Abstract
Currently, considerable information is available about how muscle-specific genes are activated during myogenesis, yet little is known about how non-muscle genes are down-regulated. The intermediate filament protein vimentin is known to be “turned off” during myogenesis to be replaced by desmin, the muscle-specific intermediate filament protein. Here, we demonstrate that vimentin down-regulation is the result of the combined effect of several transcription factors. Levels of the positive activators, Sp1/Sp3, which are essential for vimentin expression, decrease during myogenesis. In addition, c-Jun and Stat3, two additional positive-acting transcription factors for vimentin gene expression, are also down-regulated. Over-expression via adenoviral approaches demonstrates that the up-regulation of the repressor ZBP-89 is critical to vimentin down-regulation. Elimination of ZBP-89 via siRNA blocks the down-regulation of vimentin and Sp1/Sp3 expression. From these studies we conclude that the combinatorial effect of the down-regulation of positive-acting transcription factors such as Sp1/Sp3, c-Jun and Stat3 versus the up-regulation of the repressor ZBP-89 contributes to the “turning off” of the vimentin gene during myogenesis.
Keywords: ZBP-89, vimentin, myogenesis, Sp1, Sp3, c-Jun, Stat3, C2C12
Introduction
Intermediate filaments comprise a heterogeneous family of over 50 proteins (IFPs) expressed in a tissue- and developmental-dependent manner (Parry and Steinert, 1992; Steinert and Liem, 1990). Common IFPs include vimentin, desmin, GFAP, keratins, neurofilaments and lamins. Initially, vimentin is highly expressed in the embryo, but becomes progressively restricted to fewer cell-types during development (Duprey and Paulin, 1995). For example, it is expressed in early stages of muscle or astrocyte development, but is selectively down-regulated during differentiation to make way for the tissue-specific IFPs, desmin and GFAP, respectively (Moura-Neto et al., 1996; Sax et al., 1989). It has been proposed that negative regulation is involved in the down-regulation of vimentin during differentiation. However, a mechanism for the loss of vimentin during myogenesis has not been elucidated, although a role for the transcriptional repressor, ZBP-89 has been proposed (Wieczorek et al., 2000).
Several skeletal myogenic cell lines exist, which duplicate embryonic myogenesis in vitro, and thus represent an excellent model system for investigating mechanisms of genetic control (McKinsey et al., 2001). C2C12 cells are capable of continued proliferation; however upon removal of serum myoblasts withdraw from cell cycle, fuse, activate muscle-specific genes and differentiate into myotubes. Analysis of the muscle program has identified two important families of myogenic factors (Pownall et al., 2002). One family is the basic Helix-Loop-Helix proteins (bHLH), which consists of Myf5, MRF4, MyoD and myogenin. The temporal and spatial expression of these proteins contributes to terminal differentiation of skeletal muscle. The bHLH proteins bind as heterodimers with E-box proteins to E-box sequences within muscle gene enhancers (Black and Olson, 1998). The second family of myogenic factors is the MEF2 family, which bind to AT-rich sequences. MEF2 factors are not able to activate muscle genes alone, but potentiate the effect of bHLH proteins, suggesting both families are essential for myoblast differentiation. While considerable information is known about how muscle-specific genes are activated during myogenesis, little information exists about how a non-muscle gene like vimentin is selectively deactivated, although both events are necessary to achieve terminal differentiation.
Currently, several cis-elements and associated factors are required to regulate expression of the human vimentin gene. These include a TATA-box, eight putative GC-boxes, a NF-KB site (-239 to -224), a PEA3-binding site (-173 to -159), Δ19 (-349 to -329), two copies of a negative element (proximal silencer, PS1 at -111 to -83 and PS2 at -329 to -289), tandem AP-1 binding sites, and the anti-silencer element, ASE (Chen et al., 1996; Izmailova and Zehner, 1999; Lilienbaum and Paulin, 1993; Rittling et al., 1989; Rittling and Baserga, 1987; Salvetti et al., 1993; Wieczorek et al., 2000; Wu et al., 2007b; Wu et al., 2007a). The positive factors that regulate vimentin expression are fairly well understood, i.e., Sp1/Sp3 bind GC-Box 1 (Izmailova et al., 1999), Stat1/Stat3 bind the ASE (Izmailova et al., 2000; Wu et al., 2004) while c-Jun can form either homo- or heterodimers with other AP-1 family members and bind the AP-1 sites (Rittling et al., 1989; Wu et al., 2007a). The only negative factor identified to repress vimentin gene expression is ZBP-89 and its family member ZBP-99 (Zhang et al., 2003).
ZBP-89 (ZNF-148) is a Krüppel-like, zinc-finger transcription factor that binds to a GC-rich region and can activate or repress transcription. ZBP-89 activates human stromelysin (Reizis and Leder 1999; Ye et al., 1999), p21waf1/cip1 (Hasegawa et al., 1999) and lck (Yamada et al., 2001) genes. However, ZBP-89 functions as a repressor for the human gastrin (Merchant et al., 1996), ODC (Law et al., 1999), rat β-enolase (Passantino et al., 1998), bovine adrenodoxin (Cheng et al., 2000) human ENA-78 (Keates et al., 2001), and vimentin (Wieczorek et al., 2000) genes. When ZBP-89 acts as an activator, it recruits p300 to the p21 promoter (Bai and Merchant, 2000). When ZBP-89 functions as a repressor, it either competes with Sp1 for binding on the gastrin gene promoter (Merchant et al., 1996) or binds separate elements as found for the vimentin gene and recruits HDAC1 (Wieczorek et al., 2000) (Wu et al., 2007b).
Originally, ZBP-89 was proposed to be ubiquitously expressed (Merchant et al., 1996). However, it is now apparent that ZBP-89 levels may change during myogenesis serving to repress vimentin and perhaps other non-muscle genes (Passantino et al., 1998). In mouse, there appears to be less ZBP-89 mRNA in the adult heart and testis, intermediate levels in skeletal muscle and spleen, and higher levels in brain, kidney, lung and liver. Conversely, in adult rat equal levels of ZBP-89 were found in heart, brain, spleen, lung, liver, skeletal muscle, kidney and testis (Merchant et al., 1996). A comparison of the mouse and human ZBP-89 promoters revealed multiple Sp1 (8), MEF-2 (2), MyoD and ZBP-89 binding sites, which could support its up-regulation during myogenesis (Feo et al., 2001). In fact, a ZBP-89 promoter-reporter construct including the MyoD site displayed a 3-fold increase in expression in C2C12 myoblasts (MBs) versus myotubes (MTs) (Feo et al., 2001). Here, we propose that the up-regulation of ZBP-89 expression together with the decreased expression of vimentin's positive-acting transcription factors coordinates the down-regulation of vimentin during muscle differentiation.
Materials and methods
Cell Cultures, Plasmid Constructions and Reagents
C2C12 skeletal myoblast (MB) cells (ATCC CRL-1772), a sub-culture derived from murine skeletal cells, were grown as described previously (Wu et al., 2007a). Sub-confluent cells were switched to differentiation medium (DM) containing 1% FBS to initiate differentiation into myotubes.
Various 5′-deletion constructs of the human vimentin promoter fused to the CAT reporter gene (-261/+72, -319/+72, -353/+72, -353-Sp1/+72, -353-DMPS/+72, -725/+72, -775/+72, -775-AP1/+72, -815/+72, -815-ASE/+72) were created as described (Wu et al., 2007a). Adenovirus containing flag-tagged rat ZBP-89 (Ad-ZBP-89) was provided by Dr. Juanita Merchant (Univ. of Michigan, Ann Arbor, MI). Protease inhibitor cocktail (P-8340), monoclonal anti-vimentin antibody (V6630) and anti-Flag antibody (F-3165) were purchased from Sigma. Anti-MyoD (sc-6246), anti-tubulin (sc-9104), anti-MHC (sc-20641), and anti-myogenin (sc-576) were from Santa Cruz Biotechnology.
