 | 1.000 |
| rs4646301 | SLC22A5 | G | A | 0.236 | 0.253 | 0.614 | 1.000 |
| rs274571 | SLC22A5 | T | C | 0.365 | 0.379 | 0.719 | 1.000 |
| rs2073642 | SLC22A5 | C | T | 0.241 | 0.254 | 0.707 | 1.000 |
| rs183898 | SLC22A5 | G | C | 0.361 | 0.381 | 0.597 | 1.000 |
| rs4646305 | SLC22A5 | G | A | 0.237 | 0.251 | 0.678 | 1.000 |
| rs274559 | SLC22A5 | C | T | 0.348 | 0.299 | 0.181 | 0.985 |
| rs274558 | SLC22A5 | C | T | 0.353 | 0.299 | 0.144 | 0.965 |
| rs274554 | SLC22A5 | A | G | 0.837 | 0.887 | 0.059 | 0.748 |
| rs274553 | SLC22A5 | G | C | 0.163 | 0.113 | 0.059 | 0.748 |
| rs274551 | SLC22A5 | C | T | 0.161 | 0.113 | 0.072 | 0.804 |
| rs274549 | SLC22A5 | G | T | 0.163 | 0.113 | 0.059 | 0.748 |
| rs274547 | SLC22A5 | T | A | 0.839 | 0.887 | 0.072 | 0.804 |
| rs2285673 | LOC441108 | C | T | 0.227 | 0.199 | 0.388 | 1.000 |
| rs2269822 | LOC441108 | Cb | T | 0.282 | 0.281 | 1.000 | 1.000 |
| rs3749834 | C | T | 0.236 | 0.238 | 0.929 | 1.000 | |
| rs2070730 | IRF1 | C | T | 0.418 | 0.407 | 0.765 | 1.000 |
| rs2070727 | IRF1 | G | T | 0.42 | 0.406 | 0.725 | 1.000 |
| rs2070723 | IRF1 | C | T | 0.582 | 0.593 | 0.765 | 1.000 |
| rs2070722 | IRF1 | G | T | 0.596 | 0.587 | 0.823 | 1.000 |
| rs739718a | A | G | 0.257 | 0.261 | 0.903 | 1.000 | |
| rs2069812a | IL5 | T | C | 0.241 | 0.302 | 0.083 | 0.848 |
| rs2299015 | RAD50 | A | C | 0.111 | 0.186 | 0.008 | 0.193 |
| rs2299014 | RAD50 | T | G | 0.074 | 0.07 | 0.838 | 1.000 |
| rs2243677 | RAD50 | G | A | 0.887 | 0.804 | 0.004 | 0.107 |
| rs2522414 | RAD50 | G | C | 0.887 | 0.804 | 0.004 | 0.107 |
| rs2299013 | RAD50 | Cc | G | 0.104 | 0.186 | 0.004 | 0.101 |
| rs2252775 | RAD50 | A | C | 0.11 | 0.186 | 0.007 | 0.174 |
| rs2522394 | RAD50 | A | G | 0.887 | 0.804 | 0.004 | 0.107 |
| rs2245460 | RAD50 | A | T | 0.112 | 0.189 | 0.006 | 0.160 |
| rs2301713 | RAD50 | T | C | 0.113 | 0.186 | 0.010 | 0.242 |
| rs3798135 | RAD50 | G | A | 0.113 | 0.186 | 0.010 | 0.242 |
| rs2237060 | RAD50 | A | C | 0.04 | 0.055 | 0.351 | 1.000 |
| rs2074369 | RAD50 | C | T | 0.883 | 0.804 | 0.006 | 0.158 |
| rs2240032 | RAD50 | C | T | 0.106 | 0.183 | 0.006 | 0.152 |
| rs1881457 | IL13 | A | C | 0.124 | 0.219 | 0.002 | 0.046d |
| rs1800925 | IL13 | T | C | 0.894 | 0.826 | 0.014 | 0.296 |
| rs2066960 | IL13 | A | C | 0.628 | 0.636 | 0.830 | 1.000 |
| rs20541a | IL13 | C | T | 0.33 | 0.389 | 0.115 | 0.924 |
| rs2070874 | IL4 | T | C | 0.273 | 0.274 | 0.975 | 1.000 |
