Abstract
Mutation in p53 tumor suppressor gene is a hallmark of human cancers. Six major mutational hotspots in p53 contain methylated CpG (mCpG) sites, and C →T transition is the most common mutation at these sites. It was hypothesized that the formation of 5-methylcytosine glycol induced by reactive oxygen species, its spontaneous deamination to thymine glycol and the miscoding property of the latter may account, in part, for the ubiquitous C →T mutation at CpG site. Here, we assessed the kinetics of deamination for two diastereomers of 5-methylcytosine glycol in duplex DNA. Our results revealed that the half-lives for the deamination of the (5S,6S) and (5R,6R) diastereomers of 5-methylcytosine glycol in duplex DNA at 37°C were 37.4 ± 1.6 and 27.4 ± 1.0 h, respectively. The deamination rates were only slightly lower than those for the two diastereomers in mononucleosides. Next, we assessed the formation of 5-methyl-2′-deoxycytidine glycol in the form of its deaminated product, namely, thymidine glycol (Tg), in methyl-CpG-bearing duplex DNA treated with Cu(II)/H2O2/ascorbate. LC-MS/MS quantification results showed that the yield of Tg is similar as that of 5-(hydroxymethyl)-2′-deoxycytidine. Together, our data support that the formation and deamination of 5-methylcytosine glycol may contribute significantly to the C →T transition mutation at mCpG dinucleotide site.
INTRODUCTION
Mutation in p53 tumor suppressor gene is common in human cancers, and the cytosines at CpG sites in the p53 gene are methylated. Although CpGs are underrepresented by 5-fold of their expected frequency in mammalian DNA, six major mutational hotspots in p53, i.e. codons 175, 213, 245, 248, 273 and 282, contain CpGs (1). The methylated CpGs (mCpG), therefore, are the most important mutation targets in p53.
Reactive oxygen species (ROS) constitute an important endogenous source of DNA damage (2), and an array of ROS-induced DNA lesions have been identified and characterized (3). The main species responsible for the ROS-induced DNA damage in cells appear to be hydroxyl radical generated by a Fenton-type reaction involving the reduction of H2O2 by transition metal ions (e.g. Fe2+ and Cu+) (4). In this context, ROS can damage 5-methylcytosine to give methyl group oxidation products and lead to the saturation of the C5 = C6 double bond, the latter gives 5-methyl-5,6-dihydroxy-5,6-dihydro-2′-deoxycytidine (or 5-methyl-2′-deoxycytidine glycol, mdCg; Scheme 1). The saturation of the C5 = C6 double bond in mdCg renders it susceptible to deamination, which affords 5,6-dihydroxy-5,6-dihydrothymidine (or thymidine glycol, Tg; Scheme 1) (5). Thymidine glycol is mostly a blocking lesion for replicative DNA polymerases (6). It, however, can be bypassed by translesion synthesis DNA polymerases including yeast DNA polymerases η and ζ as well as human DNA polymerase κ, and all three polymerases preferentially insert a dAMP opposite the lesion (7–9). Therefore, the formation and subsequent deamination of mdCg are thought to be one of the pathways leading to the C →T transition mutation at the CpG dinucleotide site. However, it remains unexplored how cytosine methylation affects the formation and deamination of the ring-saturated glycol derivative in duplex DNA.
Scheme 1.
ROS-induced DNA lesions of 5-methyl-2′-deoxycytidine and the deamination of 5-methyl-2′-deoxycytidine glycol.
Previous studies showed that the half-lives for the deamination of one diastereomer of 2′-deoxycytidine glycol and cytosine glycol (Cg) in calf thymus DNA at 37°C are 50 min and 28 h, respectively (10). On the other hand, the half-life for the deamination of the two diastereomers of mdCg in mononucleoside at 37°C is ∼20 h (5). If the difference in deamination rate of Cg in mononucleoside and duplex DNA can be extrapolated to that for 5-methylcytosine glycol, then the deamination of 5-methylcytosine glycol in DNA may not contribute significantly to the C →T mutation at the CpG dinucleotide site. Therefore, understanding the implications of the deamination of 5-methylcytosine glycol in the ubiquitous C →T transition mutation at CpG sites necessitates the measurement of the rate of deamination of 5-methylcytosine glycol in duplex DNA.
