Abstract
Vitamin B12 (cobalamin) is among the largest known non-polymeric natural products, and the only vitamin synthesized exclusively by microorganisms1. The biosynthesis of the lower ligand of vitamin B12, 5,6-dimethylbenzimidazole (DMB), is poorly understood1–3. Recently, we discovered that a Sinorhizobium meliloti gene, bluB, is necessary for DMB biosynthesis4. Here we show that BluB triggers the unprecedented fragmentation and contraction of the bound flavin mononucleotide cofactor and cleavage of the ribityl tail to form DMB and D-erythrose 4-phosphate. Our structural analysis shows that BluB resembles an NAD(P)H-flavin oxidoreductase, except that its unusually tight binding pocket accommodates flavin mononucleotide but not NAD(P)H. We characterize crystallographically an early intermediate along the reaction coordinate, revealing molecular oxygen poised over reduced flavin. Thus, BluB isolates and directs reduced flavin to activate molecular oxygen for its own cannibalization. This investigation of the biosynthesis of DMB provides clarification of an aspect of vitamin B12 that was otherwise incomplete, and may contribute to a better understanding of vitamin B12-related disease.
The lower ligand of vitamin B12, DMB, is the only component of B12 for which no biosynthetic enzymes have been identified1–3. Previous studies using cell extracts have demonstrated the oxygen-dependent transformation of flavin mononucleotide (FMN) to DMB (Fig. 1a); however, the mechanism of conversion and enzymes involved in this pathway remain unknown3,5,6. Recently, a DMB auxo-troph of the symbiotic nitrogen-fixing bacterium Sinorhizobium melilotiwas identified4. This mutant, bluB, exhibited abnormal exo-polysaccharide structure, as indicated by a calcofluor-bright phenotype, and was deficient in free-living growth and symbiosis with its plant host. A putative B12-binding riboswitch immediately upstream of the bluB gene indicated a B12 biosynthetic role7; addition of vitamin B12 or DMB reversed all of the bluB mutant phenotypes4. BluB homologues exist in phylogenetically and metabolically diverse prokaryotic genomes, including notable pathogens of the genera Mycobacterium, Brucella and Vibrio, as well as the eukaryotic mouse malarial agent Plasmodium yoelii, indicating a ubiquitous role in DMB synthesis4,7.
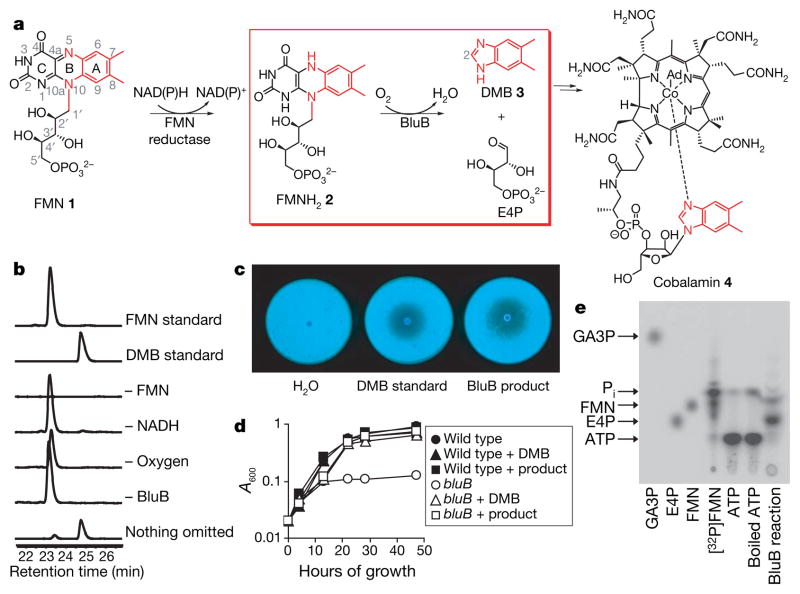
Figure 1. BluB catalyses DMB production.
a, Overall scheme for DMB biosynthesis and B12 formation. The atoms converted to DMB are shown in red; the reaction catalysed by BluB is in the red box. b, HPLC traces (280 nm) of reactions containing 10 μM BluB, 20 mM NADH and 100 μM FMN, or lacking the indicated components. Similar conversion was observed in reactions containing NADPH instead of NADH (not shown). In the absence of NAD(P)H, trace amounts of product formed, probably through adventitious photoreduction of FMN followed by turnover in the active site. c, Calcofluor fluorescence of the bluB mutant. d, Growth of wild-type S. meliloti and the bluB mutant. e, Radio-TLC of the BluB reaction containing [32P]FMN. Arrows represent migration of standards. The same result was obtained using two additional eluant systems (data not shown).
