Summary
Infection of legumes by Rhizobium sp. NGR234 and subsequent development of nitrogen-fixing nodules are dependent on the coordinated actions of Nod factors, proteins secreted by a type III secretion system (T3SS) and modifications to surface polysaccharides. The production of these signal molecules is dependent on plant flavonoids which trigger a regulatory cascade controlled by the transcriptional activators NodD1, NodD2, SyrM2 and TtsI. TtsI is known to control the genes responsible for T3SS function and synthesis of a symbiotically important rhamnose-rich lipo-polysaccharide, most probably by binding to cis elements termed tts boxes. Eleven tts boxes were identified in the promoter regions of target genes on the symbiotic plasmid of NGR234. Expression profiles of lacZ fusions to these tts boxes showed that they are part of a TtsI-dependent regulon induced by plant-derived flavonoids. TtsI was purified and demonstrated to bind directly to two of these tts boxes. DNase I footprinting revealed that TtsI occupied not only the tts box consensus sequence, but also upstream and downstream regions in a concentration-dependent manner. Highly conserved bases of the consensus tts box were mutated and, although TtsI binding was still observed in vitro, gfp fusions were no longer transcribed in vivo. Random mutagenesis of a tts box-containing promoter revealed more nucleotides critical for transcriptional activity outside of the consensus.
Introduction
Symbioses between legumes and rhizobia which result in the formation of nitrogen-fixing root nodules are the result of a complex signal exchange between both partners. Initially, flavonoids exuded by the plant trigger synthesis of Nod factors (NF) that are secreted from the bacteria and are critical for rhizobial infection (Broughton et al., 2000; Perret et al., 2000). Nevertheless, establishment of functional nitrogen-fixing nodules requires other bacterial signals such as surface polysaccharides and secreted proteins (Fraysse et al., 2003; Broughton et al., 2006; Soto et al., 2006; Jones et al., 2007). Rhizobium sp. NGR234 (hereafter NGR234) is the most promiscuous known microsymbiont, capable of establishing symbioses with more than 112 genera of legumes (Pueppke and Broughton, 1999). Its genome is partitioned into three replicons: the chromosome and two large plasmids, pNGR234a and pNGR234b (Viprey et al., 2000). pNGR234a is also called the symbiotic plasmid as it contains all genes necessary for NF synthesis and nitrogen fixation. Sequencing of this 536-kb plasmid showed that it also contains orthologues of a type III protein secretion system (T3SS) (Freiberg et al., 1997). Until this discovery, such secretion systems were thought to be characteristic of pathogenic bacteria, where they play important roles in host infection. T3SS form an apparatus that injects bacterial proteins directly into eukaryotic cells to disrupt normal functioning of the cell, facilitating infection (Hueck, 1998). The T3SS of NGR234 is capable of secreting Nops (nodulation outer proteins), and is an important determinant of host range (Viprey et al., 1998).
Production of NF requires flavonoids and the LysR-type regulator NodD1, which binds to conserved motifs (termed nod boxes) found in the promoter regions of genes/operons responsible for NF synthesis. Nop secretion also requires flavonoids, NodD1 and another regulatory protein TtsI (Viprey et al., 1998; Marie et al., 2004). A nod box is located in the promoter region of ttsI, and NodD1 is thought to activate TtsI which in turn initiates transcription of T3SS genes (Kobayashi et al., 2004). TtsI shares characteristics with the DNA-binding response regulators of two-component regulatory systems (Viprey et al., 1998; Marie et al., 2004). Usually, such regulators are activated by their partner sensor, histidine protein kinases, which auto-phosphorylate at a histidine residue upon sensing an environmental signal. The phosphoryl group is subsequently transferred to an aspartate residue in the response regulator, inducing a conformational change that leads to its activation. Once phosphorylated, response regulators act as transcriptional activators by binding to cis elements in the promoters of genes required to process the initial environmental signal. TtsI, however, has a glutamate residue instead of the conserved aspartate. In other bacterial response regulators, such a substitution leads to constitutive activation, bypassing the requirement for the sensor kinase partner. It is thus possible that TtsI functions as a transcriptional activator independent of phosphorylation and a sensor kinase partner. Instead, transcription of ttsI and therefore function(s) regulated by TtsI, are modulated by NodD1 in a flavonoid-dependent manner (Kobayashi et al., 2004; Marie et al., 2004).
Sequencing numerous rhizobial genomes has revealed that T3SS and TtsI control are relatively common. A comparison of the promoter regions of T3SS genes from several rhizobia identified a putative cis-regulatory element termed a tts box (TB). TtsI is thought to bind TB and stimulate transcription of downstream genes (Krause et al., 2002). Using this consensus sequence, 11 putative TBs (TB1–TB11) were identified on pNGR234a (Marie et al., 2004). Five of the TBs are located upstream of genes/operons involved in the assembly of the type III secretion machine; others precede genes encoding possible secreted proteins. TBs are also found in the promoters of genes encoding proteins not directly related to T3SS functions (Fig S1). Thus TtsI potentially regulates more than the T3SS, and indeed mutation of ttsI leads to different symbiotic phenotypes compared to a T3SS mutant alone (Viprey et al., 1998). As well as being impaired in protein secretion, the ttsI mutant failed to produce a rhamnose-rich lipo-polysaccharide (LPS) known to be important for successful nodulation (Marie et al., 2004; Reuhs et al., 2005; Broughton et al., 2006). A flavonoid-inducible operon encoding enzymes responsible for rhamnose synthesis was shown to require TtsI for activation and to contain a TB in its promoter region. Evidence that the TB is essential for the activity of this operon was obtained by deleting a small region containing the TB which abolished TtsI-mediated induction (Marie et al., 2004).
In this work, we determined whether the promoter regions containing the 11 predicted TB are inducible in a TtsI- and flavonoid-dependent manner. We then tested if TtsI could physically bind to TB-containing promoters and mapped the actual binding site in vitro by DNaseI footprinting. We also mutated key residues in the TB consensus sequence and randomly mutated a TB-containing promoter to identify further important residues. It seems likely that the TtsI/TB regulatory system is a basic feature of rhizobial T3SS as Bradyrhizobium japonicum USDA110, Mesorhizobium loti MAFF303099 and Sinorhizobium fredii USDA257 all possess T3SS, TtsI homologues, as well as predicted TB sequences (Krause et al., 2002; Krishnan et al., 2003; Hubber et al., 2004). Thus our findings are applicable to multiple genera of rhizobia.
Results
NGRΔttsI, a non-polar deletion mutant of ttsI
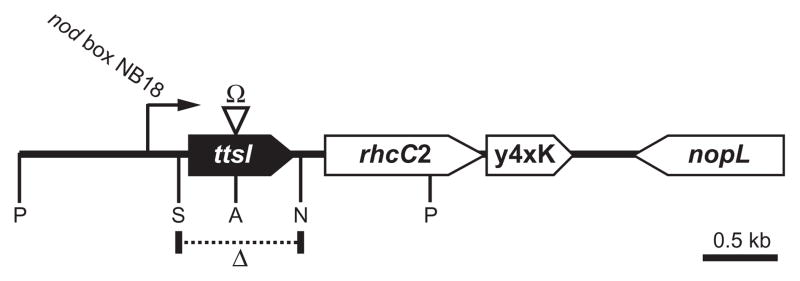
Polar mutation of ttsI (NGRΩttsI) demonstrated that TtsI is required for Nop secretion and synthesis of a rhamnose-rich polysaccharide, as well as the transcriptional activation of two of the 11 TBs, TB2 and TB8. Although introduction of a plasmid-born copy of ttsI into NGRΩttsI allowed complementation of rhamnan synthesis, it failed to restore Nop secretion (Viprey et al., 1998; Marie et al., 2004). As ttsI, rhcC2 and y4xK are predicted to form an operon (Perret et al., 2003), the insertion of an Ω cassette into ttsI most probably blocked transcription of these downstream genes. In other bacteria, rhcC2 encodes an essential component of the T3SS: its addition in trans (possibly y4xK as well) would thus be required for complementation of Nop secretion by NGRΩttsI. Additional regulatory controls of T3SS gene expression have been shown in some bacteria: if the machinery fails to assemble correctly (because of mutation of a key gene for example), then expression of genes encoding secreted proteins is suppressed (Wei et al., 2000). It is thus possible that the block in TB expression in NGRΩttsI is due to the absence of RhcC2, causing the T3SS assembly machine to fail. In this case, TB expression was probably suppressed by another regulator. For this reason, we constructed a non-polar deletion mutant of ttsI (NGRΔttsI) to maintain transcription of rhcC2 and y4xK from nod box 18 and thus avoid this possibility (Fig. 1). To characterize the new mutant, secreted proteins and surface polysaccharides were isolated from NGRΔttsI and NGR234. Nop secretion and de novo synthesis of rhamnose-rich LPS were blocked in NGRΔttsI (Fig. S2), consistent with our previous observations of the polar mutant NGRΩttsI (Marie et al., 2004). To complement NGRΔttsI, ttsI with its own promoter region was subcloned into pRG960 (Van den Eede et al., 1992), giving rise to pttsI-2. Introduction of pttsI-2 into NGRΔttsI restored both Nop secretion and production of rhamnose-rich LPS. Thus NGRΔttsI does not appear to dramatically affect transcription of rhcC2 (or y4xK) and this mutant was used in all subsequent work.