Recently, we evaluated the formation of single-nucleobase lesions, i.e. 5-(hydroxymethyl)-2′-deoxycytidine (5-HmdC) and 5-formyl-2′-deoxycytidine (5-FodC), and tandem intrastrand cross-link lesions, i.e. G[8-5m]mC and mC[5m-8]G, emanating from the 5-methyl radical of mC at CpG sites (11). We found that the yield of single-nucleobase lesions is ∼100 times more than intrastrand cross-links when methyl-CpG-bearing DNA was treated with Fenton-type reagents (11). A recent theoretical study on 5-methylcytosine predicted that the addition of •OH radical to the double bond is more facile than hydrogen atom abstraction from the 5-methyl group (12), which is in agreement with the scarce experimental data (13). It has not been examined how efficiently the mdCg can be formed relative to the methyl group oxidation products in duplex DNA.
8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) is an abundant and well-studied oxidatively induced lesion of 2′-deoxyguanosine when DNA was treated with Fenton-type reagents (11,14). The tandem lesions, in addition to single-nucleobase lesions, with a Tg residing on the 5′-side of an 8-oxodG [i.e. Tg-(8-oxodG)] could also arise from ROS attack at mCpG site and contribute to CpG mutagenesis. In this respect, vicinal or cross-linked base damage at mCpG site has been proposed to account for the abnormally high frequency of CG →TT double mutation observed after CpG-methylated and Cu(II)/H2O2/ascorbate-treated pSP189 shuttle vector was propagated in nucleotide excision repair (NER)-deficient human XPA cells (15).
In the present study, we employed LC-MS/MS with the standard isotope dilution technique to quantify the Tg and Tg-(8-oxodG) lesions formed at mCpG site in synthetic duplex oligodeoxyribonucleotides (ODNs) treated with Cu(II)/H2O2/ascorbate. We demonstrated that the yield of Tg is comparable to those of the methyl group oxidation products of mC. In addition, we developed an LC-MS method to measure the rate of deamination of mdCg in duplex DNA. Our results revealed that the deamination of mdCg in double-stranded DNA is slightly slower than that in the free nucleoside, which is significantly different from what was observed for Cg.
MATERIALS AND METHODS
Materials
CuCl2, L-methionine, L-ascorbic acid and nuclease P1 were from Sigma–Aldrich (St. Louis, MO, USA). Hydrogen peroxide (30%) was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Common reagents for solid-phase DNA synthesis were obtained from Glen Research (Sterling, VA, USA). ODNs incorporated with an isolated Tg or Tg-(8-oxodG) tandem lesion were synthesized previously (16).
Treatment of synthetic ODNs with Fenton-type reagents
A self-complementary ODN, d(AXGTAXGTAXGTAXGT) (‘X’ represents 5-methylcytosine), was annealed in a buffer containing 50 mM NaCl and 20 mM phosphate (pH 6.9) by heating the solution to 80°C and cooling slowly to room temperature. Aliquots of ODNs (2.5 nmol) were incubated with CuCl2 (6.25–100 µM), H2O2 (50–800 µM) and ascorbate (0.5–8 mM) in a 100 µl buffer (Table 1), which contained 50 mM NaCl and 20 mM phosphate (pH 6.9), at room temperature for 60 min. All chemicals were freshly dissolved in doubly distilled water and the reactions were carried out under aerobic conditions. The reaction was terminated by adding an excess amount of L-methionine, and the ODN samples were desalted by ethanol precipitation.
Table 1.
Concentrations of Fenton-type reagents employed for the treatment of DNAa
| Control | A | B | C | D | E | |
|---|---|---|---|---|---|---|
| Cu(II) (µM) | 100 | 6.75 | 12.5 | 25 | 50 | 100 |
| H2O2 (µM) | 0 | 50 | 100 | 200 | 400 | 800 |
| Ascorbate (mM) | 0 | 0.5 | 1.0 | 2.0 | 4.0 | 8.0 |
aAll reactions were carried out in a 100 µl solution containing 25 μM ODN, 25 mM NaCl and 50 mM phosphate (pH 6.9).