The protein sequence of BluB has ~16% identity to several enzymes of the nitroreductase/flavin oxidoreductase family. These enzymes catalyse the NAD(P)H-dependent reduction of flavin for diverse redox reactions8,9. On the basis of this sequence similarity and our previous findings, we proposed that BluB might catalyse the transformation of FMN to DMB. Indeed, high-performance liquid chromatography (HPLC) analysis reveals the BluB-dependent consumption of FMN and formation of DMB, as confirmed by co-elution, ultraviolet/visible spectroscopy, 1H NMR spectroscopy and mass spectrometry, by comparison with an authentic standard (Fig. 1b; see also Supplementary Fig. 1). Furthermore, two of the phenotypes originally observed for the bluB mutant—the calcofluor fluorescence and poor growth in minimal media—are also reversed by the BluB reaction product (Fig. 1c, d). The catalysed reaction requires oxygen, consistent with previous ex vivo data3,5,10 (Fig. 1b). BluB also consumes FMNH2 as a substrate in the absence of NAD(P)H (data not shown). Thus, FMNH2 is the true substrate for BluB, whereas molecular oxygen drives oxidative fragmentation of the heterocycle. NAD(P)H is only required initially to reduce FMN.
Kinetic characterization of BluB reveals a non-Michaelis–Menten profile, as the reaction is inhibited by high concentrations of FMN (Supplementary Fig. 2)11,12. The apparent Km for FMN (64 μM) is close to its physiological concentration, whereas inhibition occurs at non-physiological concentrations13 (Table 1). The apparent Km values for NADH and NADPH (5.1 and 4.4 mM, respectively) are 10–100-fold higher than their physiological concentrations and imply an unfavourable reductase activity13. These data suggest an alternative kinetic model in which FMNH2 is the substrate and NAD(P)H does not saturate the active site, but rather reduces FMN in solution.
Table 1.
Kinetic parameters of BluB-catalysed DMB synthesis
| Compound tested | kcat (apparent) (h−1)* | Km (apparent) (μM)* | Ki (μM)* |
|---|---|---|---|
| FMN† | 15 ± 3 | 64 ± 23 | 231 ± 83 |
| NADH‡ | 7.2 ± 0.7 | 5,100 ± 1,700 | NA |
| NADPH‡ | 7.6 ± 0.5 | 4,400 ± 1,100 | NA |
kcat, catalytic rate constant; Ki, inhibition constant; NA, not applicable.
Initial velocity data for DMB formation were fitted to the Michaelis–Menten equation using KaleidaGraph (Synergy Software). Parameters for FMN were calculated using a modified equation that takes into account substrate inhibition (Supplementary Fig. 2). Errors represent standard deviation.
Reactions contained 10 μM BluB, 40 mM NADH and FMN at concentrations ranging from 10 μM to 2 mM.
Reactions contained 10 μM BluB, 200 μM FMN and NAD(P)H at concentrations ranging from 1 to 40 mM.
BluB, unlike the closely related oxidoreductase enzyme family, does not catalyse NAD(P)H-dependent FMN reduction, whereas Escherichia coli SsuE, a well-characterized flavin reductase11, accelerates FMN reduction by at least 60-fold above background levels (Supplementary Table 1). Significant non-enzymatic reduction of FMN by NAD(P)H (Supplementary Table 1) enables BluB to turn over in vitro, whereas an accessory reductase probably delivers FMNH2 to BluB in vivo. Indeed, addition of SsuE to the BluB reaction increases the initial rate ~20-fold (Supplementary Table 2). The rate of DMB formation increases markedly (kobs = 3 min −1) in reactions containing SsuE and a concentration of FMN (1 μM) below BluB’s dissociation constant (Kd ≈ 2 μM) (Supplementary Fig. 3). These data suggest that, despite its apparent sequence homology, BluB is not a flavin reductase but may require a separate reductase for delivery of FMNH2 (ref. 9).