Fig. 1.
Physical map of the ttsI locus of pNGR234a. Restriction sites are as follows: ApaI (A); NheI (N); PstI (P); SacII (S). Location of the deleted region in NGRΔttsI is shown by Δ and the site of the omega cassette insertion in NGRΩttsI shown by Ω.
Promoter activities of TB-containing loci
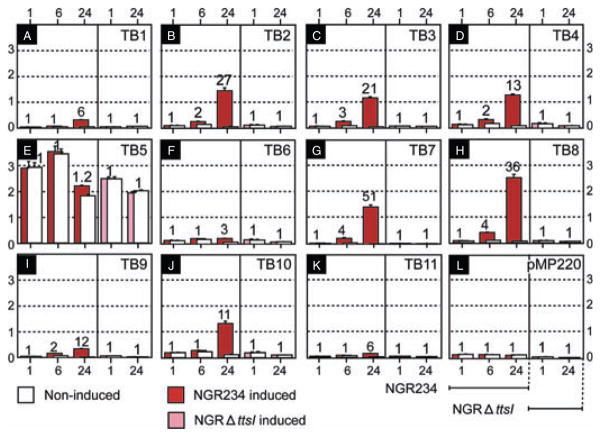
To examine the function of all TBs, we subcloned the 11 predicted TBs into pMP220, a broad-host-range transcriptional-lacZ reporter system (Spaink et al., 1987), thus creating pMP-TB1 to pMP-TB11 (see Table 1). To assess the flavonoid and TtsI dependence on transcription of the TB, tri-parental matings were used to introduce each of the constructs into NGR234 and NGRΔttsI. Liquid cultures were grown to an OD600 of 0.1 in RMS and the flavonoid daidzein added at 2 × 10−7 M. At 1, 6 and 24 h post induction (hpi), β-galactosidase activities were monitored in transconjugants of NGR234 (red bars in Fig. 2) or at 1 and 24 hpi for NGRΔttsI transconjugants (pink bars in Fig. 2). In the absence of an inducer, and with the exception of pMP-TB5, only low promoter activities were observed with the different constructs (open bars in Fig. 2), but addition of daidzein caused significant increases in β-galactosidase activities 24 hpi (Fig. 2). At 24 hpi, TB8, which controls expression of the nopBrhcJnolUVrhcNy4yJrhcQRSTU operon, is the strongest promoter (2500 ± 130 Miller units), whereas the lowest activity was recorded with TB11 (157 ± 5 units). Although expression of TB11 appears to be low, it represents a sixfold induction over that found in the NGRΔttsI mutant (Fig. 2K). By mobilizing the different TB–lacZ fusions into the mutant strain NGRΔttsI, the role of TtsI in the activation of each individual TB was assessed. All TBs lost flavonoid inducibility in NGRΔttsI (Fig. 2). Introduction of pttsI-2 into four randomly selected NGRΔttsI (pMP-TB2, TB4, TB8 and TB10) transconjugants restored flavonoid-dependent induction (closed bars, Fig. S3).
Table 1.
Bacterial strains and plasmids and primers.
| Relevant characteristics | Reference | |
|---|---|---|
| Strains | ||
| NGR234 | RifR derivative of the isolate NGR234 | Lewin et al. (1990) |
| NGRΔttsI | ttsI deletion mutant of NGR234 RifR | This work |
| NGRΔrmlB-wbgA | Rhamnose synthesis mutant of NGR234, RifR, KmR | Marie et al. (2004) |
| Plasmids | ||
| pBluescript II | KS+ Phage f1, lacZ, ApR | Stratagene |
| pUC::ttsI | pUC18 containing ttsI and its promoter as a 2.4-kb PstI fragment | Marie et al. (2004) |
| pJQ200mp18 | Suicide vector used in directed mutagenesis, GmR | Quandt and Hynes (1993) |
| pJQΔttsI | As above, but carrying ΔttsI fragment, subcloned into PstI site, GmR | This work |
| pRK2013 | Tra+ helper plasmid for mobilisation, KmR | Ditta et al. (1980) |
| pRG960 | IncQ vector containing promoter-less uidA, SpR | Van den Eede et al. (1992) |
| pttsI-2 | pRG960 containing ttsI and its promoter as a 2.4 kb PstI fragment | This work |
| pMP220 | IncP vector containing promoter-less lacZ, TetR | Spaink et al. (1987) |
| pMP-TB1 | TB1 cloned in pMP220 as a XbaI-PstI fragment | This work |
| pMP-TB2 | Also called pMP220-rmlB; TB2 cloned as a BamHI fragment in pMP220 | Marie et al. (2004) |
| pMP-TB3 | TB3 cloned in pMP220 as a XbaI-PstI fragment | This work |
| pMP-TB4 | TB4 cloned in pMP220 as a XbaI-PstI fragment | This work |
| pMP-TB5 | TB5 cloned in pMP220 as a XbaI-PstI fragment | This work |
| pMP-TB6 | TB6 cloned in pMP220 as a XbaI-PstI fragment | This work |
| pMP-TB7 | TB7 cloned in pMP220 as a XbaI-PstI fragment | This work |
| pMP-TB8 | Also called pMP220-B; TB8 cloned as a XbaI-PstI fragment in pMP220 | Marie et al. (2004) |
| pMP-TB9 | TB9 cloned in pMP220 as a XbaI-PstI fragment | This work |
| pMP-TB10 | TB10 cloned in pMP220 as a XbaI-PstI fragment | This work |
| pMP-TB11 | TB11 cloned in pMP220 as a XbaI-PstI fragment | This work |
| pPROBE-GT | pVS1/p15a vector, GmR | Miller, 2000 |
| pPROBE-GT′ | As above but with inverted MCS | Miller, 2000 |
| pGT-nopB | TB8 region in pPROBE-GT′ as a HindIII fragment | This work |
| pGT-GAATG | Site-directed mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-A117 | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-A2D | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-A3G | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-A518 | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-A62 | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-A74 | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-A812 | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-A9BG | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-B312 | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-B620 | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| pGT-B1127 | Random mutation of TB8 region in pPROBE-GT as an EcoRI fragment | This work |
| Primers | ||
| TB1-F | 5′-tctcatctctagaccaatcggcg-3′ | |
| TB1-R | 5′-cctgcagcgatattgtctcctcg-3′ | |
| TB3-F | 5′-tctagagccgtcagtcatctttcg-3′ | |
| TB3-R | 5′-tgtcctgcagatcgttgatgagg-3′ | |
| TB4-F | 5′-tctagacgatggcgatgttgctc-3′ | |
| TB4-R | 5′-acgactgcaggccttcaagatgg-3′ | |
| TB5-F | 5′-tgctctagagcaccggaagc-3′ | |
| TB5-R | 5′-tcctgcaggggaaagtagcatc-3′ | |
| TB6-F | 5′-cctctagagcctgtcttttctcg-3′ | |
| TB6-R | 5′-gctgcaggcttgcgtttagtgg-3′ | |
| TB7-F | 5′-tcgtctagacgttgaacggtctac-3′ | |
| TB7-R | 5′-aaggctgcaggccgacattgtg-3′ | |
| TB9-F | 5′-tctctagatggcgtcaaatgctgc-3′ | |
| TB9-R | 5′-ttctgcagccatttgcttgctgg-3′ | |
| TB10-F | 5′-catctagactgagagagcttcacg-3′ | |
| TB10-R | 5′-acctgcagctactcctgccttag-3′ | |
| TB11-F | 5′-cgaatctagacgactttcgatcgc-3′ | |
| TB11-R | 5′-gactctgcagggcgttcgtttccc-3′ | |
| rmlB-F | 5′-aagcaccccgaaaactacct-3′ | |
| rmlB-R | 5′-gcattgcgaaatttggatgga-3′ | |
| nopB-F | 5′-ctcgtcttgataaaccaaatctgaa-3′ | |
| nopB-R | 5′-ggactcgattacttaactctttgac-3′ | |
| TB8-F mut | 5′-ggccggtagaatgcgtgtcgtcagctcgcctc-3′ | |
| TB8-R mut | 5′-ggaggcgagctgacgacacgcattctaccggc-3′ | |
Fig. 2.