RNA Extraction and Quantitative Real-time PCR (qPCR)
Total RNA was extracted from C2C12 MBs, MTs or similar cells 48 h after infection with adenovirus vector empty (Ad-empty) or containing β-gal (Ad-β-gal) or full-length rat ZBP-89 (Ad-ZBP-89). Total RNA was extracted using the TRIzol reagent (Invitrogen Corporation). qPCR data acquisition and analysis was carried out with a LightCycler instrument using a sequence-detection system [Model 7000, Applied Biosystems, Foster City, CA as described previously (Wu et al., 2007a). Primer sequences were as follows: vimentin 5′-AGGAATGGCTCGTCACCTTCGTGAATA-3′ and 5′-GGAGTGTCGGTTGTTAAGAACTAGAGCT-3′, for Sp1 5′-TGAGGCATTAATGTGCTTGG -3′ and 5′-AAATGCTGATCAAAGGGTGG-3′, for U6 5′-CTCGCTTCGGCAGCACA-3′ and 5′-AACGCTTCACGAATTTGCGT-3′, for Sp3 5′-TGCCAACATCCTCTTCATCA-3′ and 5′-CAATTTGGGCTTGACTGGTT-3′, for ZBP-89 5′-CGCATTTAGAAGATGCGTCA-3′ and 5′-CCCCTCCTGCAAATTATCAA-3′, for Stat3 5′-ACCCAACAGCCGCCGTAG-3′ and 5′-CAGACTGGTTGTTTCCATTCAGAT-3′, for cJun 5′-CCCCAAGATCCTGAAACAGA-3′ and 5′-CCGTTGCTGGACTGGATTAT-3′ for desmin 5′-TCAAGGGCACCAACGACT-3′ and 5′-GGTCTGGATCGGAAGGTTGAT-3′, and for myogenin 5′-ACTCTTCGCCCCCGT-3′ and 5′-CCGCCCTGCCACTCAT-3′. Gene amplification was carried out as follows: 95°C for 10 min, and then 50 cycles in 3 steps: 95°C for 15 s, 53°C for 30 s, and 68°C for 45 s. At the end of the amplification cycles, dissociation curves were determined to rule out signal from primer dimers and other non-specific dsDNA species. Data was normalized to U6 mRNA levels. Experiments were carried out in triplicate on three separate RNA preps and the standard error is shown. The size of the PCR products was confirmed on a 2% agarose gel by ethidium bromide staining.
Adenoviral Infection and siRNA Transfection
For adenoviral infection, C2C12 cells (5 × 104) were plated in each well of a 6 well dish, incubated overnight at 37°C and then infected with Ad-ZBP-89, Ad-empty vector or Ad-β-gal at an MOI of 5. Cells were visualized at successive time points using a light microscope and digital camera. All pictures were representative of three independent experiments. For siRNA transfection, C2C12 cells (5×104) were plated in each well of a 6 well dish, incubated overnight at 37°C and kept in GM for MBs or switched to DM to promote myotube formation. Cells were transfected with siControl 2 (20 nm) or siZBP-89 (20 nm) (Dharmacon) using Dharmacon transfection reagent 2 to further verify siRNA results. Cells were visualized at successive time points as described above.
DNA Transfection and CAT Assays
C2C12 cells (2 × 105) were plated in a 6-well plate, incubated overnight at 37°C, washed with 1×PBS, and then changed to either GM or DM. Cells were then transiently transfected with vimentin promoter plasmid DNA (1 μg) using the MIRUS DNA transfection method (Fisher) and then co-infected with Ad-ZBP-89 or Ad-β-gal. If necessary, vector DNA was added to keep the total amount of transfected plasmid constant. 24 h after transfection, cells were washed with 1× PBS, and then maintained in either GM or DM. MBs and MTs were both harvested 48 h after transient transfection. Cell harvesting and CAT assays were performed as described previously (Wu et al., 2007a). Data was normalized to protein levels using the BioRad DC Protein Assay kit (BioRad, Hercules, CA). All transfections were performed three times in triplicate with at least two different DNA preparations for each plasmid preparation.
Whole Cell Extract (WCE) Preparation and Western Blot Analysis
WCEs were prepared from C2C12 cells infected with Ad-ZBP-89, Ad-empty or Ad-β-gal. Cells were washed with 1× PBS and lysed in 4% SDS in 1× PBS with protease inhibitor cocktail (Sigma, St. Louis, MO) at 10 μl/1 ml extract. After a brief sonication, an equal volume of 1× PBS was added to reduce the SDS to 2%. Lysates were centrifuged for 10 min at 12,000 rpm, supernatants collected and protein concentration measured as described above. Equivalent amounts of protein (50 μg) were boiled in 1× SDS sample buffer for 5 min and analyzed on 4-20% gradient polyacrylamide gel (NuSep Inc., Austell, GA). Western blots with antibodies (1:1000) were carried out as previously described (Zhang et al., 2003).
Electrophoretic Mobility Shift Assays (EMSAs)
A 40 bp H4TF-1DNA fragment (-640 to -600) from the human vimentin promoter, was synthesized as 5′-CGACAGAACCTCCCCTCCCCCCAACATCTCCC-3′ and 5′-GGGAGATGTTGGGGGGAGGGGAGGTTCTGTCG-3′. Various mutant H4TF-1 fragement were constructed in which sequential three base pair substitutions replaced the underlined sequence in Figure 6. His-tagged ZBP-89 was prepared as described (Wieczorek et al., 2000). EMSAs were carried out as previously described (Wu et al., 2007a). For competition assays, a 50-fold molar excess of unlabeled dsDNA fragment was incubated with His-tagged ZBP-89 prior to adding 32P-labeled DNA.
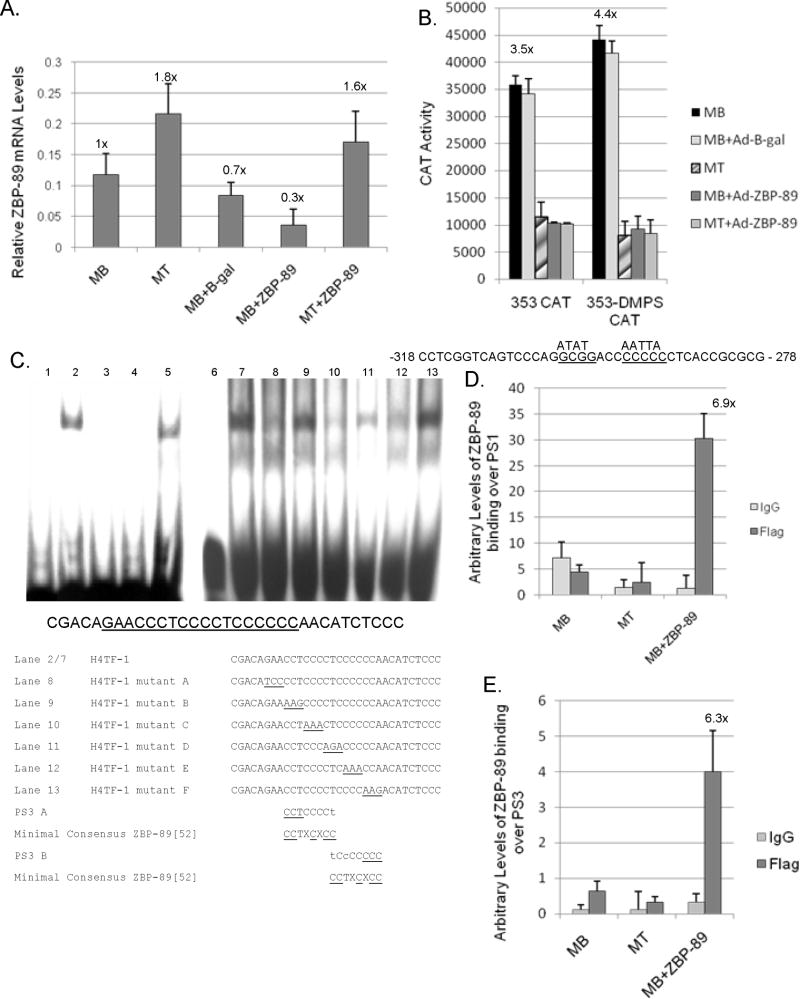
Fig. 6.
ZBP-89 is critical to vimentin down-regulation during the C2C12 myogenic program. (A) qPCR analysis of endogenous ZBP-89 mRNA levels normalized to U6 expression as described in Fig. 3A with MBs set to 1X. (B) Vimentin CAT constructs -353, and -353 containing the doubly mutated PS2 site (-353-DMPS) were transiently transfected into C2C12 cells as described in Material and methods and in Fig 2B. Mutation of PS2 is indicated as previously described (Wieczorek et al., 2000). (C) EMSA analysis of recombinant His-tagged ZBP-89 binding to the H4TF-1 element as follows: Lanes 1 and 6, 32P-labeled H4TF-1 DNA alone: Lanes 2 and 7, 32P-labeled H4TF-1 DNA plus His-tagged recombinant ZBP-89; Lanes 3 and 4 as in Lane 2 with the addition of a 50-fold excess of cold competitor PS2 and PS3 DNA, respectively; Lane 5 as in Lane 2 with a 50-fold excess of ASE DNA (non-specific DNA); Lanes 8 thru 13, as in Lane 2 with the addition of a 50-fold excess of mutant DNA consisting of 3 bases mutated in succession 5' to 3' from the underlined section and referred to as A to F in lanes 8 - 13. (D) ChIP analysis of ZBP-89 binding over PS1 element as described in Materials and methods. (E) ChIP analysis of ZBP binding over the PS3. In both panels D and E, experiments were performed in triplicate three times with the calculated increase in binding with ZBP-89 over expression shown.