The remarkable contraction of the isoalloxazine ring of FMN to form DMB requires cleavage of the ribityl tail. Numerous studies have demonstrated that carbon C2 of DMB is derived from carbon C1′ of FMN3,6,14,15, and previously proposed mechanisms have suggested that the remainder of the ribityl moiety of FMN is released as either D-glyceraldehyde 3-phosphate (GA3P) and CO2 (refs 3,16) or D-erythrose 4-phosphate (E4P)10,17. Thin-layer chromatography (TLC) analysis of reactions containing [32P]FMN shows that the product mixture contains species co-migrating with E4P and FMN, but not GA3P, indicating that E4P is the major phosphorous-containing product (Fig. 1e). 31P-NMR confirms that E4P is formed, whereas GA3P is not detectable (Supplementary Fig. 4). Therefore, BluB catalyses the elimination of the ribityl tail of FMN by cleavage between C1′ and C2′ to form the four-carbon sugar E4P.
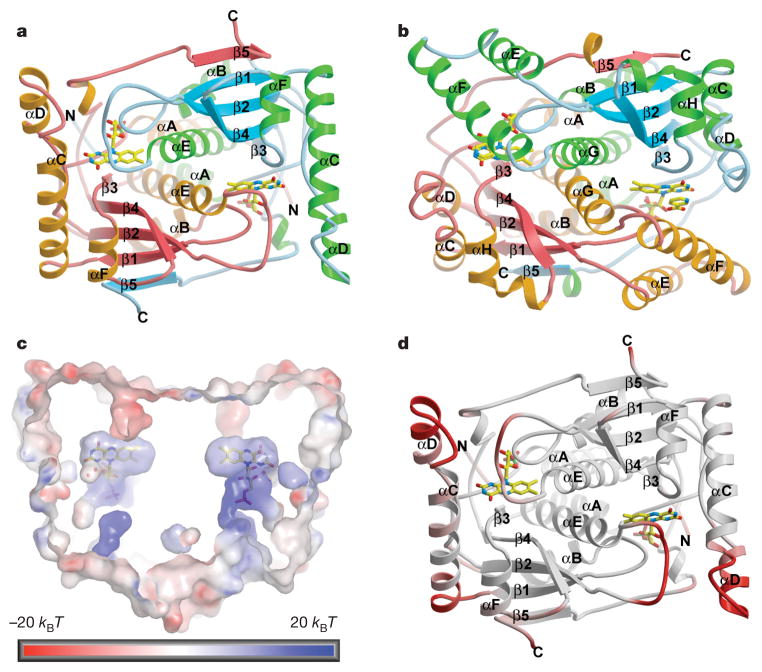
The crystal structure of BluB resembles the NAD(P)H-flavin oxido-reductases, which include flavin reductase (FRP) of Vibrio harveyi and Vibrio fischeri18,19, NADH oxidase (NOX) of Thermus thermophilus20, and the nitroreductases NfsA and NfsB of E. coli21 (Fig. 2a, b; see also Supplementary Fig. 5). The root mean square (r.m.s.) deviation between these enzymes and BluB is 2.1–3.8 Å over 60–80% of the structure, depending on the family member (Fig. 2a, b)22. Comparison of BluB to these structural homologues reveals a unique insertion in αC that significantly extends the αC-loop-αD motif to form a lid that completely buries FMN (Fig. 2a–c). In addition, there is a unique deletion after strand β2, replacing two helices (αE and αF in the typical reductase) with a short loop that further constricts the binding pocket (Fig. 2a, b; see also Supplementary Fig. 5). These unique elements appear to have flexibility, and thus provide gated access to the active site (Fig. 2d). Nevertheless, they conspire to isolate FMN from other substrates in an extremely snug, electropositive pocket (Fig. 2c; see also Supplementary Fig. 6). In contrast, the open, solvent-accessible active site in oxidoreductases facilitates promiscuous exchange of substrates (Fig. 2b)19,21. Thus, the active site of BluB carefully excludes interaction with any substrate other than dioxygen.