Expression analyses of TBs in pNGR234a. Flavonoid inducibility of TBs in the wild-type and NGRΔttsI backgrounds. A–L represent the levels of β-galactosidase activity (× 103 Miller’s units). Activity of the vector pMP220 devoid of an insert is reported in L. Assays were performed 1, 6 and 24 hpi with 2 × 10−7 M daidzein. In the absence of an inducer, basal levels of β-galactosidase activity are shown as open bars. Values obtained with induced transconjugants are coded as: NGR234 (red bars); NGRΔttsI (pink bars). The values reported represent the means of three independent experiments. Error bars are shown on top of each column. Numbers on top of each bar represent the relative increase in activity of induced- as compared with non-induced cultures.
TtsI binds to TB2 and TB8
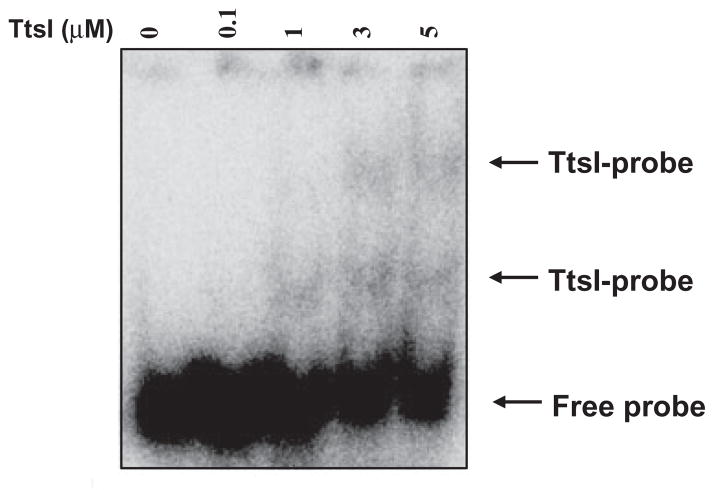
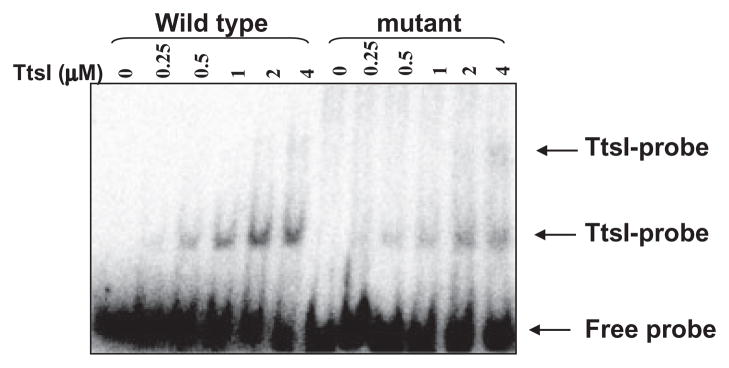
TtsI contains an N-terminal conserved receiver domain and a C-terminal helix-turn-helix (HTH) domain, both typical of response regulators of two-component systems. The HTH domain usually interacts directly with a DNA binding site present in the promoters of genes controlled by these regulators. Although the HTH structure is highly conserved, it has the ability to recognize specific bases, and thus discriminate small modifications in a binding site (Stock et al., 2000). TtsI of NGR234 and other rhizobia has been shown to control the expression of several genes containing a TB in the promoter region, but direct contact had never been shown. To further characterize the binding of TtsI to the TB-containing promoter regions, we cloned the NGR234 ttsI coding sequence into the pETBlue2 vector, then overexpressed and purified a His-tagged TtsI protein. High levels of expression were obtained, but more than 50% of the TtsI-His was insoluble, which was also observed with other members of this family (Kumagai et al., 2006). To test the ability of TtsI-His to bind to TB sequences, we initially performed electrophoretic mobility shift assays using a 150-bp region of the nopB promoter that contains TB8. When increasing amounts of TtsI-His were added to the reaction, slower migrating bands were observed, consistent with DNA–protein interactions (Fig. 3). The slowest band appeared only at increasing concentrations of TtsI, which could be a result of oligomerization of TtsI-His upon binding to DNA, or the presence of more than one TB at different locations in the DNA. Inspection of the sequence did not reveal any other candidate region, suggesting that TtsI forms oligomers upon binding TB8. Addition of up to 50 μg ml−1 poly(dI-dC) as heterologous competitor DNA did not disrupt the DNA–protein complexes, suggesting that binding of TtsI to TB8 is specific (Fig. S4). This specificity was confirmed using a double-stranded oligonucleotide specific to TB8 which disrupted TtsI–nopB promoter complexes (data not shown). A second double-stranded oligonucleotide composed of the adjacent sequence upstream of TB8 (originally designed as a control) also disrupted the complexes, however. This unexpected effect suggested that the TtsI binding region extends outside of the TB consensus sequence and, for this reason, we mapped exactly where TtsI bound to the nopB promoter.
Fig. 3.
TtsI binding to the nopB (TB8) promoter. Mobility shift assay using a 32P-labelled 150-bp PCR fragment containing the nopB promoter region. Purified TtsI-His was added at increasing concentrations, incubated for 20 min and subject to electrophoresis under native conditions.
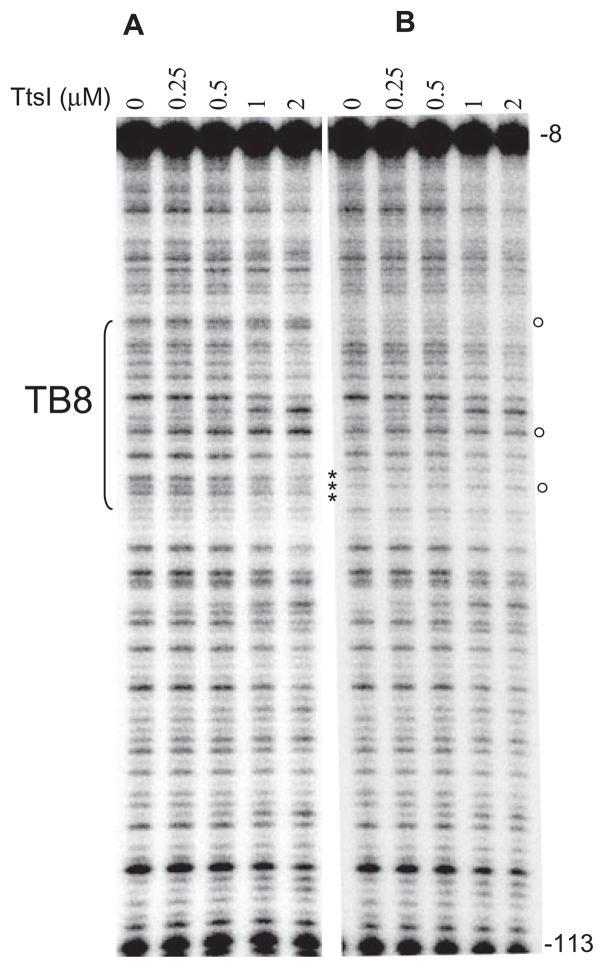
The identification of the precise interaction site of TtsI with the nopB promoter was determined by DNase I cleavage protection (footprinting) assays in which evidence of protein binding is seen as modifications of the cleavage pattern that result in both decreases and increases in the intensity of cleaved fragments. When the polymerase chain reaction (PCR) fragment containing the TB8 site used in the mobility shift assays described above was labelled and used as the probe, it clearly interacted with TtsI-His as revealed by the presence of protected and hyperreactive bands in the consensus sequence (Fig. 4A). Increasing concentrations of TtsI produced a clearer pattern of interaction, but did not reveal any new interaction sites. Furthermore, TtsI caused a modification to the cleavage pattern of bases located upstream and downstream of the TB, suggesting that it occupies a broad section of the target DNA (Fig. 4A). This result is consistent with oligomerization of TtsI upon binding to DNA, as suggested by the mobility shift assays. The hyperreactive bands observed are an indication that TtsI induces major distortions in the bound DNA, exposing these sites to DNase I cleavage.