ChIP Assays
C2C12 MBs were either harvested, allowed to differentiate for 4 days, or harvested after being infected with Ad-ZBP-89 for 48 h. ChIP assays were carried out as previously described (Wu et al., 2007a). For qPCR, extracted DNA (2/20 μl) was used in 50 cycles of amplification in 3 steps: 95°C for 15 s, 55°C for 30 s, and 68°C for 45 s and processed as described earlier for qPCR. Data was normalized to IgG immunoprecipitated DNA levels. Experiments were carried out three times in triplicate and a representative experiment is shown. Primer sequences for GC-box1 and PS1 were 5′-GCTAGGTCCCGATTGGCT-3′ and 5′-CGAGGGCGCTGTTTTTAT-3′ yielding a PCR product of 92 bp. Primer sequences for the PS3 element were 5′GCCCGTTAGGTCCC-TCGACA-3′ and 5′-TCCGGCCCGCGCCTCTGTCC-3′, yielding an 80 bp PCR product. Primer sequences for the AP-1 element were described previously (Wu et al., 2007a). Primer sequences for the ASE were 5′-TTGGTAGCACTGAGAACTAG-CAGCG-3′ and 5′-ATTGAGGGCTCGTAGCGGTTTAG-3′ giving a 100 bp PCR product.
Results
Vimentin Expression is Down-regulated During Myogenesis
Vimentin gene expression is selectively down-regulated during myogenesis to make way for the muscle-specific IFP, desmin (Moura-Neto et al., 1996; Farrell et al., 1990). To determine if the transcription factor ZBP-89 is involved, C2C12 MBs were infected with either Ad-empty vector, Ad-β-gal or Ad-ZBP-89, harvested after 48 h, and vimentin and desmin mRNA levels monitored by qPCR (Fig. 1A). As predicted, vimentin expression declined 3-fold in C2C12 MTs compared to MBs. Control MBs infected with either Ad-β-gal or Ad-empty vector retained vimentin expression at a level comparable to non-infected MBs. The over-expression of ZBP-89 in MBs kept in GM resulted in the down-regulation of vimentin expression to mRNA levels equivalent to that detected in MTs. MBs infected with Ad-ZBP-89 but promoted to differentiate to MTs by a switch to DM also displayed a similar decrease in vimentin mRNA levels. Conversely, desmin mRNA levels increased (6-fold) during myogenesis in agreement with previous results for desmin protein (Gard and Lazarides, 1980). Over-expression of ZBP-89 in MBs resulted in the up-regulation of desmin to levels equivalent to MTs while expression remained low in cells infected with either Ad-β-gal or Ad-empty vector. Within this context, we determined if Ad-ZBP-89 over expression was visually inducing the formation of myotubes (Fig. 1B). Here, Ad-ZBP-89 infected MBs began to exhibit elongated, spindle-shaped cells 18 h after infection and further elongated, spindle shaped cells were evident by 36 h similar to MTs in DM, yet these MBs were kept in GM containing high serum (Fig. 1B). Non-infected or Ad-β-gal infected myoblasts remained as single cells even after 36 h in GM. Western blots confirmed the temporal expression of Flag-tagged ZBP-89 in MBs+Ad-ZBP-89 (Fig. 1C). ZBP-89 induction during in vitro myogenesis was verified via qPCR analysis (Fig.1D). Myogenin up-regulation further confirmed addition of ZBP-89 promoted myogensis. From these observations it appeared that ZBP-89 over expression was enhancing the C2C12 myogenic program. Although details of this mechanism will be presented elsewhere, the down-regulation of vimentin will be investigated here.
Fig. 1.
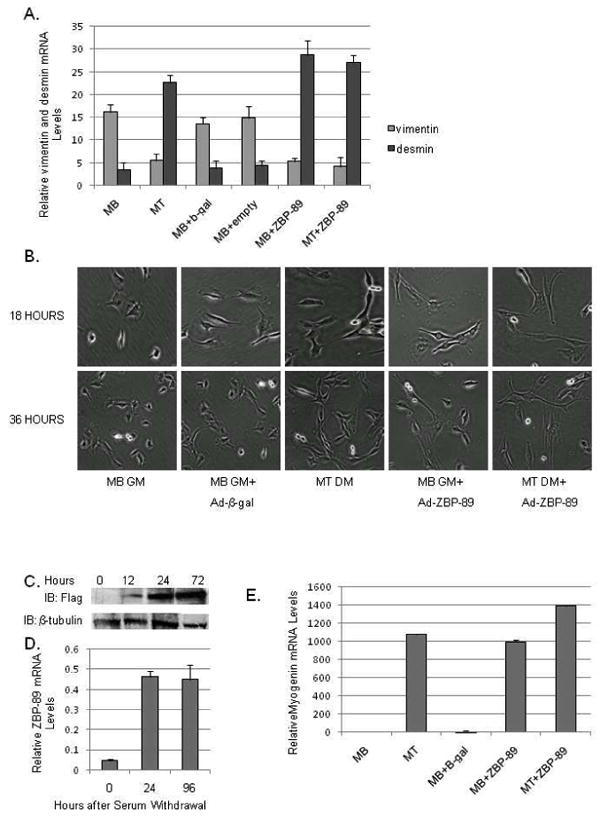
Vimentin expression is down-regulated during the C2C12 myogenic program and in response to ZBP-89 over-expression. (A) qPCR analysis of endogenous vimentin and desmin mRNA levels in myoblasts (MBs) incubated in DMEM medium plus 10% FBS (growth medium, GM), in myotubes (MTs) switched to 1% FBS (differentiation medium, DM), MBs infected with Ad-β-gal (as a negative control) and MBs or MTs infected with Ad+ZBP-89 and treated accordingly three times in triplicate. Details of qPCR conditions are discussed in Materials and Methods. The y-axis represents the relative vimentin and desmin mRNA levels normalized to U6 expression. (B) C2C12 myoblast cells (5×104) were incubated in GM or DM plus either Ad-β-gal or Ad-ZBP-89 for 18, and 36 h and visualized using a light microscope at 20X magnification. Representative fields from 3 independent experiments are shown. (C) Western blot analyses of WCEs (50 μg) isolated from C2C12 cells as grown in Fig. 1B for 12, 24, and 72 as described in Materials and Methods. β-tubulin is included as a loading control. (D) qPCR analysis of endogenous ZBP-89 mRNA levels in MBs incubated for 24 h in 10% FBS and in MTs switched to 1% FBS for 24 and 96 h. Results are the average of 3 separate experiments performed in triplicate with bars representing the standard error for panels A and D. (E) qPCR analysis of endogenous myogenin levels in MB and MT infected with Ad-β-gal and Ad-ZBP-89 for 48 h. Results are normalized to MB set as 1× and are the average of three separate experiments performed in triplicate with bars representing the standard error.
At this time the vimentin promoter was known to contain multiple DNA regulatory elements including two ZBP-89 binding sites (PS2 and PS1), which affected vimentin gene repression in other cell types (Fig. 2A). Transient transfection analyses were performed with a variety of vimentin promoter CAT constructs in C2C12 MB, MT, MB plus Ad-β-gal, and MB and MT plus Ad-ZBP-89 to ascertain which DNA elements were important for the down-regulation of vimentin during myogenic differentiation (Fig. 2B). In MB and MB plus Ad-β-gal cells, an expression pattern was detected analogous to that displayed by non-muscle cells such as HeLa (Wieczorek et al., 2000). The proximal promoter region in construct -261CAT gave substantial reporter gene activity probably due to its several GC-boxes. Inclusion of PS2 in -319CAT showed less activity with restoration of activity to near proximal promoter levels with the addition of the Δ19 enhancer element in the -353CAT construct (Moura-Neto et al., 1996;Wieczorek et al., 2000). Construct -725CAT yielded a considerable reduction in reporter gene activity perhaps due to the inclusion of additional negative element(s) (Kryszke et al., 2001). To date the transcription factor that binds to this region has not been fully characterized, although ZBP-89 binding to a third PS element has been suggested. The addition of the tandem AP-1 elements in -775CAT, which bind c-Jun and c-Fos, restored CAT activity to those levels displayed by the shorter promoter regions contained in the -353 and -261CAT constructs (Wu et al., 2007a). Further activation occurred with the addition of the ASE in the -815CAT construct, which binds Stat3 (Izmailova et al., 2000). Thus, it appeared that the positive factors, Sp1/Sp3, c-Jun and Stat3, which bind to their respective elements and promote vimentin gene expression, as well as some amount of the repressor ZBP-89 binding the PS elements were present in MBs and MBs+Ad-β-gal. However, considerably lower expression (2- to 10-fold) was noted in MTs for all of these promoter constructs. Interestingly, the expression of Ad-ZBP-89 in MBs resulted in a CAT expression pattern that mimicked MTs while addition of Ad-ZBP-89 in MT resulted in further suppression of the various CAT constructs. A number of scenarios could explain this result, 1) vimentin's positive-acting transcription factors are not present in MTs or MBs+ZBP-89, 2) the positive-acting factors are present, but not active perhaps due to excess ZBP-89 repressor, or 3) a combination of these possibilities.