Deamination of mdCg and enzymatic digestion
A 27-mer duplex ODN, 5′-ACGGGCCGAAACXGGAGGCCCGACCGT-3′/5′-ACGGTCGGGCCTCXGGTTTCGGCCCGT-3′ (‘X’ represents 5-methylcytosine, 3 nmol), was treated with Cu(II)/H2O2/ascorbate as described above (condition E in Table 1). The resulting solution was exchanged into a 100 µl buffer solution containing 10 mM phosphate (pH 6.9) by using YM-10 Centricon membrane filters. The solution was subsequently incubated at 37°C. Aliquots were removed at specific time points, frozen immediately on dry ice and stored at −80°C. Prior to mass spectrometric analysis, 0.5 U of nuclease P1 and a 1.5 µl solution containing 300 mM sodium acetate (pH 5.0) and 10 mM zinc acetate were added to 25 µl of the ODN sample. The digestion was continued at room temperature for 40 min, and the enzymes were removed by passing through a YM-10 Centricon membrane filter (Millipore, Billerica, MA, USA).
Generation of authentic Tg-containing ODNs
The ODN d(ATGGCTgGGCTAT), which contained the (5R,6S) diastereomer of the Tg, was synthesized previously (16,17). ODNs containing the two cis diastereomers of Tg were obtained by OsO4 oxidation (18). Briefly, 50 nmol d(ATGC) was dissolved in a 100 μl aqueous solution of 2% OsO4 and incubated at room temperature for 24 h before adding excess amount of NaHSO3 to reduce the OsO4. The residual OsO4 was removed by three times of extraction with equal volumes of carbon tetrachloride, and the aqueous layer was dried by using a Savant Speed-Vac (Savant Instruments Inc., Holbrook, NY, USA). The dried residue was desalted by using HPLC and digested with nuclease P1 (vide supra) for the subsequent LC-MS/MS analysis.
Synthesis of ODNs containing Tg with a 3′ neighboring dG that is uniformly labeled with 15N (ODNX)
Uniformly 15N-labeled dGTP, i.e., [2-amino-1,3,7,9-15N5]-2′-deoxyguanosine 5′-triphosphate (Cambridge Isotope Laboratories, Andover, MA, USA), was enzymatically added to the 3′-terminus of a synthetic tetrameric ODN d(GCAT) using terminal deoxynucleotidyl transferase (New England Biolabs, Ipswich, MA, USA) (19). The extension products were then treated with OsO4 to afford the Tg-containing ODNs (vide supra). The enzyme was removed by passing through YM-10 Centricon membrane filter and the ODNs were recovered by ethanol precipitation, desalted on HPLC, and used as internal standard for LC-MS quantification.
LC-MS and MS/MS analysis of p(mdCg)pdG and pTgpdG
The digestion mixture was analyzed by LC-MS/MS with an Agilent 1100 capillary HPLC system and an LTQ linear ion-trap mass spectrometer (Thermo Fisher, San Jose, CA, USA) equipped with an electrospray ionization source. A 0.5 × 150 mm Zorbax SB-C18 column (5 µm in particle size, Agilent Technologies, Palo Alto, CA, USA) was used for the separation of the above nuclease P1 digestion mixtures, and the flow rate was 6.0 µl/min. For deamination analysis, a 5-min gradient of 0–15% methanol in 400 mM 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP, pH was adjusted to 7.0 by the addition of triethylamine), followed by a 35-min linear gradient of 15–35% methanol in 400 mM HFIP, was employed for the separation. The mass spectrometer was operated in the negative-ion mode, and the instrument was set up to acquire MS in the high-resolution ‘ultra zoom scan’ mode in the m/z range of 680–688 and the tandem mass spectra (MS/MS) for the fragmentation of the [M – H]– ions of p(mdCg)pdG and pTgpdG. The temperature for the ion transport tube of the mass spectrometer was maintained at 300°C.
For quantitative analysis, the digestion mixtures, to which 4 µl of ODNX was added prior to nuclease P1 digestion, was incubated at 37°C for over 60 h to allow for the extensive deamination of p(mdCg)pdG to pTgpdG. The mass spectrometer was set up for monitoring the fragmentation of the [M – H]– ions of the stable isotope-labeled pTgpdG* as well as unlabeled dinucleotides pTgpdG and pTgp(8-oxodG). The calibration curves were constructed by subjecting, to LC-MS/MS analysis, the same amount of the isotope-labeled ODNX solution (4 µl) and different amounts of the authentic dodecameric standard d(ATGGCTgGGCTAT) or d(ATGGCTgG▵GCTAT) (‘G▵’ represents 8-oxodG) containing single Tg or tandem Tg-(8-oxodG) lesion, respectively (16). A 5-min gradient of 2–20% methanol in 400 mM HFIP, followed by a 35-min gradient of 20–50% methanol in 400 mM HFIP, was employed for the separation.