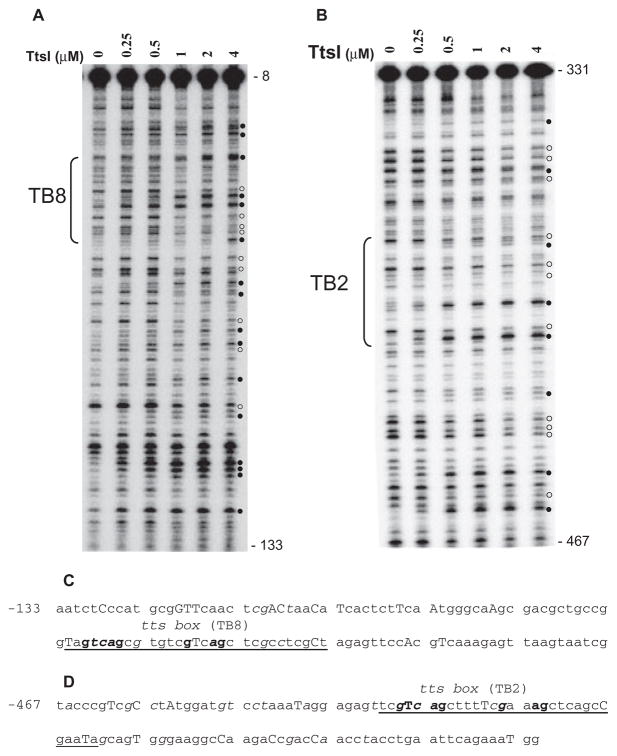
Fig. 4.
DNase I footprints of TtsI bound to the nopB (TB8) and rmlB (TB2) promoter regions. 32P-labelled PCR fragments were incubated with different concentrations of TtsI and digested using limiting concentrations of DNase I. After purification, DNA fragments were separated in a 6% sequencing gel, dried, exposed to Phosphoimager screens and scanned. The TB8- and TB2-containing PCR fragments are shown in A and B respectively. Numbers on the right refer to the DNA position compared with ATG of the downstream gene. Brackets indicate the limits of the TB. Open and closed circles pinpoint DNase I protected and hyperreactive bands respectively. Below the panels, the DNA sequences show the organization of the nopB (C) and rmlB (D) promoter regions and their reactivity to DNase I cleavage. Underlined bases delineate the TBs with the highly conserved bases in bold font. Capital letters represent DNase I hyperreactive bases and italics show protected bases. Numbers refer to positions upstream of the ATG of the downstream gene.
To test whether such a broad footprint is specific to TB8, a TB2-containing portion of the rmlB promoter region was also used as a template in identical experiments. As seen with TB8, modified bases were also observed within and outside of the TB, and increasing the quantity of TtsI in the reactions sharpened the footprint (Fig. 4B). Thus TtsI has a broader than predicted binding site on both promoter regions. Bases modified by TtsI binding in both TBs were mapped (Fig. 4C and D), but it was not possible to detect any trend of base cleavage modification which could pinpoint bases important for binding outside both TBs. Nevertheless, it is evident from the protection and hyper-reactivity of the highly conserved bases of the TB that they are clearly involved in direct interaction with TtsI. It should be noted that the modified cleavage pattern induced by TtsI towards the 5′ and 3′ ends of the templates may not necessarily indicate direct contact of these sequences with the protein, but could represent distortion of the DNA caused by the binding of the activator with the TB.
Site-directed mutagenesis of a TB
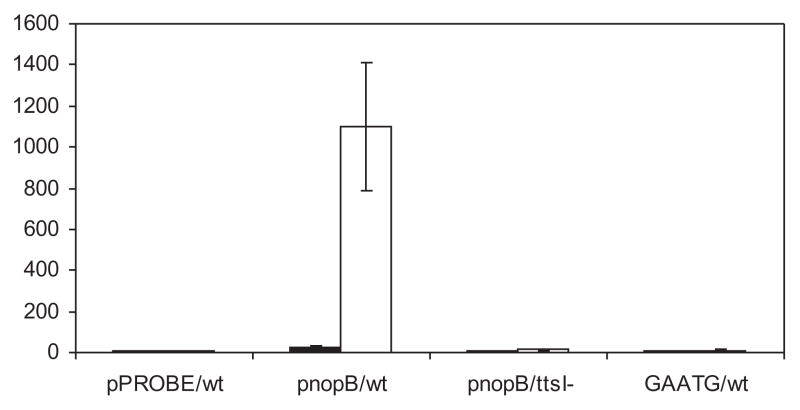
Specific point mutations were introduced to three highly conserved bases of the TB8 consensus sequence (specifically GTCAG to GAATG), and the ability of TtsI to bind and activate transcription was tested in vitro and in vivo. Mobility shift assays showed that TtsI binds in vitro to both wild-type and mutant TBs at similar concentrations (Fig. 5). The ability of the mutated promoter to activate transcription of a reporter gene was then tested using the green fluorescent protein (GFP) to facilitate reporter assays. First, the non-mutated TB8 promoter was sub-cloned upstream of a promoter-less gfp gene in plasmid pPROBE-GT′ (Miller et al., 2000) and then introduced into NGR234 as well as NGRΔttsI by tri-parental matings. Cultures were induced with flavonoids for 40 h and GFP production measured in terms of cellular fluorescence. As expected, a strong GFP signal was observed in NGR234 containing the TB8–gfp fusion, whereas no expression was observed from NGRΔttsI (Fig. 6). The mutated TB8 promoter was also subcloned upstream of gfp in plasmid pPROBE-GT (Miller et al., 2000). When the activity of this plasmid was assayed in NGR234 (under the same conditions of induction), no GFP signal was observed (Fig. 6). To answer the question of why the mutated TB8 promoter was inactive in vivo but apparently unaffected in its binding in vitro led us to test the ability of TtsI to bind to the GAATG TB8 by the more sensitive assay of DNase I footprinting. Using the same experimental conditions described earlier, differences between TB8 and GAATG TB8 were found, especially near the mutated bases, but also elsewhere in the TB8 consensus (Fig. 7). Thus although the GAATG TB8 mutation is clearly insufficient to block TtsI binding, it alters the nature of the binding which in turn may well lead to the inability of GAATG TB8 to be transcribed, as observed in vivo.
Fig. 5.
TtsI binds to the site-directed TB8 mutant. Mobility shift assays were performed as before, with 32P-labelled 150-bp PCR fragments containing the nopB promoter regions.
Fig. 6.
In vivo expression of the site-directed TB8 mutant. The mutated nopB promoter region was fused to a promoter-less gfp gene and assayed after 40-h induction with 2 × 10−6 M apigenin. Numbers shown (× 103) are an average of at least three replicate experiments. pPROBE/wt, the vector control in NGR234; pnopB/ttsI-, the wild-type nopB promoter in NGRΔttsI; pnopB/wt, the wild-type nopB promoter in NGR234; GAATG/wt, the site-directed mutant nopB promoter in NGR234.
Fig. 7.
DNase I footprint of the site-directed TB8 mutant.32P-labelled PCR fragments (A, wild-type and B, site-directed mutant) were incubated with increasing concentrations of TtsI and then digested with limiting concentration of DNase I. After purification, DNA fragments were separated in a 6% sequencing gel, dried, exposed to Phosphoimager screens and scanned. Numbers on the right refer to DNA positions compared with ATG of downstream genes. Asterisks show the mutated bases; open circles highlight the differences in DNase I activity observed (as compared with the wild-type TB8).
Random mutagenesis of a TB
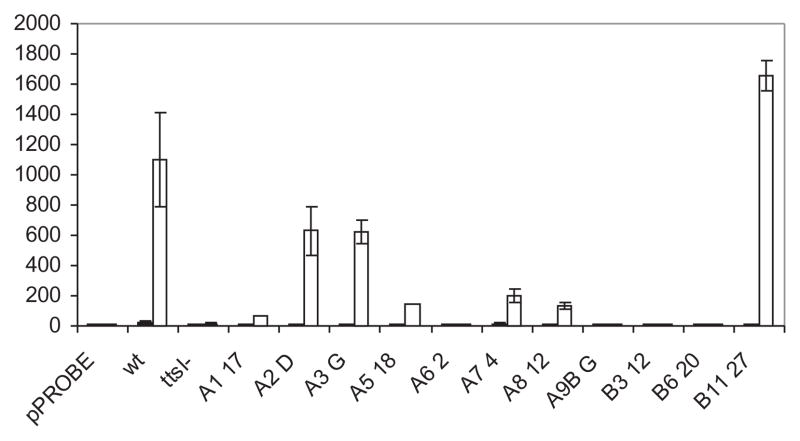
As TtsI bound to regions of TB-containing promoters outside of the TB, we used a random mutagenesis approach to identify other potential bases important for transcriptional control by TtsI. Error-prone PCR was performed on the nopB promoter, the products cloned into pGEM-T, and then sequenced. Because of the mutagenic conditions used, several bases were often changed in each mutant. A number of these PCR products were selected according to the location of the mutations and subcloned upstream of the promoter-less gfp gene and mobilized into NGR234. Most of the mutants analysed had no activity, some had partial activity when induced by apigenin, but none had constitutive activity in the absence of flavonoid (Fig. 8). Three mutants had no mutations within the TB (A62, A117 and A74) and had zero, 6% and 18% of the wild-type TB8 activity, suggesting that either the consensus sequence is longer than predicted (in agreement with the large footprint), or the mutated bases altered binding of other components of the transcriptional machinery. Interestingly, one mutant exhibited higher activities than the wild type upon induction. Attempts to delimit the nopB promoter by footprinting assays using the holoenzyme Escherichia coli RNA polymerase were not successful, possibly as a result of differences in the housekeeping sigma factors of both species. However, the nopB promoter region of S. fredii USDA257 is identical to NGR234, and the exact position of the start of transcription was mapped to a cytidine residue located at the −30 position from the ATG (Kovács et al., 1995). Inspection of the upstream sequence using the proposed consensus sequence for rhizobial promoters (MacLellan et al., 2006) allowed the identification of a compatible −10 RNA polymerase binding site (Fig. 9). Although the −10 sequence is fairly well conserved (four out of six bases), the −35 region does not resemble the consensus, and indeed overlaps the highly conserved GTCAG bases of the TB. The majority of inactive mutants had mutations either in the TB or the putative −10 site (Fig. 9). On the other hand, the three mutants that remained active (B1127) or partially active (A2D andA3G) did not have mutations in the TB consensus sequence or the putative −10 promoter. Interestingly, the A518 mutant possessed four mutations, none of which mapped to either of binding sites, but it still exhibited very low transcriptional activity. This indicates that bases outside of the TB are important for nopB transcription, presumably by modulating binding of TtsI.