Fig. 2.
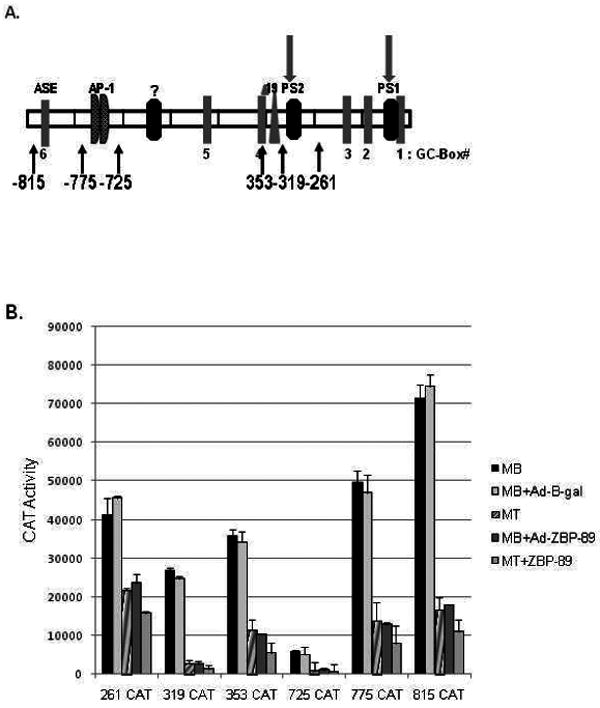
Vimentin gene expression is down-regulated in response to the C2C12 myogenic program and ZBP-89 over-expression. (A) Schematic representation of the vimentin promoter depicting the location of its various regulatory elements as follows: GC-boxes are numbered 1-6; proximal silencer elements (PS1 and PS2), which binds ZBP-89; Δ19 whose binding protein is unknown; tandem AP-1 elements, which bind AP-1 family members; and the anti-silencer element, ASE, which binds Stat3 (Wieczorek et al., 2000; Wu et al., 2007b; Zhang et al., 2003). An uncharacterized silencer element (H4TF-1) is represented as a ? (Kryszke et al., 2001) (B) Vimentin CAT constructs (containing from -261,-319,-353, -725, -775 or -815 to +72) were transiently transfected into C2C12 cells as described in Material and methods. Reporter gene activity was measured in MBs, MTs, and MBs infected with either Ad-β-gal or Ad-ZBP-89, 48 h after transfection. Bars represent the standard error of the mean. Transient transfections were performed three times in triplicate on at least two separate plasmid preps.
Myogenesis Induces the Down-regulation of Vimentin's Positive-acting Factors
To determine if vimentin's positive-acting factors were lower in MTs, qPCR analysis was performed on RNA isolated from MBs, MTs and similar cells infected with Ad-empty vector, Ad-β-gal, or Ad-ZBP-89 (Fig. 3A). Sp1 mRNA levels were high in MBs and declined (>2-fold) in MTs. ZBP-89 expression produced a similar decline in Sp1 mRNA levels in both MB and MT cells infected with Ad-ZBP-89 whereas infection with Ad-empty vector or Ad-β-gal had minimal effects. An even more striking decrease (50-fold) was noted for Sp3 mRNA levels (Fig. 3B) with little effect from infection with Ad-empty vector or Ad-β-gal. Again ZBP-89 expression mimicked the decline in Sp3 mRNA levels even for MBs kept in GM. These results agreed with previous findings that Sp1/Sp3 protein levels decline considerably during C2C12 myogenesis via Western blots, EMSAs and reporter construct activity (deLeon et al., 2005).
Fig. 3.
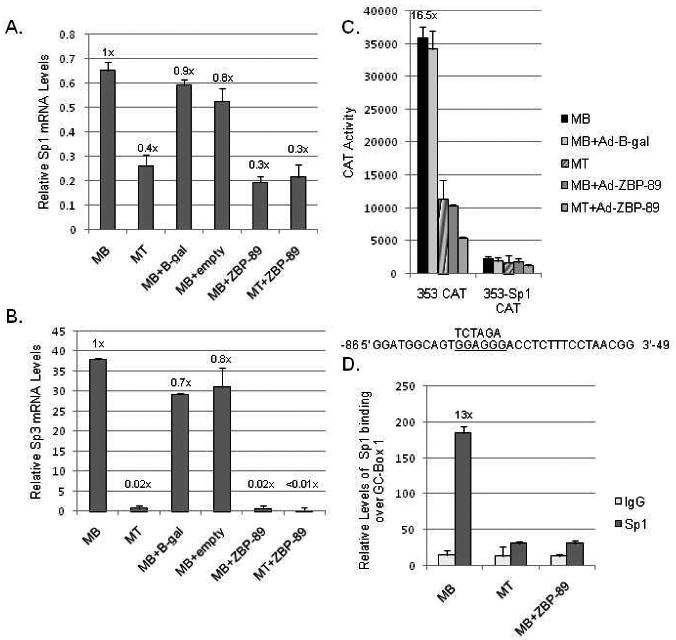
The effect of Sp1/Sp3 elimination during C2C12 myogenesis. (A) qPCR analysis of endogenous Sp1 mRNA levels in C2C12 MBs, MTs, MBs+Ad-empty vector, Ad-β-gal or Ad-ZBP-89, or MTs+Ad-ZBP-89 isolated 48 h after infection as described in Materials and methods. The y-axis represents the relative Sp1 mRNA levels normalized to U6 expression. Results are the average of three separate experiments performed in triplicate with bars representing the standard error of the mean. The numbers above each bar are calculated fold-values compared to MBs set as 1X. (B) qPCR analysis of endogenous Sp3 mRNAs levels prepared as mentioned in part A. (C) Vimentin CAT constructs -353 and -353 with GC-box1 mutated (-353-Sp1) were transiently transfected into C2C12 cells and reporter gene activity in MBs, MTs and MBs+Ad-β-gal or Ad-ZBP-89 (48 h after infection) monitored as described in Material and Methods and in Fig. 2B. The numbers above each bar are calculated fold-values compared to MTs set as 1X. Mutation of GC-box 1 is indicated as previously described (Izmailova et al., 1999; Zhang et al., 2003). (D) ChIP analysis of Sp1 binding over GC-box 1 in MBs, MTs, and MBs+Ad-ZBP-89 (48 h after infection) as described in Materials and methods.
Previously, transient transfection assays in HeLa cells showed that a mutation in vimentin's GC-box 1 (-Sp1) resulted in reduced activity of the -353-Sp1 CAT construct despite its inclusion of upstream elements (Izmailova et al., 1999). Here, the activity of this construct was compared when transfected into C2C12 MBs, MTs or MBs infected with Ad-β-gal or Ad-ZBP-89 (Fig. 3C). MBs and MBs+Ad-β-gal yielded considerable CAT activity, which was reduced 16-fold in MTs or MBs+Ad-ZBP-89. Mutation of GC-box 1 yielded low activity in all samples. This result confirmed the importance of GC-box 1 to vimentin promoter activity and suggested that most of the activity displayed by this proximal promoter region was due to Sp1/Sp3 binding as reporter gene activity was reduced when this site was mutated. ChIP analysis confirmed a decrease in Sp1 binding to the endogenous vimentin gene in MTs or MBs+Ad-ZBP-89 compared to untreated MBs (Fig. 3D). A consistent low level of binding was detected for IgG.
c-Jun protein levels were also found to naturally decline during myogenesis (Wu et al., 2007a). qPCR analysis indicated a concomitant 5-fold reduction in c-Jun mRNA levels during myogenesis. Although less of a decline was noted for Ad-ZBP-89 infected MBs, a comparable decrease was detected in MTs infected with Ad-ZBP-89 (Fig. 4A). Reporter gene activity of the wild-type -775 construct was compared to a known mutant -775-AP-1 construct by transient transfection into the C2C12 MB, MT and MB infected with Ad-ZBP-89 or Ad-β-gal cell lines (Fig. 4B). Mutation of the tandem AP-1 elements resulted in a significant decrease (>3-fold) of vimentin promoter activity. It is notable that some activity remains for the -775-AP-1 mutant construct in MBs probably due to the GC-boxes binding Sp1/Sp3 that surround these AP-1 sites (Wu et al., 2007a). The reduced activity of the -775-AP-1 construct in MTs and MBs+ZBP-89 further supported the fact that functional Sp1/Sp3 levels decline during myogenesis. It also suggested that little c-Jun was present on the vimentin promoter after the myogenic program was induced as further verified via ChIP assays (Fig. 4C).