Figure 2. Structure of BluB.
a, Ribbon diagram of BluB with FMN in the binding pocket (stick representation). The two-fold axis is perpendicular to the plane of the figure. b, E. coli nitroreductase NfsB (Protein Data Bank 1ICR)21 with FMN and nicotinic acid (stick representation) in the binding pocket. c, Cross-section of BluB’s molecular surface. The two-fold axis lies along the y axis such that the si-face of FMN is viewed on the left and re-face on the right. The surface is coloured according to electrostatic potential, where blue is electropositive, red is electronegative and kB is Boltzmann’s constant. The back and front of the surface are cut away to reveal the FMN binding pocket buried in the dimer interface. The pocket wraps snugly around FMN, preventing interaction with other substrates. d, The BluB ribbon diagram has been coloured according to B-factor. Red represents flexible regions that may control or gate access to the active site.
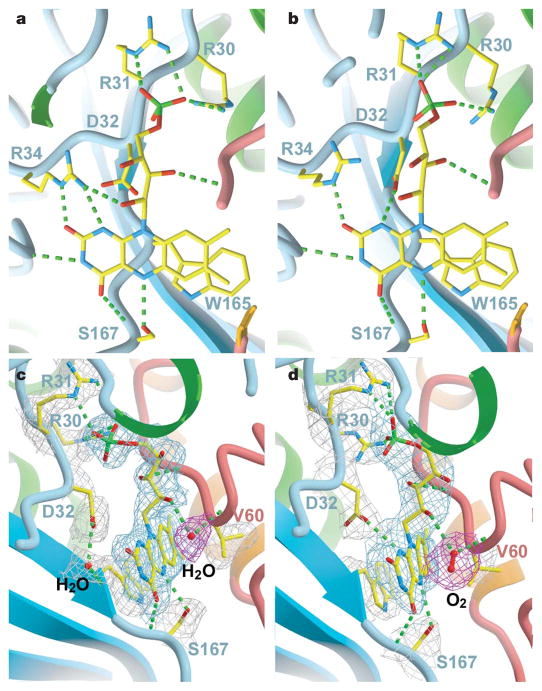
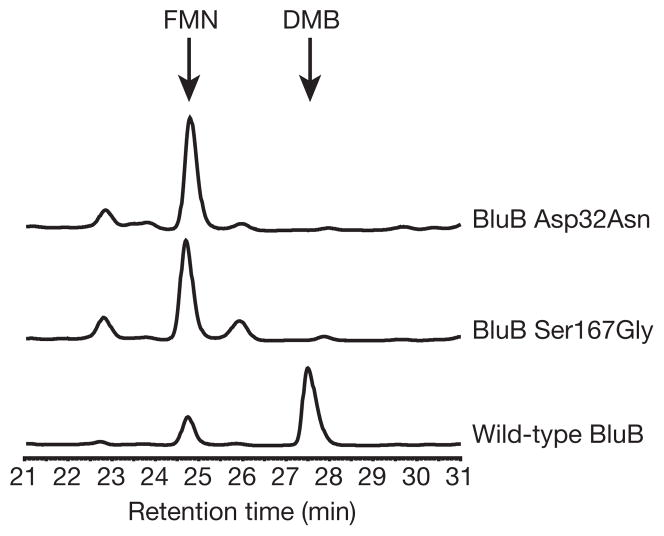
We obtained the crystal structure of BluB bound to substrate (FMNH2) and molecular oxygen by soaking crystals in the reducing agent dithionite and then back-soaking in oxygenated mother liquor. The slow turnover of BluB (Table 1) is retarded further because of the acidic crystallization buffer, enabling capture of a crystallographic snap-shot along the reaction coordinate. Notably, the dithionite-soaked crystals turn purple, probably representing a non-physiological, charge-transfer complex with the reduced flavin anion (Supplementary Fig. 7). On reintroduction of oxygen, the purple crystals become pale yellow or clear, with the spectroscopic signature of FMNH2 (Supplementary Fig. 7). These clear crystals disintegrate rapidly; evidently, turnover in the crystal is coupled to a structural rearrangement that disrupts the lattice. We successfully froze one such exceptionally large, pale-yellow crystal and collected diffraction data to 2.9 Å. The conformational difference between oxidized and reduced FMN is subtle (Fig. 3a, b). Notably, some of the H-bonds rearrange at the N1 position, which is protonated in the reduced form at acidic pH (Fig. 3; see also Supplementary Table 3). Asp 32 also shows conformational mobility, and is able to form a close contact with C1′ of the ribityl tail and N1 (Fig. 3c, d; see also Supplementary Table 3). This residue is conserved in BluB homologues4, but is not observed in oxidoreductases. Mutation of Asp 32 to Ala, Asn or Ser abrogates DMB formation but retains flavin binding, indicating that it may be essential for catalysis (Fig. 4).