Fig. 8.
In vivo expression levels of the randomly mutated nopB promoter regions. Mutated nopB promoter regions were fused to promoter-less gfp genes, transferred to NGR234 and NGRΔttsI and assayed after 40-h induction with 2 × 10−6 M apigenin. The results (× 103) are averages of at least three replicate experiments. pPROBE, vector only in NGR234; wt, the wild-type nopB promoter in NGR234; ttsI-, the wild type nopB promoter in NGRΔttsI; the code names refer to randomly mutated nopB promoters.
Fig. 9.
Positions of the mutated bases in the nopB promoter region. Top line of the DNA sequence shows the wild-type nopB promoter with the probable transcription start, TB8 underlined. The highly conserved bases of TB8 are also underlined. The putative −35/−10 promoter (labelled as POL) is shown, with bases not matching the consensus sequence (labelled as POL CON) in lower case. Dots represent unchanged bases, whereas mutated bases are lettered, code names are indicated at the left. Dotted lines below represent specific mutated nopB promoter regions (labelled with code names), mutated bases are lettered. Numbers refer to positions upstream of the nopB ATG.
Discussion
Phytopathogenic bacteria use T3SS to facilitate infection or conversely to trigger plant defences (Mudgett, 2005; Grant et al., 2006). Many different effector proteins can be secreted by such T3SS and they have been catalogued based upon homology searches or by assessing the potential for co-regulation with other T3SS genes (Cunnac et al., 2004; Lindeberg et al., 2006). Specific plant compounds that induce expression of T3SS genes are not known and, as a consequence, the proteins used as sensors by phytopathogens have not been identified. Nevertheless, many players in the intermediate stages have been identified through mutagenesis and two broad classes of gene regulation are now recognized (Mole et al., 2006; Tang et al., 2006): in group I (represented by Erwinia spp., Pantoea spp. and Pseudomonas syringae), the activator is a sigma factor, termed HrpL which is thought to bind directly to a cis element (hrp boxes) in promoter regions of T3SS genes. In group II, the direct activators are members of the AraC family of transcriptional regulators, called HrpX in Xanthomonas spp. and HrpB in Ralstonia solanacearum, both of which are proposed to bind to cis elements in the promoter regions of T3SS genes. HrpL and AraC-type transcriptional activators are encoded by genes in the T3SS loci.
In contrast, flavonoids trigger induction of T3SS (along with other symbiotic genes) in rhizobia (Krause et al., 2002; Krishnan et al., 2003; Kobayashi et al., 2004; Viprey et al., 1998). Members of the NodD/LysR family of transcriptional regulators act at the top of this cascade, and by binding to nod boxes activate both genes encoding the synthesis of symbiotic signalling molecules along with other regulatory proteins (Schlaman et al., 1998). The gene encoding one such regulator, TtsI, is found within the T3SS loci of several rhizobia and in all cases is preceded by a nod box. ttsI is the only gene encoding a transcriptional regulator in all T3SS loci and, when translated, is proposed to recognize a further cis element, the TB, found in the promoters of T3SS-related genes. The TtsI/TB regulatory system is found in for all rhizobia possessing T3SS (Krause et al., 2002; Krishnan et al., 2003; Marie et al., 2004). Thus the demonstration that the response regulator transcriptional activator TtsI binds directly to TB suggests that the regulation of rhizobial T3SS is different from that found in phytopathogens.
TtsI binding is not specific to the TB consensus sequence, however, and the DNase I footprint extended up to 100 bp up- and downstream of the TB. Furthermore, a double-stranded oligonucleotide (located from −111 to −79) paired to sequences adjacent to the TB and competed with TtsI binding to the nopB promoter region. It should be noted that other members of the response regulator family of transcriptional activators also demonstrate large footprints despite specific consensus sequence predictions (Cullen et al., 1996). It is possible that other proteins bind TtsI and, in doing so, increase specificity of the interaction with TB. Mixing the soluble protein fraction of cell extracts with TtsI and TB-containing promoter regions had no effect on binding. Furthermore, TtsI did not bind to promoter regions lacking TB, nor did it bind to its own promoter (containing a nod box – data not shown), demonstrating that TtsI recognizes specific DNA sequences and that auto-regulation is absent.
Site-specific mutation of TB8 blocked promoter activity but did not abolish TtsI binding. A similar result was observed with HrpL of Pantoea agglomerans where direct binding was unaffected by mutations of the hrp box consensus sequence that did not permit transcription (Nissan et al., 2005). Given that TtsI has a large footprint, it may well bind to or interact with a relatively large stretch of DNA, and thus site-directed mutants would not be expected to prevent binding. However, application of the more sensitive DNase I cleavage protection assay to promoter regions containing the site-specific mutations revealed subtle differences compared with the non-mutated promoter. Modulation of cleavage protection was observed at the sites of mutation, but also at a second location in the TB, suggesting that the interaction between TtsI and the TB is altered and transcription thus blocked. It should also be noted that the nature of the mutations may have indirectly affected the ability of DNase I to digest the target DNA as this ability is very dependent on the nature of the sequence (Herrera and Chaires, 1994).
Random mutagenesis of the nopB promoter region identified further bases that are important for TtsI-dependent transcriptional activity. In one case, transcriptional activity was higher than wild-type levels, although flavonoids and TtsI were always required. No constitutively active mutations were generated. The mutants A117, A518 and A74, in which activity was blocked, were particularly revealing as none of the mutated residues were within the TB consensus sequence. As the nopB promoter regions of S. fredii USDA257 and NGR234 have identical DNA sequence, we used the experimentally proven transcription start of USDA257 to identify a putative rhizobial RNA polymerase binding site. Although the nopB promoter is not highly similar to the consensus sequences of S. meliloti (MacLellan et al., 2006), the −10 box is a good match, whereas the −35 region is poorly conserved. However the sequence encompassing the −35 region overlaps with numerous highly conserved residues of the TB. It is possible that the high activity of the nopB promoter may be due to the transition from a normally inactive state to one in which the changed conformation upon TtsI binding, exposes the promoter to RNA polymerase. Indeed, other transcriptional activators are known to interact with RNA polymerase and bind to sites located very close to the promoter (Browning and Busby, 2004), some by binding to sites which partially overlap the −35 region of the promoter and interact directly with the σ70 subunit of RNA polymerase (Dhiman and Schleif, 2000; Wickstrum and Egan, 2004). Transcriptional control of genes by the response regulator PhoB, where the conserved −35 hexamer in the RNA polymerase binding site is absent and replaced by a pho box to which PhoB binds (Makino et al., 1993), might be a good model for the TtsI/TB system. In these promoters, RNA polymerase is only able to bind in the presence of phosphorylated PhoB.
Following comparison of promoter regions of T3SS genes from several rhizobial species, Krause et al. (2002) predicted a consensus TB sequence. Using this sequence to search the symbiotic plasmid of NGR234, we identified 11 TBs, the majority of which are involved in regulating T3SS functions. Although we had previously demonstrated that TB2 and TB8 are active (Marie et al., 2004), it is now clear that 10 of the 11 known TB loci of pNGR234a are functional flavonoid- and TtsI-dependent promoters. TB3, which is upstream of y4gJ, an open reading frame (ORF) without obvious homologues (but part of a cluster of genes involved in LPS modification, some of which are controlled by TB2), is also induced to relatively high levels in a TtsI-dependent manner. Taken together, these data suggest that genes downstream of TB3 (e.g. y4gJ which is involved in modifying LPS) may also have a role in symbiosis.