Fig. 4.
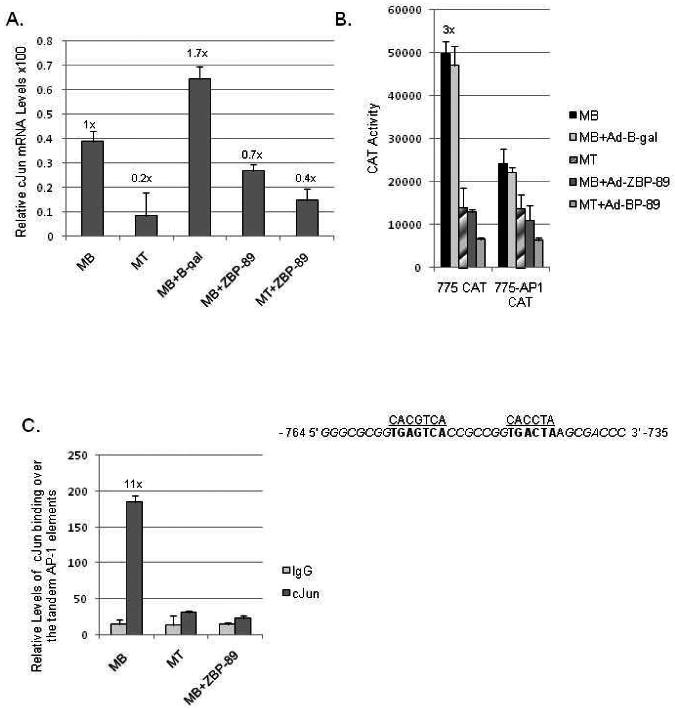
The effect of c-Jun elimination during C2C12 myogenesis. (A) qPCR analysis of endogenous c-Jun mRNA levels prepared and quantitated as described in Fig. 3A. (B) Vimentin CAT constructs -775 and -775 with its tandem AP-1 elements mutated (-775-AP1) were transiently transfected into C2C12 cells and reporter gene activity measured as described in Materials and methods. Mutations in the AP-1 binding sites are indicated as previously described plus the location of surrounding GC-rich elements thought to bind Sp1 are noted in italics (Wu et al., 2007a). (C) ChIP analysis of c-Jun binding over the tandem AP-1 elements in MBs, MTs, and MBs+Ad-ZBP-89 as described in Materials and methods. The calculated fold-value for MBs versus MTs or MBs +Ad-ZBP-89 is shown.
Finally, qPCR analysis indicated that Stat3 mRNA was severely down-regulated (>50-fold) during C2C12 myogenesis (Fig. 5A). Stat3 mRNA levels were high in MBs and MBs+Ad-β-gal and then significantly decreased in MTs. Interestingly, Stat3 mRNA levels in MBs or MTs infected with Ad-ZBP-89 were roughly equivalent to MT levels. Transient transfection analysis of an ASE Stat3 binding site mutant (-815-ASE) resulted in a significant decline in vimentin promoter activity in MB and MBs+Ad-β-gal cells compared to the wild type -815CAT construct (Fig. 5B). CAT activity was minimal with or without the known ASE mutation in MTs and MBs+Ad-ZBP-89; thus, confirming the lack of active Stat3 in MTs. The remaining activity of the -815-ASE construct in MBs or MBs+Ad-β-gal was probably due to the retention of intact binding sites for Sp1/Sp3 and AP-1 proteins as discussed earlier. These results were verified via ChIP analysis of Stat3 binding on the endogenous vimentin promoter (Fig. 5C).
Fig. 5.
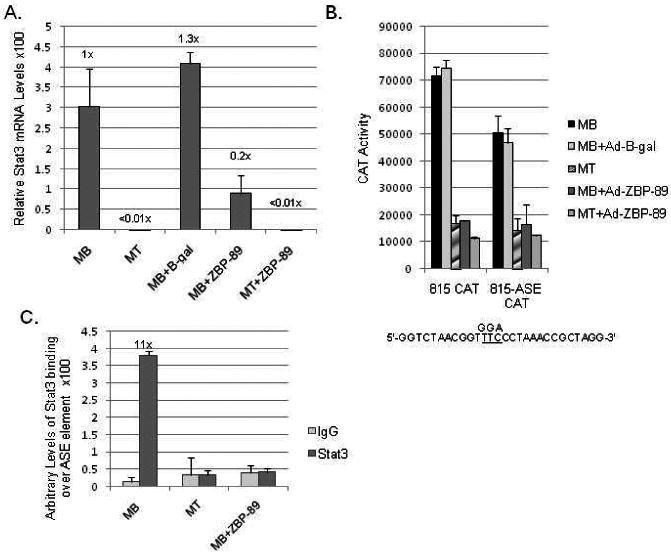
The effect of Stat3 elimination during C2C12 myogenesis. (A) qPCR analysis of endogenous Stat3 mRNA levels normalized to U6 expression was carried out as described in Fig. 3A. (B) Vimentin CAT constructs -815 and -815 with its Stat3 site mutated (-815-ASE) were transiently transfected into C2C12 cells and reporter gene activity was measured as described in Material and methods and in Fig. 2B. Mutation of the Stat3 site is indicated as previously described (Wu et al., 2004). (C) ChIP analysis of Stat3 binding over the ASE element in MBs, MTs, and MBs+ZBP-89 as described in Materials and methods. The calculated fold-value for MB compared to MT or MBs +Ad-ZBP-89 is shown.
ZBP-89 Contributes to Vimentin Down-regulation during Myogenesis
With primers specific to mouse ZBP-89, we verified via qPCR that the 1.8-fold increase in endogenous ZBP-89 mRNA levels during C2C12 myogenesis was not signifcantly affected by Ad-B-gal infection (Fig. 6A). Interestingly, the expression of rat Ad-ZBP-89 induced a slight decrease (0.3X) in expression of endogenous mouse ZBP-89 in MBs. Perhaps ZBP-89 acts via an auto-regulatory loop to decrease MB expression where developmentally expression should be limited.
Two ZBP-89 binding sites, PS1 and PS2, are known within the vimentin promoter (Fig. 2A) as confirmed by transient transfection analysis (Fig. 6B). Mutation of PS2 within the -353CAT construct (-353-DMPS) resulted in a slight increase in CAT activity in C2C12 MBs and MBs+Ad-β-gal. Upon differentiation to MTs or MBs infected with Ad-ZBP-89, the -353-DMPS construct exhibited decreased CAT activity. This result was probably due to the retention of the intact PS1 site and the documented decrease of Sp1/Sp3 protein during C2C12 myogenesis both of which would result in the lack of vimentin expression (Fig. 3C) (deLeon et al. 2005).
Previously, another silencer element (H4TF-1) was proposed within the vimentin promoter (at -620 to -610), but the protein binding this element had not been identified (Kryszke et al. 2001). EMSAs using recombinant His-tagged ZBP-89 demonstrated that ZBP-89 can bind to the H4TF-1 element (Fig. 6C: lanes 2 and 7) as shown for PS2 (Wieczorek, et al. 2000). The addition of a 50-fold excess of unlabeled PS2 or H4TF-1 DNA (lanes 3 and 4) completely eliminated ZBP-89 binding, but not the addition of non-specific ASE DNA (lane 5) verifying that ZBP-89 binding was specific. Competition assays with unlabeled DNA containing sequential mutations that substitute GC-bases for A-residues (mutants A-F) within the proposed ZBP-89 binding site (Fig. 6C, lanes 8-13), suggested that there are two regions within the H4TF-1 element important for ZBP-89 binding. The regions contained within mutants B (PS3A) and F (PS3B) did not compete as well as mutants A, C, D, and E for 32P-DNA binding. Interestingly, each region contained a partial match (7/8 or 6/8, respectively) to the ZBP-89 consensus site, which might explain the strong repression exhibited by the -725CAT construct (Fig. 2B). Endogenous binding of Ad-ZBP-89 to the vimentin promoter was verified over both PS1 at -111 to -83 (Fig. 6D) and the new PS3 site (H4TF-1) at -620 to -610 (Fig. 6E) via ChIP assays using Flag-tagged antibody. Binding to PS1 confirms why despite mutation of PS2 in the -353-DMPS mutant, repression was not relieved. Since a commercial ZBP-89 antibody suitable for ChIP analysis is not available, we could not monitor ZBP-89 binding to the endogenous vimentin promoter in non-infected cells. Flag-tagged antibody binding was equivalent to background levels of control IgG binding in MBs and MTs.