| rs2243270 | IL4 | G | A | 0.263 | 0.266 | 0.913 | 1.000 |
| rs2243289 | IL4 | A | G | 0.729 | 0.736 | 0.825 | 1.000 |
| rs1468215 | KIF3A | T | A | 0.26 | 0.247 | 0.725 | 1.000 |
| rs3798132 | KIF3A | A | G | 0.352 | 0.336 | 0.685 | 1.000 |
| rs3798130 | KIF3A | G | A | 0.671 | 0.677 | 0.882 | 1.000 |
| rs2299007 | KIF3A | T | C | 0.383 | 0.431 | 0.219 | 0.994 |
| rs2237057 | KIF3A | T | C | 0.742 | 0.75 | 0.818 | 1.000 |
| rs2299006 | KIF3A | G | C | 0.654 | 0.667 | 0.738 | 1.000 |
| rs2299005 | KIF3A | T | C | 0.661 | 0.668 | 0.836 | 1.000 |
| rs3798129 | KIF3A | A | T | 0.336 | 0.337 | 0.975 | 1.000 |
| rs3756754 | KIF3A | C | T | 0.022 | 0.013 | 0.366 | 1.000 |
| rs256871 | SEPT8 | C | T | 0.609 | 0.635 | 0.490 | 1.000 |
| rs30534 | SEPT8 | G | A | 0.391 | 0.365 | 0.490 | 1.000 |
| rs30533 | SEPT8 | T | C | 0.386 | 0.371 | 0.686 | 1.000 |
| rs39588 | SEPT8 | C | G | 0.383 | 0.369 | 0.717 | 1.000 |
| rs256875 | SEPT8 | T | C | 0.385 | 0.379 | 0.888 | 1.000 |
| rs392916 | SEPT8 | T | A | 0.532 | 0.561 | 0.459 | 1.000 |
| rs30527 | SEPT8 | C | T | 0.62 | 0.639 | 0.620 | 1.000 |
| rs30524 | SEPT8 | G | T | 0.383 | 0.362 | 0.573 | 1.000 |
| rs757537 | ANKRD43 | T | C | 0.131 | 0.157 | 0.357 | 1.000 |
a SNP genotyped by TaqMan assay.
b Allelic state could not be inferred.
c Allelic state inferred based on the sequence of rhesus macaque.
d Permutation P value < 0.05.
Statistical analysis
The allele frequency at each SNP locus was compared between mild and severe malaria patients using the chi-square test and a permutation P value was calculated from 100000 permutations. A permutation P value of less than 0.05 was considered to be statistically significant. The pairwise LD coefficients (r2) between SNPs were calculated to evaluate the structure of LD on 5q31-33 in 368 Thai malaria patients. The frequencies of haplotypes consisting of rs2069812, rs2299015, rs2299014, rs2243677, rs2522414, rs2299013, rs2252775, rs2522394, rs2245460, rs2301713, rs3798135, rs2237060, rs2074369, rs2240032, rs1881457, and rs1800925 were estimated only for this haplotype block. All the statistical analyses were performed by using the Haploview software version 4.0 [10]. The allelic state for each SNP (i.e., ancestral or derived) was inferred based on the genome sequence of Pan troglodytes (chimpanzee), obtained from the NCBI BLAST database (Table 1). When the genomic sequence of chimpanzee was not available, one of Macaca mulatta (rhesus macaque) was used.