In NGR234, at least 20 genes are thought to be involved in T3SS, including those that encode NopA, NopB, NopC, NopL, NopP and NopX (Viprey et al., 1998; Marie et al., 2003; Ausmees et al., 2004; Deakin et al., 2005; Saad et al., 2005). NopA, B and X are associated with pilus-like cell surface appendages, and are therefore thought to be a part of extracellular component of the T3SS machine (Krishnan et al., 2003; Deakin et al., 2005; Saad et al., 2005). NopL and NopP are known effectors, which are probably injected into the cytoplasm of host cells (Bartsev et al., 2003; 2004; Marie et al., 2003; Ausmees et al., 2004; Skorpil et al., 2005). Generally, genes involved in the formation of the membrane-spanning secretion apparatus, such as nopBrhcJnolUVrhcNy4yJrhcQRSTU (TB8), nopX (TB7) and nopCAy4yQrhcVy4yS (TB10), are under the control of moderate to strong promoters. In contrast, TB6 and TB9 that control transcription of two effector proteins (NopL and NopP) are only weakly induced (threefold to 12-fold induction as compared with non-induced levels) 24 hpi (Fig. 2F and I). Furthermore, the ORFs y4fR (TB1), y4lO (TB4) and y4zC (TB11) are all homologous to known T3SS effector proteins (Marie et al., 2001) and their expression profiles are lower than TB controlling the machinery (sixfold, 13-fold and sixfold respectively). That these promoters are active in a TtsI-dependent manner and after flavonoid induction suggests that they may well be upstream of genes encoding functional effector proteins. Generally, promoters upstream of potential effector proteins have variable expression levels. As an example, the expression profile of pMP-TB5 differs significantly from those of other constructs. Although a slight induction (1.2-fold) by daidzein was observed 24 hpi, TB5 had basal transcriptional activities as high as 3440 ± 190 Miller units even under non-induced conditions. As a null mutation in ttsI had no affect on its basal activity, TB5 is probably under additional regulatory control. ORF downstream of TB5 are not obviously related to T3SS or LPS functions, and seem to be involved in stabilization of pNGR234a (Dombrecht et al., 2001).
Use of the TB consensus sequence in a bio-informatics screen to identify TtsI-regulated genes on pNGR234a was largely successful as 10 of the 11 predicted TBs are active. Indeed, extending such screens to the other replicons of NGR234 as well as other rhizobia should be feasible, although the predictions will need to be verified on a case-by-case base as the example of TB5 shows. Differences in the TtsI regulon could be specific to T3SS function, as rhizobia possess and may well secrete different effector proteins, which TB-based searches can help identify. Alternatively, production of diverse signalling molecules could also be controlled by TtsI, such as the rhamnose-rich LPS synthesized by NGR234 but not by B. japonicum USDA110 or S. fredii USDA257(Marie et al., 2004). Control of rhamnose synthesis is brought about by genes recruited into the TtsI regulon out of temporal necessity, as all are induced relatively late, or as a functional requirement linked to T3SS activity. Over 30 predicted TB sequences are present in the B. japonicum USDA110 genome (Suss et al., 2006). Completion of more rhizobial genomes, particularly those with T3SS and TB, will allow ever more powerful comparative studies into the roles of flavonoid-inducible genes in symbiosis.
Conclusions
We have extended the inventory of members of the flavonoid-regulatory cascade of NGR234 which bring about the exceptionally broad host range of this Rhizobium. Methods used here to analyse the TtsI regulon of NGR234 are applicable to other TB-possessing rhizobia, although it is very probable that genes modulated by TtsI will vary considerably. What remains is to test TB expression levels in planta, as the potential cocktail of inducing compounds and environments may well reveal more mechanisms of regulatory control.
Experimental procedures
Microbiological techniques
Escherichia coli recombinants were grown at 37°C on Luria–Bertani medium (Sambrook et al., 1989). NGR234 and its derivatives were raised at 28°C in Rhizobium minimal medium containing succinate as the carbon source (RMS) (Broughton et al., 1986) or TY (Beringer, 1974). Ampicillin (for Bluescript/pGem), gentamycin, kanamycin (for pRK2013), rifampicin, spectinomycin and tetracycline were added at concentrations of 50, 20, 50, 100, 50 and 15 μg ml−1 respectively. The flavonoids, apigenein or daidzein were used to induce ttsI, as both have been shown to induce NB18 (Viprey et al., 1998; Kobayashi et al., 2004; Marie et al., 2004).
Construction of NGRΔttsI and pttsI-2
To obtain a mutant with a deletion in ttsI, the 2.4-kb PstI fragment from pUC::ttsI (Marie et al., 2004), which carries ttsI as well as its flanking region (Fig. 1), was digested with SacII and NheI, purified by electrophoresis, treated with the Klenow fragment, and self-ligated. Then, the remaining 0.8-kb PstI fragment, lacking ttsI, was purified and sub-cloned into the PstI site of pJQ200-mp18 (Quandt and Hynes, 1993). The resulting plasmid was mobilized into NGR234 by tri-parental matings using the pRK2013 helper plasmid (Ditta et al., 1980). Marker exchange in NGRΔttsI was confirmed by Southern hybridization. To complement NGRΔttsI, ttsI along with its own nod box-containing promoter region, the 2.4-kb PstI fragment of pUC::ttsI (Fig. 1), were subcloned into the PstI site of pRG960 (Van den Eede et al., 1992), a vector which is compatible with pMP220-based lacZ fusion constructs described below, giving rise to pttsI-2.
Extraction and analysis of Nops and LPS
After 40-h induction with 10−6 M apigenin, secreted proteins and LPS were recovered from the various NGR234 strains as described in Viprey et al. (1998) and Marie et al. (2004) respectively. Aliquots of purified proteins were separated on SDS-polyacrylamide (PAGE) gels and stained with silver (Ausubel et al., 1991) or, for immuno-detection, separated proteins were transferred to PVDF Immobilon-P membranes (Millipore Corporation, Bedford, Massachusetts, USA) and probed with antibodies against NopL, NopP and NopX. Horseradish peroxidase-labelled goat anti-rabbit immunoglobulin antibodies of the ECL kit (GE Life-sciences Amersham Pharmacia Biotech, Uppsala, Sweden) were used as secondary antibodies. Reactions were visualized by enhanced chemi-luminescence. Extracted LPS samples were separated on 16% deoxycholic acid (DOC)-PAGE and stained with silver nitrate (Tsai and Frasch, 1982).
Cloning of the 11 TBs in pMP220
Predicted promoter regions containing TB1, TB3–TB11 were amplified by PCR using the primer pairs listed in Table 1. The amplified products were subcloned into pBluescript KS+ (Stratagene, La Jolla, California) and verified by sequencing the inserts. Then, the inserts were excised and cloned into pMP220. pMP220 constructs containing TB2 (pMP-TB2) and TB8 (pMP-TB8) correspond to plasmids pMP220-rmlB and pMP220B (Marie et al., 2004) respectively. Promoter constructs cloned into the broad-host-range reporter vector pMP220 (Spaink et al., 1987) were mobilized into NGR234 and its derivatives by tri-parental matings using pRK2013 as the helper plasmid (Ditta et al., 1980). Flavonoid induction was performed as described previously (Kobayashi et al., 2004): rhizobial cultures grown to a density of 1cmOD600 0.5–0.6 were diluted to 1cmOD600 0.1 in RMS medium and induced with 2 × 10−7 M daidzein or 1 × 10−6 M apigenin. β-Galactosidase activity was assayed according to Miller (1972).
DNase I footprinting
DNA fragments were amplified with polynucleotide kinase-end-labelled primers. The rmlB and nopB promoter regions were amplified with the following primers (respectively): rmlB-F and rmlB-R, and nopB for and nopB rev (Table 1). DNase I footprinting assays were performed in a total volume of 50 μl of TAP buffer without polyethyleneglycol (50 mM Tris-acetate pH 8.0, 100 mM potassium acetate, 8 mM magnesium acetate, 27 mM ammonium acetate, 1 mM dithiothreitol). Labelled fragments were added at a final concentration of approximately 5 nM and incubated with the proteins for 30 min at room temperature. After incubation, 0.05 U of freshly diluted DNase I (Invitrogen, Carlsbad, CA) was added, and the reaction was allowed to run for 2 min at room temperature. Then, the samples were extracted with phenol, precipitated with ethanol, washed, re-suspended in loading buffer and loaded on a 6% DNA sequencing gel. Images were obtained by a Cyclone imager (Packard Institute, Downers Grove, Illinois) after exposing the dried gel to a Phosphoimager screen or by exposing the gels to X-ray film and scanning. Sequence ladders were prepared using the Sequenase Quick Denature Plasmid DNA Sequencing Kit as described by the manufacturer (USB, Cleveland, Ohio).