Elimination of ZBP-89 Results in the Maintenance of Vimentin Expression
To further evaluate the role of ZBP-89 in the myogenic program, ZBP-89 was eliminated from C2C12 MBs by transfection of a mouse-specific siRNA. MBs were allowed to differentiate to MTs by serum withdrawal and examined for visual fusion of MTs compared to MBs and MTs treated with a siControl (Fig. 7A). MBs continued to proliferate and remained as single, spindle-shaped cells whereas MTs formed long, fused cells in culture. MBs transfected with siZBP-89 remained proliferative, since more cells were prevalent at 36 h than at 18 h. Since a proven, commercial ZBP-89 antibody is lacking, ZBP-89 mRNA levels were assessed via qPCR analysis (Fig. 7B). Here, ZBP-89 mRNA levels were down-regulated after treatment with siRNA in both MB and MT cells while control cells exhibited normal patterns of up-regulation (≅1.8-fold) during myogenesis. qPCR analysis confirmed a substantial induction (>1000-fold) of myogenin mRNA during C2C12 myogenesis, which was eliminated in the presence of siZBP-89 whereas the siControl showed little effect (Fig. 7C). Western blots confirmed that eliminating ZBP-89 blocked the expression of the muscle-specific proteins myogenin, MHC or MyoD even after 96 h in culture (Fig. 7D). Tubulin levels remained constant serving as a loading control. MT differentiated for 24 h then transfected with siZBP-89 further confirmed ZBP-89's importance to myogenesis for elimination of ZBP-89 at 24 h down-regulated myogenin mRNA levels despite initiation of myogenesis.
Fig. 7.
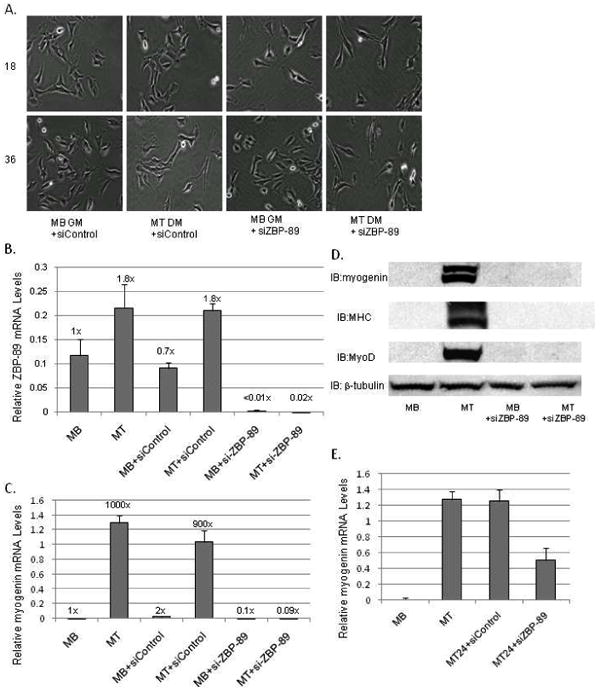
The effect of eliminating ZBP-89 expression via ZBP-89 siRNA on the C2C12 myogenic program. (A) MBs (5×104) were incubated in GM, DM or transfected with either si-ZBP-89 or siControl in GM then visualized with a light microscope for 18 and 36 h as described in Fig. 1B. (B) qPCR analysis of endogenous ZBP-89 mRNA levels normalized to U6 expression isolated at 48 h as described in Fig. 3A (C) qPCR analysis of endogenous myogenin mRNA levels normalized to U6 expression isolated at 48 h as prepared and described in panel B. (D) Western blots analysis of WCEs (50 μg) isolated from C2C12 cells as grown in panel A for 96 h as described in Materials and methods. β-tubulin serves as a loading control. (E) MT cells (5×104) were allowed to differentiate for 24 h and then transfected with either siZBP-89 or siControl. Cells were harvested 48 h later and qPCR analysis was performed as mentioned in panel B. Experiments were performed in triplicate three times with bars representing the standard error of the mean.
Since ZBP-89 is a known repressor for the vimentin promoter, elimination of ZBP-89 could result in continued vimentin expression despite the reduction in serum. In fact this was the case, as vimentin mRNA levels remained high with ZBP-89 elimination (Fig 8A). However, vimentin mRNA levels decreased in MTs or MTs treated with the siControl. ZBP-89 elimination gave the opposite result for desmin expression. Retention of vimentin protein in MBs treated with siZBP-89 was quite evident in Western blots superceding that level displayed by untreated MBs (Fig. 8B).
Fig. 8.
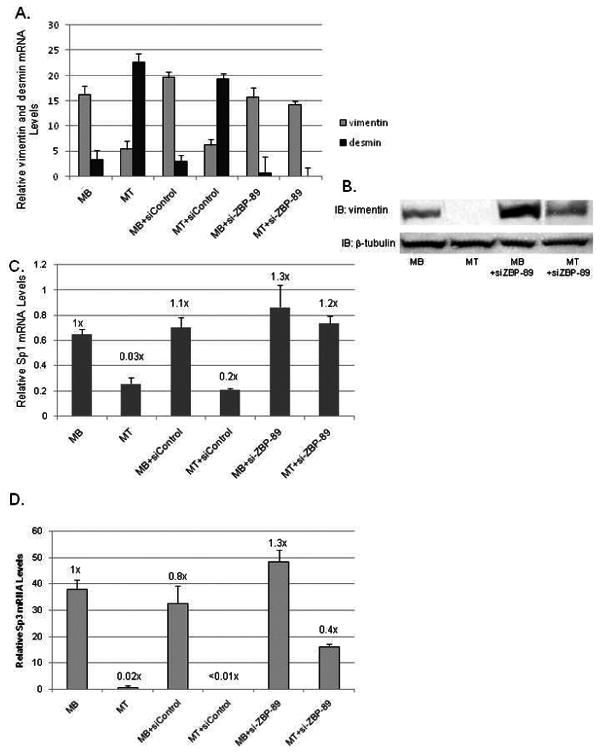
ZBP-89 elimination via siRNA prevents vimentin down-regulation and affects Sp1/Sp3 expression during C2C12 myogenesis. (A) qPCR analysis of endogenous vimentin and desmin mRNA levels normalized to U6 expression as described in Figs. 7B and 3A. (B) Western blots analysis of WCEs (50 μg) isolated from C2C12 cells grown as in Fig. 7A for 96 h and described in Fig. 7D. β-tubulin serves as a loading control. (C) qPCR analysis of endogenous Sp1 mRNA levels isolated at 48 h, normalized to U6 levels and prepared as described in Figs. 3A and 7B. (D) qPCR analysis of endogenous Sp3 mRNA levels prepared as in panel A.
Since vimentin is present in MBs treated with siZBP-89, we determined if expression of vimentin's essential positive-acting factors Sp1 and Sp3 were affected by siZBP-89. qPCR analysis showed that Sp1 mRNA levels were high in MBs, but declined (3-fold) in MTs (Fig. 8C). MB and MT cells treated with siZBP-89 exhibited slightly higher levels of Sp1 mRNA compared to untreated MBs. Similar results were seen for Sp3 mRNA levels except MTs+siZBP-89 declined ≅ 2-fold, but not 50-fold as for untreated MTs (Fig. 8D).