Results
Association test
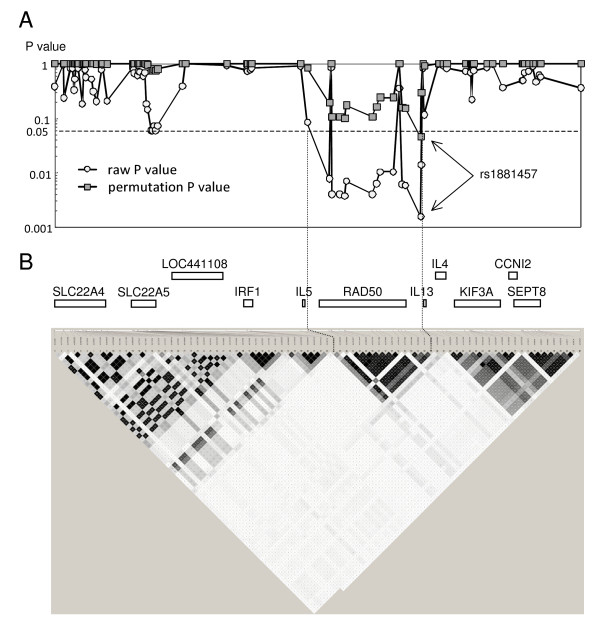
Eighty-two SNPs including rs1800925 were analysed to evaluate the association of the 5q31 region with severity of malaria (Table 1). The permutation P value as well as the raw P value was calculated for each SNP to avoid any false positive findings due to multiple testing (Table 1 and Figure 1A). Only rs1881457 showed a significant association with severe malaria (raw P value = 0.002 and permutation P value = 0.046) and no SNPs in the other genes showed any such association (permutation P value > 0.05). When a derived allele is focused on in association test, rs1881457-C may be referred to as a protective allele against severe malaria.
Figure 1.
Association P values and LD structure of 82 SNPs on 5q31. Association P values and LD structure of 82 SNPs on 5q31. (A) The raw P value (open circle) and permutation P value (shaded square) for each SNP. (B) Pairwise LD measured by r2 between 82 SNPs. White, shades of grey, and black squares indicate no LD (r2 = 0), intermediate LD (0 < r2 < 1), and strong LD (r2 = 1), respectively.
LD structure
In previous study, rs1800925 was found to be associated with severe malaria [7]. Since rs1881457, showing the strongest association in the present study, is closely located to rs1800925, these SNPs may be in LD. In addition, a number of SNPs near rs1881457 and rs1800925 showed also raw P values of less than 0.05 (Table 1 and Figure 1), thus suggesting that some, if not all, of these SNPs are in the same haplotype block. To clarify the structure of the LD around rs1881457 and rs1800925, r2 values between the 82 SNPs were calculated. The LD analysis for the 5q31 region revealed that all the SNPs showing low P values were in a distinct haplotype block spanning 97 kb from rs2069812 to rs2240032 (Figure 1B). This block contained the RAD50 gene and the promoter of IL13, but none of the other candidate genes such as IL4, IL5, and IRF1.
Six frequent haplotypes were observed in the detected block and two of which, haplotypes 1 and 4, bore rs1881457-C (Table 2). Both haplotypes showed a decreased frequency in severe malaria patients in comparison to those with mild malaria, thus suggesting that the association of rs1881457-C with the protection against severe malaria was not caused by a specific haplotype.
Table 2.
Estimated haplotype frequencies in malaria patients.
| Haplotypea | Estimated frequency | |
| Severe malaria | Mild malaria | |
| 1: CCTGGGCATCAACTCT | 0.095 | 0.163 |
| 2: TATACCAGATGATCAC | 0.723 | 0.637 |
| 3: CAGACCAGATGATCAC | 0.03 | 0.018 |
| 4: TATACCAGATGATCCC | 0.025 | 0.04 |
| 5: CATACCAGATGATCAC | 0.057 | 0.045 |
| 6: CAGACCAGATGCTCAC | 0.038 | 0.048 |
aThe haplotype consists of rs2069812, rs2299015, rs2299014, rs2243677, rs2522414, rs2299013, rs2252775, rs2522394, rs2245460, rs2301713, rs3798135, rs2237060, rs2074369, rs2240032, rs1881457, and rs1800925. Only haplotypes with the frequency of more than 0.02 either in severe malaria or in mild malaria were presented.