Electrophoretic mobility shift assays
32P-labelled PCR fragments were incubated for 30 min in TAP buffer (50 mM Tris-acetate, 100 mM potassium acetate, 8 mM magnesium acetate, 3.5% polyethylene glycol 8000, 1 mM DTT, pH 7.9) with the indicated concentrations of TtsI and reactions loaded into a 4% native PAGE gel. Running buffer contained 50 mM Tris, 400 mM glycine, 2 mM EDTA, 8 mM MgSO4 at pH 8.5. Dried gels were exposed to X-ray films and images obtained by scanning the film.
Site-directed mutagenesis of TB8
The non-mutated TB8 promoter was excised from pMP-TB8 as a HindIII fragment and subcloned upstream of a promoter-less gfp gene in plasmid pPROBE-GT′. Point mutations in TB8 were introduced by PCR using a pair of primers that replace the highly conserved triplet TCA for AAT. Two independent PCR reactions were carried out using primers TB8-F mut (5′-ggccggtagaatgcgtgtcgtcagctcgcctc-3′) versus nopB rev (5′-ggactcgattacttaactctttgac-3′) and TB8-R mut (5′-ggaggcgagctgacgacacgcattctaccggc-3′) versus nopB for (5′-ctcgtcttgataaaccaaatctgaa-3′). PCR products were pooled, amplified, cloned into pGEM-T (Promega, Madison, Wisconsin) and sequenced. The mutated promoters were subsequently transferred into pPROBE-GT as an EcoRI insert and orientation was checked. The constructs were mobilized into NGR234 or NGRΔttsI as described previously.
Random mutagenesis of TB8
Mutagenesis of the nopB promoter region was accomplished using mutagenic PCR as described by Vartanian et al. (1996). PCR reactions contained 1× taq buffer, 2.5 mM MgCl2, 0.5 mM MnCl2, 50 μM dATP and dCTP, 1 mM dGTP and dTTP, 2.5 μM each primer, template DNA and taq polymerase (Eppendorf). PCR fragments were cloned into pGEM-T (Promega), sequenced and transferred to pPROBE-GT as an EcoRI insert and orientation was checked.
Assay of GFP intensity
Rhizobial cultures were grown to an OD600 of 0.5, diluted to an OD600 of 0.1 in RMS medium and induced with 1 × 10−6 M apigenin. Both optical density (OD595) and fluorescence (excitation filter at 485 nm and emission filter at 535 nm) were measured on 100 μl of cultures 40 hpi using a Plate Chameleon Multilabel Detection Platform (Hidex Oy, Turku, Finland). Optical densities and fluorescence were corrected to background levels using un-inoculated media, and the results represent the means of at least three independent experiments.
Supplementary Material
Acknowledgments
This work was supported by the Fonds National Suisse de la Recherche Scientifique (Projects 3100AO-104097 and 3100A0-116858 to W.J.B. and W.J.D.), the Département de l’Instruction Publique du Canton de Genève (to W.J.B. and W.J.D.), the Universitè de Genève (to W.J.B.) and a grant from the National Institute of Health (GM31010 to G.C.W.). G.C.W. is also supported by an American Cancer Society Research Professorship and a Howard Hughes Medical Institute Professorship. R.W. was supported by a CAPES postdoctoral fellowship. H.K. is supported by JSPS Postdoctoral Fellowships for Research Abroad. We wish to thank Y.-Y. Aung and D. Gerber for their unstinting help, M.M. Saad for useful discussions on Nop preparation and K.E. Gibson for kind help with LPS preparation.
Footnotes
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2008.06187.x
(This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Ausmees N, Kobayashi H, Deakin WJ, Marie C, Krishnan HB, Broughton WJ, Perret X. Characeterisation of NopP, a type III secreted effector of Rhizobium sp. NGR234. J Bacteriol. 2004;186:4774–4780. doi: 10.1128/JB.186.14.4774-4780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1991. [Google Scholar]
- Bartsev AV, Boukli NM, Deakin WJ, Staehelin C, Broughton WJ. Purification and phosphorylation of the effector protein NopL from Rhizobium sp. NGR234. FEBS Lett. 2003;554:271–274. doi: 10.1016/s0014-5793(03)01145-1. [DOI] [PubMed] [Google Scholar]
- Bartsev AV, Deakin WJ, Boukli NM, McAlvin CB, Stacey G, Malnoë P, et al. NopL, an effector protein of Rhizobium sp. NGR234 thwarts activation of plant defence reactions. Plant Physiol. 2004;134:871–879. doi: 10.1104/pp.103.031740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beringer JE. R-factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Broughton WJ, Wong CH, Lewin A, Samrey U, Myint H, Meyer zAH, et al. Identification of Rhizobium plasmid sequences involved in recognition of Psophocarpus, Vigna, and other legumes. J Cell Biol. 1986;102:1173–1182. doi: 10.1083/jcb.102.4.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Jabbouri S, Perret X. Keys to symbiotic harmony. J Bacteriol. 2000;182:5641–5652. doi: 10.1128/jb.182.20.5641-5652.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton WJ, Hanin M, Relić B, Kopcinska J, Golinowski W, Şimşek S, et al. Flavonoid-inducible modifications to rhamnan O antigens are necessary for Rhizobium sp. strain NGR234-legume symbioses. J Bacteriol. 2006;188:3654–3663. doi: 10.1128/JB.188.10.3654-3663.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning DF, Busby SJW. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:1–9. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Bowman WC, Kranz RG. In Vitro reconstitution and characterization of the Rhodobacter capsulatus NtrB and NtrC two-component system. J Biol Chem. 1996;271:6530–6536. doi: 10.1074/jbc.271.11.6530. [DOI] [PubMed] [Google Scholar]
- Cunnac S, Occhialini A, Barberis P, Boucher C, Genin S. Inventory and functional analysis of the large Hrp regulon in Ralstonia solanacearum: identification of novel effector proteins translocated to plant host cells through the type III secretion system. Mol Microbiol. 2004;53:115–128. doi: 10.1111/j.1365-2958.2004.04118.x. [DOI] [PubMed] [Google Scholar]
- Deakin WJ, Marie C, Saad MM, Krishnan HB, Broughton WJ. NopA is associated with cell surface appendages produced by the type III secretion system of Rhizobium sp. strain NGR234. Mol Plant Microbe Interact. 2005;18:499–507. doi: 10.1094/MPMI-18-0499. [DOI] [PubMed] [Google Scholar]
- Dhiman A, Schleif R. Recognition of overlapping nucleotides by AraC and the sigma subunit of RNA polymerase. J Bacteriol. 2000;182:5076–5081. doi: 10.1128/jb.182.18.5076-5081.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G, Stanfield S, Corbin D, Helsinki DR. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Vanderleyden J, Michiels J. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in Gram-negative bacteria. Mol Plant Microbe Interact. 2001;14:426–430. doi: 10.1094/MPMI.2001.14.3.426. [DOI] [PubMed] [Google Scholar]
- Fraysse N, Couderc F, Poinsot V. Surface polysaccharide involvement in establishing the Rhizobium-legume symbiosis. Eur J Biochem. 2003;270:1365–1380. doi: 10.1046/j.1432-1033.2003.03492.x. [DOI] [PubMed] [Google Scholar]
- Freiberg C, Fellay R, Bairoch A, Broughton WJ, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- Grant SR, Fisher EJ, Chang JH, Mole BM, Dangl JL. Subterfuge and manipulation: type III effector proteins of phytopathogenic bacteria. 2006;60:425–449. doi: 10.1146/annurev.micro.60.080805.142251. [DOI] [PubMed] [Google Scholar]
- Herrera JE, Chaires JB. Characterization of preferred deoxyribonuclease I cleavage sites. J Mol Biol. 1994;236:405–411. doi: 10.1006/jmbi.1994.1152. [DOI] [PubMed] [Google Scholar]
- Hubber A, Vergunst AC, Sullivan JT, Hooykaas PJ, Ronson CW. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol Microbiol. 2004;54:561–574. doi: 10.1111/j.1365-2958.2004.04292.x. [DOI] [PubMed] [Google Scholar]
- Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Naciri-Graven Y, Broughton WJ, Perret X. Flavonoids induce temporal shifts in gene-expression of nod-box controlled loci in Rhizobium sp. NGR234. Mol Microbiol. 2004;51:335–347. doi: 10.1046/j.1365-2958.2003.03841.x. [DOI] [PubMed] [Google Scholar]