Figure 3. Active site of BluB.
a, b, The active site with oxidized FMN (a) and reduced FMN (b), viewed from the re-face. H-bonds are represented as dashed lines. For clarity, water molecules are not rendered in this view. The rearrangement in H-bonds around N1 reflects the change in protonation in the reduced structure. Asp 32 may also form a close contact with C1′ of the ribityl chain, suggesting a potential catalytic role for this residue. c, d, Side views of the active site in the oxidized (c) and reduced (d) structures. For clarity, Arg 34 has not been rendered in this view. The sigma-A weighted 2Fo−Fc electron density map is contoured at 1σ and coloured grey (around protein side chains), blue (around FMN/FMNH2) and red (around water/oxygen) to enhance contrast.
Figure 4. Mutation of Asp 32 or Ser 167 eliminates DMB production.
HPLC traces (280 nm) of reactions performed with BluB proteins containing the point mutations Asp32Asn or Ser167Gly. The peaks corresponding to FMN and DMB, as confirmed by retention time and ultraviolet/visible spectra, are labelled. No DMB is detected in the Asp32Asn mutant; DMB produced by the Ser167Gly mutant is ~3% of the wild-type level. No DMB is detected in reactions containing BluB with Asp32Ala, Asp32Ser, or Ser167Cys point mutations (not shown). The size of the small peaks at 22.7 and 25.8 min in the reactions with mutant enzymes is reduced by addition of catalase, indicating that H2O2 is generated in reactions with mutant BluB, probably via futile cycling through a peroxyflavin intermediate (not shown).
A strong electron density peak appears over the re-face of reduced flavin, replacing a weaker peak (modelled as water) in the oxidized structure (Fig. 3c, d). Modelling this strong peak as water results in unacceptably low B-factors, whereas refinement as molecular oxygen accounts for the extra density and is consistent with the enzyme’s dependence on oxygen (Fig. 3d; see also Supplementary Table 4). This oxygen is poised for attack at C4a and anchored by two H-bonds from the O2′ hydroxyl and the backbone amide from residue 61. These H-bonds might serve as a ‘peroxyanion hole’ for stabilization of a reactive peroxyflavin intermediate, analogous to the oxyanion hole that stabilizes the tetrahedral intermediate in serine proteases and hydrolases23 (Fig. 3d). Such stabilization would deter uncoupling of hydrogen peroxide (H2O2), which occurs favourably in solution. Mono-oxygenases direct formation of a peroxyflavin intermediate for subsequent hydroxylation reactions, and also seem to use peroxyanion holes, which masquerade as two backbone amides, Arg or Asn24–26. Finally, the peroxyanion hole of BluB is defined, in part, by the orientation of the ribityl tail, which resembles that in oxidoreductases, but differs from mono-oxygenases, where the tail is swivelled away and sheltered from the site of oxygen attack24–27. Thus, BluB is a hybrid enzyme with structural similarity to the flavin oxidoreductases and functional relatedness to the mono-oxygenases.