Discussion
In this study rs1881457 was found to be significantly associated with severe malaria, and this SNP was included in a haplotype block encompassing the whole RAD50 gene and the promoter of IL13 (Figure 1). Together with MRE11 and NBS1, RAD50 forms a conserved multiprotein complex, MRE11-RAD50-NBS1 (MRN), which plays an important role in double-strand break repair, cell cycle checkpoint control, meiotic recombination, and telomere maintenance [11-13]. In the immune system, the MRN complex is involved in B cell-specific immunoglobulin gene diversification (e.g., Ig class-switch recombination, somatic hypermutation, and gene conversion) [14,15]. The polymorphisms of RAD50 may therefore influence the affinity and/or effector functions of antibodies. The IL13 gene encodes a immunoregulatory cytokine (Th2 cytokine) produced by activated Th2 cells. The Th2 cytokines down-regulate macrophage activity, and inhibit the production of pro-inflammatory cytokines such as TNF and IL1. It has been reported that increased concentrations of TNF and IL1β in serum are observed in severe malaria patients [16]. The IL13 promoter polymorphisms may influence the expression of IL13. Thus, both RAD50 and IL13 seem to be plausible candidate genes associated with severe malaria.
The genes encoding the Th2 cytokines IL5, IL13, and IL4 are subject to coordinate regulation and are expressed in a cell lineage-specific manner [17,18]. The expressions are regulated by a locus control region (LCR) located within a 25 kb region containing the 3' portion of RAD50 [19]. Interestingly, the LCR is included in the haplotype block associated with severe malaria, raising a possibility that polymorphisms which influence the LCR activity could account for the observed association with the severity of malaria.
Since rs1881457 is located in the promoter region of IL13, the nucleotide change at this site may affect the binding ability of some transcription factor. The TFSEARCH (TFSEARCH: Searching Transcription Factor Binding Sites, http://www.rwcp.or.jp/papia/) program based on the TRANSFAC databases [20] was used to examine the possibility. The result indicated no possible binding site of transcription factor at rs1881457 regardless of alleles (rs1881457-A and rs1881457-C) with the default setting (threshold score = 0.85). Therefore, rs1881457 itself may not be a primary polymorphism associated with severe malaria, even though rs1881457 showed the strongest association observed in this study.
Among IL13 polymorphisms, rs1800925 in the IL13 promoter has been reported to be associated with various diseases [21-24]. This SNP is located within a putative primate-specific cis-regulatory element [25] and has been shown to affect the promoter activity of IL13 [25] and IL13 production [26]. In the present study rs1800925 and rs1881457 had a high r2 value (r2 = 0.73). Therefore, the possibility that rs1800925 is a primary SNP and the significant association of rs1881457 with severe malaria resulted from LD between these SNPs is not excluded. The future functional and association studies for rs1881457, rs1800925, and other polymorphisms, including those not analyzed in the present study, may thus help us to better understand the genetic susceptibility to severe malaria.
Conclusion
A haplotype block spanning 97 kb encompassing RAD50 gene and IL13 promoter region that was associated with severity of malaria was identified in a Thai population.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NI carried out the genotyping, helped to conduct statistical analyses, and wrote the manuscript. NN and KT helped to genotype the samples. JP, PN and HH collected blood samples, extracted DNA, and helped to genotype the samples. JP participated in the design of the study and coordination. NT was involved in the interpretation of the data and preparation of the manuscript. JO conceived of the study, and participated in its design, performed statistical analyses, and helped to write the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors sincerely thank all patients who kindly participated in this study. This study was supported in part by research funds KAKENHI Grant-in-Aid for Scientific Research on Priority Areas "Comprehensive Genomics" and "Applied Genomics" from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Contributor Information
Izumi Naka, Email: izumin-tky@umin.ac.jp.