- Kovács LG, Balatti PA, Krishnan HB, Pueppke SG. Transcriptional organization and expression of nolXWBTUV, a locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1995;17:923–933. doi: 10.1111/j.1365-2958.1995.mmi_17050923.x. [DOI] [PubMed] [Google Scholar]
- Krause A, Doerfel A, Göttfert M. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol Plant Microbe Interact. 2002;15:1228–1235. doi: 10.1094/MPMI.2002.15.12.1228. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Lorio L, Kim WS, Jiang G, Kim KY, DeBoer M, Pueppke SG. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol Plant Microbe Interact. 2003;16:617–625. doi: 10.1094/MPMI.2003.16.7.617. [DOI] [PubMed] [Google Scholar]
- Kumagai Y, Cheng Z, Lin M, Rikihisa Y. Biochemical activities of three pairs of Ehrlichia chaffeensis two-component regulatory system proteins involved in inhibition of lysosomal fusion. Infect Immun. 2006;74:5014–5022. doi: 10.1128/IAI.00735-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin A, Cervantes E, Wong CH, Broughton WJ. nodSU, two new nod genes of the broad host range Rhizobium strain NGR234 encode host-specific nodulation of the tropical tree Leucaena leucocephala. Mol Plant Microbe Interact. 1990;3:317–326. doi: 10.1094/mpmi-3-317. [DOI] [PubMed] [Google Scholar]
- Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact. 2006;19:1151–1158. doi: 10.1094/MPMI-19-1151. [DOI] [PubMed] [Google Scholar]
- MacLellan SR, MacLean AM, Finan TM. Promoter prediction in the rhizobia. Microbiology. 2006;152:1751–1763. doi: 10.1099/mic.0.28743-0. [DOI] [PubMed] [Google Scholar]
- Makino K, Amemura M, Kim S, Nakata A, Shinagawa H. Role of the Sigma 70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 1993;7:149–160. doi: 10.1101/gad.7.1.149. [DOI] [PubMed] [Google Scholar]
- Marie C, Broughton WJ, Deakin WJ. Rhizobium type III secretion systems: legume charmers or alarmers? Curr Opin Plant Biol. 2001;4:336–342. doi: 10.1016/s1369-5266(00)00182-5. [DOI] [PubMed] [Google Scholar]
- Marie C, Deakin WJ, Viprey V, Kopcinska J, Golinowski W, Krishnan HB, et al. Characterisation of Nops, Nodulation Outer Proteins, secreted via the type III secretion system of NGR234. Mol Plant Microbe Interact. 2003;16:743–751. doi: 10.1094/MPMI.2003.16.9.743. [DOI] [PubMed] [Google Scholar]
- Marie C, Deakin WJ, Ojanen-Reuhs T, Diallo E, Reuhs B, Broughton WJ, Perret X. TtsI, a key regulator of Rhizobium species NGR234 is required for type III-dependent protein secretion and synthesis of rhamnose-rich polysaccharides. Mol Plant Microbe Interact. 2004;17:958–966. doi: 10.1094/MPMI.2004.17.9.958. [DOI] [PubMed] [Google Scholar]
- Miller JH. Assay of β-galactosidase. In: Miller JH, editor. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- Miller WG, Leveau JH, Lindow SE. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant Microbe Interact. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- Mole BM, Baltrus DA, Dangl JL, Grant SR. Global virulence regulation networks in phytopathogenic bacteria. Trends Microbiol. 2006;15:363–371. doi: 10.1016/j.tim.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Mudgett MB. New insights to the function of phytopathogenic bacterial type III effectors in plants. Annu Rev Plant Biol. 2005;56:509–531. doi: 10.1146/annurev.arplant.56.032604.144218. [DOI] [PubMed] [Google Scholar]
- Nissan G, Manulis S, Weinthal DM, Sessa G, Barash I. Analysis of promoters recognised by HrpL, an alternative sigma-factor protein from Pantoea agglomerans pv. gypsophilae. Mol Plant Microbe Interact. 2005;18:634–643. doi: 10.1094/MPMI-18-0634. [DOI] [PubMed] [Google Scholar]
- Perret X, Kobayashi H, Collado-Vides J. Regulation of expression of symbiotic genes in Rhizobium sp. NGR234. Indian J Exp Biol. 2003;41:1101–1113. [PubMed] [Google Scholar]
- Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64:180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pueppke SG, Broughton WJ. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host-ranges. Mol Plant Microbe Interact. 1999;12:293–318. doi: 10.1094/MPMI.1999.12.4.293. [DOI] [PubMed] [Google Scholar]
- Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- Reuhs BL, Relić B, Forsberg LS, Marie C, Ojanen-Reuhs T, Stephens SB, et al. Structural characterization of a flavonoid-inducible Pseudomonas aeruginosa A-band-like O antigen of Rhizobium sp. strain NGR234, required for the formation of nitrogen-fixing nodules. J Bacteriol. 2005;187:6479–6487. doi: 10.1128/JB.187.18.6479-6487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad MM, Kobayashi H, Marie C, Brown I, Mansfield JW, Broughton WJ, Deakin WJ. NopB, a type III secreted protein of Rhizobium sp. strain NGR234, is associated with pilus-like surface appendages. J Bacteriol. 2005;187:1173–1181. doi: 10.1128/JB.187.3.1173-1181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. 2. Cold Spring Harbor, NY: Cold Spring Harbour Laboratory Press; 1989. [Google Scholar]
- Schlaman HR, Phillips DA, Kondorosi E. Genetic organization and transcriptional regulation of rhizobial nodulation genes. In: Spaink HP, Kondorosi A, Hooykaas PJJ, editors. The Rhizobiaceae. Dordrecht: Kluwer Academic Press; 1998. pp. 361–386. [Google Scholar]
- Skorpil P, Saad MM, Boukli NM, Kobayashi H, Ares-Orpel F, Broughton WJ, Deakin WJ. NopP, a phosphorylated effector of Rhizobium sp. strain NGR234, is a major determinant of nodulation of the tropical legumes Flemingia congesta and Tephrosia vogelii. Mol Microbiol. 2005;57:1304–1317. doi: 10.1111/j.1365-2958.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- Soto MJ, Sanjuan J, Olivares J. Rhizobia and plant-pathogenic bacteria: common infection weapons. Microbiology. 2006;152:3167–3174. doi: 10.1099/mic.0.29112-0. [DOI] [PubMed] [Google Scholar]
- Spaink HP, Okker RJH, Wijffelman CA, Pees E, Lugtenberg BJJ. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Suss C, Hempel J, Zehner S, Krause A, Patschkowski T, Göttfert M. Identification of genistein-inducible and type III-secreted proteins of Bradyrhizobium japonicum. J Biotechnol. 2006;126:69–77. doi: 10.1016/j.jbiotec.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Tang X, Xiao Y, Zhou JM. Regulation of the type III secretion system in phytopathogenic bacteria. Mol Plant Microbe Interact. 2006;19:1159–1166. doi: 10.1094/MPMI-19-1159. [DOI] [PubMed] [Google Scholar]
- Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Van den Eede G, Deblaere R, Goethals K, van Montagu M, Holsters M. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol Plant Microbe Interact. 1992;5:228–234. doi: 10.1094/mpmi-5-228. [DOI] [PubMed] [Google Scholar]
- Vartanian JP, Henry M, Wain-Hobson S. Hypermutagenic PCR involving all four transitions and a sizeable proportion of transversions. Nucleic Acids Res. 1996;24:2627–2631. doi: 10.1093/nar/24.14.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viprey V, Del Greco A, Golinowski W, Broughton WJ, Perret X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol Microbiol. 1998;28:1381–1389. doi: 10.1046/j.1365-2958.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- Viprey V, Rosenthal A, Broughton WJ, Perret X. Genetic snapshots of the Rhizobium species NGR234 genome. Genome Biol. 2000;1:1–17. doi: 10.1186/gb-2000-1-6-research0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Plovanich-Jones A, Deng WL, Collmer A, Huang HC, He SY. The structural protein of the Hrp pilus is required for coordinate regulation of the type III secretion system and secretion of Hrp and Avr proteins in Pseudomonas syringae pv. Tomato. Proc Natl Acad Sci USA. 2000;97:2247–2252. doi: 10.1073/pnas.040570097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstrum JR, Egan SM. Amino acid contacts between sigma 70 domain 4 and the transcription activators RhaS and RhaR. J Bacteriol. 2004;186:6277–6285. doi: 10.1128/JB.186.18.6277-6285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.