BluB has additional unique features observed in neither the oxidoreductases nor the mono-oxygenases that may have critical functions in directing DMB formation. Asp 32 and Ser 167 are completely conserved in BluB homologues but have not been observed in oxidoreductases or mono-oxygenases8,19,21,24–27. Ser 167 forms an H-bond to N5 of flavin, whereas H-bonds to N5 are fulfilled by backbone amides in oxidoreductases and are seldom observed in mono-oxygenases. Consistent with a critical catalytic role, mutation of Ser 167 to Gly reduces DMB production ~30-fold, and mutation to Cys completely abolishes DMB formation (Fig. 4). Moreover, the H-bond to N5 in oxidoreductases typically forms from one face of the flavin, with the opposite face left unhindered for interaction with substrate and oxygen8,19,21. In contrast, BluB appears to donate this H-bond and bind oxygen from the same face (Fig. 3c, d). This stereochemistry may serve as an additional control mechanism to prevent unwanted reactions from all but the smallest possible substrates, namelymolecular oxygen.
DMB formation involves controlled breakage of three bonds and formation of one, facilitating contraction of the central six-membered ring of FMNH2 to form the five-membered ring of DMB. A mechanistic proposal for this transformation must incorporate the following findings of this study: (1) requirement for molecular oxygen; (2) rupture of the ribityl side chain between C1′ and C2′ to yield E4P; (3) absence of a requirement for NAD(P)H; and (4) the critical roles of Asp 32 and Ser 167 in catalysis. Furthermore, earlier observations using cell extracts showed that C2 of DMB is derived from C1′ of FMN6,14,16, and that urea is produced from this reaction3. At this point, it is not possible to provide a detailed mechanism for this transformation. Rather, we postulate a framework to explain the fragmentation and reconstruction of FMNH2 to form DMB (see ‘Discussion’ and ‘Scheme 1’ in Supplementary Information). The closed active site strongly indicates an intramolecular route for the overall reaction, requiring not only a difficult chemical transformation but also major conformational change. Such rearrangement of the protein structure is a probable explanation for the observed fragmentation of BluB crystals on introduction of dioxygen.
BluB is thus the first characterized enzyme responsible for the formation of DMB, hence elucidating the last unknown step in the biosynthesis of B12. Moreover, this investigation has revealed a novel enzymatic activity in the fragmentation of a flavin coenzyme. Destruction of one cofactor to form another is unusual, but has parallels with the cannibalization of iron–sulphur clusters in the biosynthesis of biotin (vitamin H) and lipoamide coenzymes28,29. The biochemical and structural novelty of BluB and the widespread distribution of BluB homologues throughout both prokaryotic kingdoms call for a new enzyme classification. Therefore, we propose that this enzyme belongs to a new ‘flavin destructase’ family.
METHODS
The procedures for protein expression, purification, crystallization, structure determination and refinement are described in Supplementary Methods.
Enzymatic reactions
For endpoint assays, reactions were assembled in an anaerobic glove box, exposed to ambient O2 (except where indicated), and quenched with 6.4% trichloroacetic acid after overnight incubation at room temperature. Reaction buffer contained 50 mM HEPES pH 7.5 and 1 mM dithiothreitol. Two-hundred-microlitre samples were analysed by RP-HPLC with a C18 column (Beckman Coulter) (11% acetonitrile, 0.1% TFA in water, 1 ml min−1), monitoring absorbance at 280 and 450 nm. The peak identities were confirmed using a coupled ultraviolet/visible diode array detector. Standards contained 100 μM FMN or DMB. For rate measurements, 250 μl aliquots from 500 μl reactions were quenched at two different time points, and reaction rates for the two time points were averaged. DMB concentrations were calculated based on integrated peak areas at 280 nm compared to a standard curve. His-tagged SsuE protein was purified as described11.
Biological assays
For calcofluor analysis, 10 μl of 10 μM DMB standard, BluB reaction product, or water was added to a sterile filter disk on LB-calcofluor plates containing a lawn of an S. meliloti bluB mutant strain4. For growth measurements, M9 medium containing cobalt4 with nothing added, 100 nM DMB standard, or 100 nM BluB reaction product was inoculated with wild-type S. meliloti strain Rm1021 or the S. meliloti bluB mutant, and absorbance at 600 nm (A600) was monitored over time.