Discussion
The mechanism for the down-regulation of vimentin gene expression during C2C12 differentiation in vitro requires the careful orchestration between positive- and negative-acting regulatory factors. Vimentin expression is high in MBs and MBs+Ad-β-gal because the positive-factors that control vimentin expression, i.e. Sp1/Sp3, c-Jun and Stat3, are present and active. Conversely, as MBs differentiate into MTs, these positive-acting transcription factors are down-regulated at the mRNA level in agreement with previous reports documenting decreased protein levels (deLeon et al., 2005; Fischer and Hilfiker-Kleiner, 2007; Wu et al., 2007a). ChIP analyses confirmed decreased binding of these factors to the endogenous vimentin promoter in MTs and MBs engineered to over-express ZBP-89 compared to MBs. Thus, the down-regulation of vimentin expression is due at least in part to a significant decrease of these activators in MTs. However, this end result also requires the repressor ZBP-89, since vimentin gene expression down-regulates significantly within the first 48 h (Fig. 8A) yet Sp1/Sp3 mRNA (Fig. 3) and protein levels are still evident (deLeon et al., 2005). Moreover, Sp1/Sp3 expression must be directly or indirectly regulated by ZBP-89, since the addition of siZBP-89 blocked the normal decrease of these mRNAs.
In combination with the down-regulation of these activators, there was a concomitant up-regulation of the repressor ZBP-89. With purified recombinant ZBP-89 vimentin's H4TF-1 element was shown to contain an overlapping ZBP-89 binding site bringing the total number of sites within the human vimentin gene promoter to three in agreement with results for the chicken gene (Garzon and Zehner, 1994). By manipulating levels of ZBP-89 via adenoviral and siRNA approaches, we were able to verify its role as a repressor for vimentin expression during myogenesis. Both the down-regulation of vimentin's activators together with the up-regulation of ZBP-89 is required because the removal of serum alone could not decrease vimentin expression to MT levels in the absence of ZBP-89 (Fig. 8A). Thus, although a decline in Sp1/Sp3 levels may be necessary to down-regulate vimentin expression, it is not sufficient by itself to completely “turn off” the vimentin gene further requiring the repressor ZBP-89.
Several possibilities exist to explain how ZBP-89 might play a role in controlling myogenesis. The mouse ZBP-89 promoter displayed increased activity in MTs versus MBs, and promoter analyses indicated a number of binding sites for myogenic factors such as MEF-2 and MyoD (Feo et al., 2001). Thus, these myogenic factors could be transcriptionally activating ZBP-89 expression during the normal myogenic program and the forced overexpression of ZBP-89 recapitulating the natural developmental process for skeletal muscle cells. Further conditional gene knockout studies are warranted to further examine the developmental role of ZBP-89 during myogenesis as total gene knockout of ZBP-89 is impossible because heterozygous knockout mice are sterile (Takeuchi et al., 2003). Furthermore, ZBP-89 can be either an activator or a repressor of gene expression. ZBP-89 binds to the p21 promoter and interacts with p300 to activate gene expression while at the same time binding to the vimentin gene to down-regulate gene expression during myogenesis (Bai and Merchant, 2000; Wu el al., 2007a). Since ZBP-89 binds to GC-rich regions, a common promoter element, many additional genes could be regulated by ZBP-89. For example, the down-regulation of the dystrophin gene, Dp71, was also found to be dependent on Sp1/Sp3 protein levels. Like vimentin, this promoter is GC-rich and may also be a target for ZBP-89 regulation (deLeon et al., 2005). Thus, ZBP-89 may act as an activator for muscle-specific genes while serving as a repressor for non-muscle genes.
While there is currently little explanation as to what controls ZBP-89's ability to function as an activator versus a repressor, this mechanism must be promoter-specific and most likely depends on post-translational modifications, since ZBP-89 is able to display both functions concurrently in a single cell-type. It is known that ZBP-89 contains multiple bi-functional regulatory domains. For example, ZBP-89 recruits p300 to the p21 promoter via its N-terminal, acidic domain to form a large activation complex including ZBP-89, Sp1 and p300 (Bai and Merchant, 2001). Interestingly, this same acidic domain is also required for repression of vimentin gene expression (Zhang et al., 2003). Many other transcription factors such as Sp1 or ZNF-76 can act as bi-functional regulators (Chu and Ferro, 2005). ZNF-76 is regulated by a complex interplay of acetylation/de-acetylation, sumoylation and alternative splicing. When acetylated ZNF-76 serves as an activator, but when sumoylated it interacts with TBP to function as a repressor. This regulatory circuit is further complicated by splicing variants with altered affinities for TBP (Zheng and Yang, 2006). Thus, we speculate that post-translational modifications such as phosphorylation and/or sumoylation may regulate the ability of ZBP-89 to function as an activator or repressor of transcription. In vivo 32P-labelling followed by immunoprecipitation revealed that ZBP-89 is indeed a phosphorylated protein (Wu unpublished observation). Furthermore, ZBP-89 has recently been shown to be modified by sumoylation (Chupreta et al., 2007). When sumoylated, ZBP-89 functions as a strong repressor. Mutation of two sumo sites within the N-terminus of ZBP-89 resulted in the conversion to a weak activator. Thus, ZBP-89's role as an activator versus a repressor may consist of a complex regulatory feedback loop that includes a distinct combination of post-translational modifications such as phosphorylation, acetylation/deacetylation or sumoylation. Further experiments will be required to completely elucidate this mechanism with reference to the myogenic program.
Acknowledgments
This work was supported by NHLBI, National Institutes of Health (NIH) Grant HL-45422 and an American Heart Mid-Atlantic pre-doctoral fellowship (0415464U) to M.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreucci JJ, Grant D, Cox DM, Tomc LK, Prywes R, Goldhamer DJ, Rodrigues N, Bedard PA, McDermott JC. Composition and function of AP-1 transcription complexes during muscle cell differentiation. J Biol Chem. 2002;277:16426–16432. doi: 10.1074/jbc.M110891200. [DOI] [PubMed] [Google Scholar]
- Bai L, Merchant JL. Transcription factor ZBP-89 cooperates with histone acetyltransferase p300 during butyrate activation of p21waf1 transcription in human cells. J Biol Chem. 2000;275:30725–30733. doi: 10.1074/jbc.M004249200. [DOI] [PubMed] [Google Scholar]
- Bai L, Merchant JL. ZBP-89 promotes growth arrest through stabilization of p53. Mol Cell Biol. 2001;21:4670–4683. doi: 10.1128/MCB.21.14.4670-4683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF-2) proteins. Ann Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- Chen JH, Vercamer C, Li Z, Paulin D, Vandenbunder B, Stehelin D. PEA3 transactivates vimentin promoter in mammary epithelial and tumor cells. Oncogene. 1996;13:1667–1675. [PubMed] [Google Scholar]
- Cheng PY, Kagawa N, Takahashi Y, Waterman MR. Three zinc finger nuclear proteins, Sp1, Sp3, and a ZBP-89 homologue, bind to the cyclic adenosine monophosphate-responsive sequence of the bovine adrenodoxin gene and regulate transcription. Biochemistry. 2000;39:4347–4357. doi: 10.1021/bi992298f. [DOI] [PubMed] [Google Scholar]
- Chu S, Ferro T. Sp1: regulation of gene expression by phosphorylation. Gene. 2005;348:1–11. doi: 10.1016/j.gene.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Chupreta S, Brevig H, Bai L, Merchant JL, Iniguez-Lluhi JA. Sumoylation-dependent control of homeoptypic and heterotypic synergy by the krüppel-type zinc finger protein ZBP-89. J Biol Chem. 2007;282:36155–36166. doi: 10.1074/jbc.M708130200. [DOI] [PubMed] [Google Scholar]
- deLeon MB, Montanez C, Gomez P, Morales-Lazaro SL, Tapia-Ramirez V, Valadez-Graham V, Recillas-Targa F, Yaffe D, Nudel U, Cisneros B. Dystrophin Dp71 gene expression is down-regulated during myogenesis: Role of Sp1 and Sp3 on the Dp71 promoter activity. J Biol Chem. 2005;280:5290–5299. doi: 10.1074/jbc.M411571200. [DOI] [PubMed] [Google Scholar]