RESULTS
Kinetics of deamination for the two cis diastereomers of mdCg in duplex ODNs
To measure the kinetics of deamination of 5-methylcytosine glycol in double-stranded DNA, we first produced the 5-methylcytosine glycol-carrying duplex DNA by treating 3.0 nmol of a 27-mer duplex ODN,
5′-ACGGGCCGAAACXGGAGGCCCGACCGT-3′
3′-TGCCCGGCTTTGGXCTCCGGGCTGGCA-5′,
where ‘X’ is a 5-methylcytosine, with Fenton-type reagents (100 μM CuCl2, 800 μM H2O2 and 8 mM ascorbate) in a buffer at room temperature for 60 min. The reaction was quenched with methionine, and the solution was buffer-exchanged into a 10-mM phosphate buffer and incubated at 37°C to allow for the deamination to occur. Aliquots were removed at different time points, frozen immediately on dry ice and stored in a –80°C freezer. After a quick digestion with nuclease P1, samples were subjected immediately to on-line LC-MS analysis, where the dinucleotide p(mdCg)pdG and its deaminated counterpart, i.e., pTgpdG, were monitored in a high-resolution ‘zoom-scan’ mode on an LTQ linear ion-trap mass spectrometer. In this respect, it was found that the bulky nature of thymidine glycol and its lack of aromaticity prohibit the nuclease P1-mediated cleavage of its 3′ phosphodiester linkage (18–20); thus, the digestion with nuclease P1 produces a dinucleotide containing the lesion along with its flanking 3′ unmodified nucleotide. Indeed HPLC analysis of the nuclease P1 digestion mixture of a dodecameric ODN harboring a single Tg showed that ∼89% of the Tg was released as a dinucleotide pTgpdG (Figure S1). It is of note that the digestion with both nuclease P1 and alkaline phosphatase has been employed by Bailey et al. (19) for the LC-MS/MS identification of Tg formed in cultured mouse fibroblast cells. We, however, observed that only a small fraction of Tg is liberated as a dinucleoside monophosphate after the lesion-carrying ODN had been treated with both nuclease P1 and alkaline phosphatase; the majority of the lesion is released as a mononucleoside, which might be attributed to the presence of residual endonuclease activity in the commercial preparation of the alkaline phosphatase (data not shown). As there is no TG site in the 27-mer duplex sequence, the nuclease P1-produced dinucleotide pTgpdG has to arise from the deamination of mdCg at methyl-CpG site.
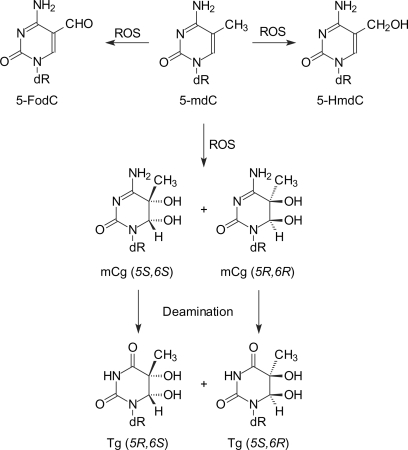
The LC-MS/MS data revealed two diastereomers of mdCg and the two diastereomers of the corresponding deaminated product, i.e. Tg (Figure 1). The earlier and later eluting fractions of the deaminated products were assigned as the dinucleotides pTgpdG housing the (5R,6S) and (5S,6R) diastereomers of Tg, respectively. The stereochemistry was established based on the LC-MS/MS analysis of dinucleotides formed from the nuclease P1 digestion of Tg-containing ODNs obtained from chemical synthesis [the (5R,6S) diastereomer of Tg] (16) or from the OsO4-induced oxidation of d(ATGC) (The two cis diastereomers, Scheme 1. LC-MS/MS results are shown in Figure 1 and Figures S2 and S3 in the Supplementary Data.) The two diastereomers gave almost identical fragmentation patterns with four major daughter ions at m/z 346, 426, 541 and 586, where the ion of m/z 541 is slightly more abundant for the dinucleotide containing the (5S,6R) diastereomer than that bearing the (5R,6S) diastereomer of Tg (Figures S2 and S3).