Radio-TLC
The procedure for [32P]FMN production by riboflavin kinase is described in Supplementary Methods. Reactions containing 10 μM [32P]FMN, 5 mM NADPH, 1 μM SsuE and 10 μM BluB were incubated for 2 h at 30 °C and quenched by passing the reactions through 30,000 MWCO filters (Microcon). The reactions were spotted on cellulose-TLC plates and developed with 3:1:6 n-butanol:acetic acid:water. The unlabelled GA3P and E4P standards included on the plate were stained with ceric ammonium molybdate, and ~1 nCi [γ-32P]ATP was overlaid onto the stained spots. Plates were visualized by phosphorimaging.
Supplementary Material
Acknowledgments
This work was supported by NIH grants to G.C.W. and C.T.W. and postdoctoral fellowships from the Jane Coffin Childs Memorial Fund for Medical Research to M.E.T. and N.A.L. G.C.W. is an American Cancer Society Research Professor. We are grateful to E. Yeh for providing purified SsuE and H. Zhang for purified riboflavin kinase. We thank C. Sheahan and G. Heffron for their assistance with 31P-NMR, and A. Haykov for assistance with protein purification. We acknowledge the Advanced Light Source for beam time. We thank S. Harrison, T. Begley, C. Drennan and members of the Walsh and Walker laboratories for helpful discussions.
Footnotes
Author Information The coordinates and structure factors for BluB-FMN, BluB-FMNA (flavin anion) and BluB-FMNH2 have been deposited in the Protein Data Bank under accession codes 2ISJ, 2ISK and 2ISL, respectively.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 2.Warren MJ. Finding the final pieces of the vitamin B12 biosynthetic jigsaw. Proc Natl Acad Sci USA. 2006;103:4799–4800. doi: 10.1073/pnas.0601030103. published online 27 March 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renz P. In: Chemistry and Biochemistry of B12. Banerjee R, editor. John Wiley & Sons; New York: 1999. pp. 557–575. [Google Scholar]
- 4.Campbell GRO, et al. Sinorhizobium meliloti bluB is necessary for production of 5,6-dimethylbenzimidazole, the lower ligand of B12. Proc Natl Acad Sci USA. 2006;103:4634–4639. doi: 10.1073/pnas.0509384103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renz P. Riboflavin as precursor in the biosynthesis of the 5,6- dimethylbenzimidazole-moiety of vitamin B12. FEBS Lett. 1970;6:187–189. doi: 10.1016/0014-5793(70)80053-9. [DOI] [PubMed] [Google Scholar]
- 6.Renz P, Weyhenmeyer R. Biosynthesis of 5,6-dimethylbenzimidazole from riboflavin. Transformation of C-1′ of riboflavin into C-2 of 5,6-dimethylbenzimidazole. FEBS Lett. 1972;22:124–126. doi: 10.1016/0014-5793(72)80236-9. [DOI] [PubMed] [Google Scholar]
- 7.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. published online 17 July 2003. [DOI] [PubMed] [Google Scholar]
- 8.Fraaije MW, Mattevi A. Flavoenzymes: diverse catalysts with recurrent features. Trends Biochem Sci. 2000;25:126–132. doi: 10.1016/s0968-0004(99)01533-9. [DOI] [PubMed] [Google Scholar]
- 9.Tu SC. Reduced flavin: donor and acceptor enzymes and mechanisms of channeling. Antioxid Redox Signal. 2001;3:881–897. doi: 10.1089/15230860152665046. [DOI] [PubMed] [Google Scholar]
- 10.Horig JA, Renz P. Biosynthesis of vitamin B12. Some properties of the 5,6- dimethylbenzimidazole-forming system of Propionibacterium freudenreichii and Propionibacterium shermanii. Eur J Biochem. 1980;105:587–592. doi: 10.1111/j.1432-1033.1980.tb04536.x. [DOI] [PubMed] [Google Scholar]
- 11.Eichhorn E, van der Ploeg JR, Leisinger T. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J Biol Chem. 1999;274:26639–26646. doi: 10.1074/jbc.274.38.26639. [DOI] [PubMed] [Google Scholar]