Nao Nishida, Email: nishida-75@umin.ac.jp.
Jintana Patarapotikul, Email: tmjpt@mahidol.ac.th.
Pornlada Nuchnoi, Email: mtpnn@staff1.mahidol.ac.th.
Katsushi Tokunaga, Email: tokunaga@m.u-tokyo.ac.jp.
Hathairad Hananantachai, Email: tmhhn@mahidol.ac.th.
Naoyuki Tsuchiya, Email: tsuchiya@md.tsukuba.ac.jp.
Jun Ohashi, Email: juno-tky@umin.ac.jp.
References
- Flori L, Kumulungui B, Aucan C, Esnault C, Traore AS, Fumoux F, Rihet P. Linkage and association between Plasmodium falciparum blood infection levels and chromosome 5q31-q33. Genes Immun. 2003;4:265–268. doi: 10.1038/sj.gene.6363960. [DOI] [PubMed] [Google Scholar]
- Garcia A, Marquet S, Bucheton B, Hillaire D, Cot M, Fievet N, Dessein AJ, Abel L. Linkage analysis of blood Plasmodium falciparum levels: interest of the 5q31-q33 chromosome region. Am J Trop Med Hyg. 1998;58:705–709. doi: 10.4269/ajtmh.1998.58.705. [DOI] [PubMed] [Google Scholar]
- Rihet P, Traore Y, Abel L, Aucan C, Traore-Leroux T, Fumoux F. Malaria in humans: Plasmodium falciparum blood infection levels are linked to chromosome 5q31-q33. Am J Hum Genet. 1998;63:498–505. doi: 10.1086/301967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuntabhai A, Ndiaye R, Casademont I, Peerapittayamongkol C, Rogier C, Tortevoye P, Tall A, Paul R, Turbpaiboon C, Phimpraphi W, Trape JF, Spiegel A, Heath S, Mercereau-Puijalon O, Dieye A, Julier C. Genetic determination and linkage mapping of Plasmodium falciparum malaria related traits in Senegal. PLoS ONE. 2008;3:e2000. doi: 10.1371/journal.pone.0002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquet S, Abel L, Hillaire D, Dessein H, Kalil J, Feingold J, Weissenbach J, Dessein AJ. Genetic localization of a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33. Nat Genet. 1996;14:181–184. doi: 10.1038/ng1096-181. [DOI] [PubMed] [Google Scholar]
- Jeronimo SM, Holst AK, Jamieson SE, Francis R, Martins DR, Bezerra FL, Ettinger NA, Nascimento ET, Monteiro GR, Lacerda HG, Miller EN, Cordell HJ, Duggal P, Beaty TH, Blackwell JM, Wilson ME. Genes at human chromosome 5q31.1 regulate delayed-type hypersensitivity responses associated with Leishmania chagasi infection. Genes Immun. 2007;8:539–551. doi: 10.1038/sj.gene.6364422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi J, Naka I, Patarapotikul J, Hananantachai H, Looareesuwan S, Tokunaga K. A single-nucleotide substitution from C to T at position -1055 in the IL-13 promoter is associated with protection from severe malaria in Thailand. Genes Immun. 2003;4:528–531. doi: 10.1038/sj.gene.6364010. [DOI] [PubMed] [Google Scholar]
- Nishida N, Tanabe T, Takasu M, Suyama A, Tokunaga K. Further development of multiplex single nucleotide polymorphism typing method, the DigiTag2 assay. Anal Biochem. 2007;364:78–85. doi: 10.1016/j.ab.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Nuchnoi P, Ohashi J, Naka I, Nacapunchai D, Tokunaga K, Nishida N, Patarapotikul J. Linkage disequilibrium structure of the 5q31-33 region in a Thai population. J Hum Genet. 2008;53:850–856. doi: 10.1007/s10038-008-0309-8. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Petrini JH, Stracker TH. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 2003;13:458–462. doi: 10.1016/S0962-8924(03)00170-3. [DOI] [PubMed] [Google Scholar]