- Duprey P, Paulin D. What can be learned from intermediate filament gene regulation in the mouse embryo? Int J Dev Biol. 1995;39:443–457. [PubMed] [Google Scholar]
- Farrell FX, Sax CM, Zehner ZE. A negative element involved in vimentin gene expression. Mol Cell Biol. 1990;10:2349–2358. doi: 10.1128/mcb.10.5.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feo S, Antona V, Cammarata G, Cavaleri F, Passantino R, Rubino P, Giallongo A. Conserved structure and promoter sequence similarity in the mouse and human genes encoding the zinc finger factor BERF-1/BFCOL1/ZBP-89. Biochem Biophys Res Commun. 2001;283:209–218. doi: 10.1006/bbrc.2001.4753. [DOI] [PubMed] [Google Scholar]
- Fischer P, Hilfiker-Kleiner D. Survival Pathways in hypertrophy and heart failure: the gp130-STAT3 axis. Basic Res Cardiol. 2007;102:393–411. doi: 10.1007/s00395-007-0674-z. [DOI] [PubMed] [Google Scholar]
- Gard DL, Lazarides E. The synthesis and distribution of desmin and vimentin during myogenesis in vitro. Cell. 1980;19:263–275. doi: 10.1016/0092-8674(80)90408-0. [DOI] [PubMed] [Google Scholar]
- Garzon RJ, Zehner ZE. Multiple silencer elements are involved in regulating the chicken vimentin gene. Mol Cell Biol. 1994;14:934–943. doi: 10.1128/mcb.14.2.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa T, Xiao H, Isobe K. Cloning of a GADD34-like gene that interacts with the zinc-finger transcription factor which binds to the p21WAF promoter. Biochem Biophys Res Commun. 1999;256:249–254. doi: 10.1006/bbrc.1999.0275. [DOI] [PubMed] [Google Scholar]
- Izmailova ES, Snyder SR, Zehner ZE. A Stat1α factor regulates the expression of the human vimentin gene by IFN-γ. J Interferon Cytokine Res. 2000;20:13–20. doi: 10.1089/107999000312694. [DOI] [PubMed] [Google Scholar]
- Izmailova ES, Wieczorek E, Perkins EB, Zehner ZE. A GC-box is required for expression of the human vimentin gene. Gene. 1999;235:69–75. doi: 10.1016/s0378-1119(99)00209-7. [DOI] [PubMed] [Google Scholar]
- Izmailova ES, Zehner ZE. An antisilencer element is involved in the transcriptional regulation of the human vimentin gene. Gene. 1999;230:111–120. doi: 10.1016/s0378-1119(99)00046-3. [DOI] [PubMed] [Google Scholar]
- Keates AC, Keates S, Kwon JH, Arseneau KO, Law DJ, Bai L, Merchant JL, Wang TC, Kelly CP. ZBP-89, Sp1, and nuclear factor-κB regulate epithelial neutrophil-activating peptide-78 gene expression in caco-2 human colonic epithelial cells. J Biol Chem. 2001;276:43713–22. doi: 10.1074/jbc.M107838200. [DOI] [PubMed] [Google Scholar]
- Kryszke MH, Moura-Neto V, Lilienbaum A, Paulin D, Auclair C. Involvement of histone H4 gene transcription factor 1 in downregulation of vimentin gene expression during skeletal muscle differentiation. FEBS Lett. 2001;491:30–34. doi: 10.1016/s0014-5793(01)02142-1. [DOI] [PubMed] [Google Scholar]
- Law DJ, Du M, Law L, Merchant JL. ZBP-99 defines a conserved family of transcription factors and regulates ornithine decarboxylase gene expression. Biochem Biophys Res Commun. 1999;262:113–120. doi: 10.1006/bbrc.1999.1180. [DOI] [PubMed] [Google Scholar]
- Lilienbaum A, Paulin D. Activation of the human vimentin gene by the Tax human T-cell leukemia virus I: Mechanisms of regulation by the NF-kB transcription Factor. J Biol Chem. 1993;268:2180–2188. [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- Merchant JL, Iyer GR, Taylor BR, Kitchen JR, Mortensen ER, Wang Z, Flintoft RJ, Michel JB, Bassel-Duby R. ZBP-89, a krüppel-like zinc finger protein, inhibits epidermal growth factor induction of the gastrin promoter. Mol Cell Biol. 1996;16:6644–6653. doi: 10.1128/mcb.16.12.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura-Neto V, Kryszke MH, Li Z, Vicart P, Lilienbaum A, Paulin D. A 28-bp negative element with multiple factor-binding activity controls expression of the vimentin-encoding gene. Gene. 1996;168:261–266. doi: 10.1016/0378-1119(95)00789-x. [DOI] [PubMed] [Google Scholar]
- Parry DA, Steinert PM. Intermediate filament structure. Curr Opin Cell Biol. 1992;4:94–98. doi: 10.1016/0955-0674(92)90064-j. [DOI] [PubMed] [Google Scholar]
- Passantino R, Antona V, Barbieri G, Rubino P, Melchionna R, Cossu G, Feo S, Giallongo A. Negative regulation of β-enolase gene transcription in embryonic muscle is dependent upon a zinc finger factor that binds to the G-rich box within the muscle-specific enhancer. J Biol Chem. 1998;273:484–494. doi: 10.1074/jbc.273.1.484. [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP. Myogenic Regulatory Factors and the Specification of Muscle Progenitors in Vertebrate Embryos. Annr Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- Reizis B, Leder P. Expression of the mouse pre-T cell receptor α gene is controlled by an upstream region containing a transcriptional enhancer. J Exp Med. 1999;189:1669–1678. doi: 10.1084/jem.189.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittling SR, Baserga R. Functional analysis and growth factor regulation of the human vimentin promoter. Mol Cell Biol. 1987;7:3908–3915. doi: 10.1128/mcb.7.11.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittling SR, Coutinho L, Amram T, Kolbe M. AP-1/jun binding sites mediate serum inducibility of the human vimentin promoter. Nucleic Acids Res. 1989;17:1619–1633. doi: 10.1093/nar/17.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti A, Lilienbaum A, Li Z, Paulin D, Gazzolo L. Identification of a negative element in the human vimentin promoter: modulation by the human T-cell leukemia virus type I Tax protein. Mol Cell Biol. 1993;13:89–97. doi: 10.1128/mcb.13.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax CM, Farrell FX, Zehner ZE. Down-regulation of vimentin gene expression during myogenesis is controlled by a 5′-flanking sequence. Gene. 1989;78:235–242. doi: 10.1016/0378-1119(89)90226-6. [DOI] [PubMed] [Google Scholar]
- Steinert PM, Liem RK. Intermediate filament dynamics. Cell. 1990;60:521–553. doi: 10.1016/0092-8674(90)90651-t. [DOI] [PubMed] [Google Scholar]
- Takeuchi A, Mishina Y, Miyaishi O, Kojima E, Hasegawa T, Isobe K. Heterozygosity with respect to zfp148 causes complete loss of fetal germ cells during mouse embryogenesis. Nature Genetics. 2003;33:172–176. doi: 10.1038/ng1072. [DOI] [PubMed] [Google Scholar]
- Wieczorek E, Lin Z, Perkins EB, Law DJ, Merchant JL, Zehner ZE. The zinc finger repressor, ZBP-89, binds to the silencer element of the human vimentin gene and complexes with the transcriptional activator, Sp1. J Biol Chem. 2000;275:12879–12888. doi: 10.1074/jbc.275.17.12879. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang X, Salmon M, Lin X, Zehner ZE. TGFβ1 regulation of vimentin gene expression during differentiation of the C2C12 skeletal myogenic cell line requires Smads, AP-1 and Sp1 family members. Biochim et Biophys Acta Mole Cell Res. 2007a;1773:427–439. doi: 10.1016/j.bbamcr.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang X, Salmon M, Zehner ZE. The zinc finger repressor, ZBP-89, recruits histone deacetylase 1 to repress vimentin gene expression. Genes to Cells. 2007b;12:905–918. doi: 10.1111/j.1365-2443.2007.01104.x. [DOI] [PubMed] [Google Scholar]
- Wu Y, Diab I, Zhang X, Izmailova ES, Zehner ZE. Stat3 enhances vimentin gene expression by binding to the antisilencer element and interacting with the repressor protein, ZBP-89. Oncogene. 2004;23:168–178. doi: 10.1038/sj.onc.1207003. [DOI] [PubMed] [Google Scholar]
- Yamada A, Takaki S, Hayashi F, Georgopoulos K, Perlmutter RM, Takatsu K. Identification and characterization of a transcriptional regulator for the lck proximal promoter. J Biol Chem. 2001;276:18082–18089. doi: 10.1074/jbc.M008387200. [DOI] [PubMed] [Google Scholar]
- Ye S, Whatling C, Watkins H, Henney A. Human stromelysin gene promoter activity is modulated by transcription factor ZBP-89. FEBS Lett. 1999;450:268–272. doi: 10.1016/s0014-5793(99)00509-8. [DOI] [PubMed] [Google Scholar]
- Zhang X, Diab IH, Zehner ZE. ZBP-89 represses vimentin gene transcription by interacting with the transcriptional activator, Sp1. Nucl Acids Res. 2003;31:2900–2914. doi: 10.1093/nar/gkg380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Yang YC. Acetylation and alternative splicing regulate ZNF76-mediated transcription. Biochem Biophys Res Commun. 2006;339:1069–1075. doi: 10.1016/j.bbrc.2005.11.122. [DOI] [PubMed] [Google Scholar]