- 12.Lei B, Tu SC. Mechanism of reduced flavin transfer from Vibrio harveyi NADPH-FMN oxidoreductase to luciferase. Biochemistry. 1998;37:14623–14629. doi: 10.1021/bi981841+. [DOI] [PubMed] [Google Scholar]
- 13.Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 14.Lingens B, Schild TA, Vogler B, Renz P. Biosynthesis of vitamin B12. Transformation of riboflavin 2H-labeled in the 1′R position of 1′S position into 5,6-dimethylbenzimidazole. Eur J Biochem. 1992;207:981–985. doi: 10.1111/j.1432-1033.1992.tb17133.x. [DOI] [PubMed] [Google Scholar]
- 15.Keck B, Munder M, Renz P. Biosynthesis of cobalamin in Salmonella typhimurium: transformation of riboflavin into the 5,6-dimethylbenzimidazole moiety. Arch Microbiol. 1998;171:66–68. doi: 10.1007/s002030050679. [DOI] [PubMed] [Google Scholar]
- 16.Kolonko B, Horig JA, Renz P. Transformation of [5′-3H]riboflavin into 5,6-dimethylbenzimidazole. Z Naturforsch C. 1992;47:171–176. doi: 10.1515/znc-1992-3-401. [DOI] [PubMed] [Google Scholar]
- 17.Maggio-Hall LA, Dorrestein PC, Escalante-Semerena JC, Begley TP. Formation of the dimethylbenzimidazole ligand of coenzyme B12 under physiological conditions by a facile oxidative cascade. Org Lett. 2003;5:2211–2213. doi: 10.1021/ol034530m. [DOI] [PubMed] [Google Scholar]
- 18.Tanner JT, Lei B, Tu SC, Krause KL. Flavin reductase P: structure of a dimeric enzyme that reduces flavin. Biochemistry. 1996;35:13531–13539. doi: 10.1021/bi961400v. [DOI] [PubMed] [Google Scholar]
- 19.Koike H, et al. 1.8 Å crystal structure of the major NAD(P)H:FMN oxidoreductase of a bioluminescent bacterium, Vibrio fischeri: overall structure, cofactor and substrate-analog binding, and comparison with related flavoproteins. J Mol Biol. 1998;280:259–273. doi: 10.1006/jmbi.1998.1871. [DOI] [PubMed] [Google Scholar]
- 20.Hecht HJ, Erdmann H, Park HJ, Sprinzl M, Schmid RD. Crystal structure of NADH oxidase from Thermus thermophilus. Nature Struct Mol Biol. 1995;2:1109–1114. doi: 10.1038/nsb1295-1109. [DOI] [PubMed] [Google Scholar]
- 21.Lovering AL, Hyde EI, Searle PF, White SA. The structure of Escherichia coli nitroreductase complexed with nicotinic acid: three crystal forms at 1.7 Å, 1.8 Å and 2.4 Å resolution. J Mol Biol. 2001;309:203–213. doi: 10.1006/jmbi.2001.4653. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 23.Larsen NA, Lin H, Wei R, Fischbach MA, Walsh CT. Structural characterization of enterobactin hydrolase IroE. Biochemistry. 2006;45:10184–10190. doi: 10.1021/bi060950i. [DOI] [PubMed] [Google Scholar]
- 24.Eswaramoorthy S, Bonanno JB, Burley SK, Swaminathan S. Mechanism of action of a flavin-containing monooxygenase. Proc Natl Acad Sci USA. 2006;103:9832–9837. doi: 10.1073/pnas.0602398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gatti DL, Entsch B, Ballou DP, Ludwig M. L pH-dependent structural changes in the active site of p-hydroxybenzoate hydroxylase point to the importance of proton and water movements during catalysis. Biochemistry. 1996;35:567–578. doi: 10.1021/bi951344i. [DOI] [PubMed] [Google Scholar]
- 26.Malito E, Alfieri A, Fraaije MW, Mattevi A. Crystal structure of a Baeyer–Villiger monooxygenase. Proc Natl Acad Sci USA. 2004;101:13157–13162. doi: 10.1073/pnas.0404538101. published online 24 August 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong C, et al. Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science. 2005;309:2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cicchillo RM, Booker SJ. Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide. J Am Chem Soc. 2005;127:2860–2861. doi: 10.1021/ja042428u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.