- Bosch M van den, Bree RT, Lowndes NF. The MRN complex: coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracker S, Bergmann Y, Demuth I, Frappart PO, Hildebrand G, Christine R, Wang ZQ, Sperling K, Digweed M, Radbruch A. Nibrin functions in Ig class-switch recombination. Proc Natl Acad Sci USA. 2005;102:1584–1589. doi: 10.1073/pnas.0409191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki M, Fujii MM, Maizels N. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat Immunol. 2005;6:730–736. doi: 10.1038/ni1215. [DOI] [PubMed] [Google Scholar]
- Kwiatkowski D, Hill AV, Sambou I, Twumasi P, Castracane J, Manogue KR, Cerami A, Brewster DR, Greenwood BM. TNF concentration in fatal cerebral, non-fatal cerebral, and uncomplicated Plasmodium falciparum malaria. Lancet. 1990;336:1201–1204. doi: 10.1016/0140-6736(90)92827-5. [DOI] [PubMed] [Google Scholar]
- Kelly BL, Locksley RM. Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J Immunol. 2000;165:2982–2986. doi: 10.4049/jimmunol.165.6.2982. [DOI] [PubMed] [Google Scholar]
- Smale ST, Fisher AG. Chromatin structure and gene regulation in the immune system. Annu Rev Immunol. 2002;20:427–462. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- Lee GR, Fields PE, Griffin TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/S1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, Podkolodny NL, Kolchanov NA. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Li Y, Yan C, Callis-Duffin KP, Matsunami N, Garcia VE, Cargill M, Civello D, Bui N, Catanese JJ, Leppert MF, Krueger GG, Begovich AB, Schrodi SJ. Variants in the 5q31 cytokine gene cluster are associated with psoriasis. Genes Immun. 2008;9:176–181. doi: 10.1038/sj.gene.6364451. [DOI] [PubMed] [Google Scholar]
- Black S, Teixeira AS, Loh AX, Vinall L, Holloway JW, Hardy R, Swallow DM. Contribution of functional variation in the IL13 gene to allergy, hay fever and asthma in the NSHD longitudinal 1946 birth cohort. Allergy. 2009;64:1172–1178. doi: 10.1111/j.1398-9995.2009.01988.x. [DOI] [PubMed] [Google Scholar]
- Nedoszytko B, Niedoszytko M, Lange M, van Doormaal J, Glen J, Zablotna M, Renke J, Vales A, Buljubasic F, Jassem E, Roszkiewicz J, Valent P. Interleukin-13 promoter gene polymorphism -1112C/T is associated with the systemic form of mastocytosis. Allergy. 2009;64:287–294. doi: 10.1111/j.1398-9995.2008.01827.x. [DOI] [PubMed] [Google Scholar]
- Llanes E, Quiralte J, Lopez E, Sastre B, Chacartegui M, del Pozo V, Palomino P, Lahoz C, Cardaba B. Analysis of polymorphisms in olive pollen allergy: IL13, IL4RA, IL5 and ADRB2 genes. Int Arch Allergy Immunol. 2009;148:228–238. doi: 10.1159/000161583. [DOI] [PubMed] [Google Scholar]
- Cameron L, Webster RB, Strempel JM, Kiesler P, Kabesch M, Ramachandran H, Yu L, Stern DA, Graves PE, Lohman IC, Wright AL, Halonen M, Klimecki WT, Vercelli D. Th2 cell-selective enhancement of human IL13 transcription by IL13-1112C>T, a polymorphism associated with allergic inflammation. J Immunol. 2006;177:8633–8642. doi: 10.4049/jimmunol.177.12.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pouw Kraan TC, van Veen A, Boeije LC, van Tuyl SA, de Groot ER, Stapel SO, Bakker A, Verweij CL, Aarden LA, van der Zee JS. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun. 1999;1:61–65. doi: 10.1038/sj.gene.6363630. [DOI] [PubMed] [Google Scholar]