Figure 1.
The SICs for monitoring the [M – H]– ions of p(mdCg)pdG [m/z 683, (a)] and pTgpdG [m/z 684, (c)], as well as for monitoring the m/z 683 →524, 567 transitions of p(mdCg)pdG (b) and for monitoring the m/z 684 →541 transition of pTgpdG (d). The sample was the nuclease P1 digestion mixture of 27-mer mC-containing duplex DNA after it was treated with Fenton reagent and deaminated at 37°C for 24 h (details are described in ‘Materials and Methods’ section). Shown also are the SICs for the m/z 684 →541 transition for the analyses of the nuclease P1 digestion mixtures of a ODN, d(ATGGCTgGGCTAT), which contains the (5R,6S) diastereomer of Tg (e) and the nuclease P1 digestion mixture of d(ATGC) that has been treated with OsO4 (f). The MS/MS of the deprotonated ions of p(mdCg)pdG and pTgpdG are shown in Figures S2–S4.
Tg in mononucleoside is known to undergo epimerization at C6 carbon atom to give a mixture of cis and trans diastereomers (21). A recent nuclear magnetic resonance (NMR) study showed that the cis-(5R,6S) diastereomer of Tg, when paired with an adenine in duplex DNA, can undergo ring opening at the C6–N1 bond and the subsequent ring closure can give rise to a mixture containing the original cis-(5R,6S) diastereomer of Tg and the trans-(5R,6R) diastereomer with a ratio of 70 : 30 (cis/trans) at 30°C (22). The cis-(5R,6S) diastereomer of Tg, however, remains stable in duplex DNA when paired with a guanine residue, and the corresponding trans-(5R,6R) diastereomer is below the detection limit of the NMR method (22). In this context, the Tg arising from the deamination of mdCg is paired with guanine residue in duplex DNA. It has not been assessed whether a similar cis–trans equilibrium exists for mdCg in isolated nucleoside or duplex DNA. Nevertheless, we were able to detect only the two components exhibiting the same retention time and MS/MS as the two authentic cis diastereomers of Tg even after the nuclease P1 digestion mixture had been incubated at room temperature for a prolonged period of time, suggesting that the trans diastereomers might not be formed in duplex DNA upon treatment with Cu(II)/H2O2/ascorbate, although we cannot exclude formally the possibility that the trans diastereomer may co-elute with the cis diastereomer under the chromatographic conditions that we used.
The tandem mass spectra of the [M – H]– ions of p(mdCg)pdG differ substantially from those of pTgpdG. The former spectra show abundant fragment ions of m/z 665, 585, 567 and 524, which are attributed to the eliminations of H2O, H3PO4, [H2O + H3PO4], and 5-methylcytosine glycol, respectively (Figure S4). It is worth noting that, although all the major fragment ions observed in Figure S4 could be assigned, the lack of pure ODN harboring a site-specifically inserted mdCg prevents us from excluding the possibility of the presence of co-eluting isobaric impurities. Aside from the two major peaks at 19.5 and 20.3 min, we also observed a weak peak at 18.3 min in the selected-ion chromatogram (SIC) for monitoring the formation of the m/z-683 ion (Figure 1a). This peak is attributed to the [2M + Na – 2H]– ion of 2′-deoxyadenosine 5′-phosphate (pdA), which elutes at the same retention time (i.e. 18.3 min). This argument is substantiated by the product-ion spectrum of the m/z-683 ion averaged from the 18.3-min peak, which showed the formation of a dominant fragment ion of m/z 432, arising from the neutral loss of a 2′-deoxyadenosine, and an abundant fragment ion of m/z 330, being the [M – H]– ion of pdA.
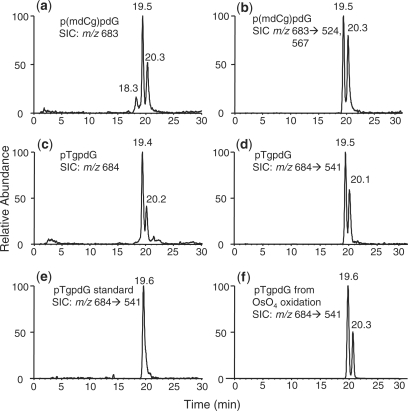
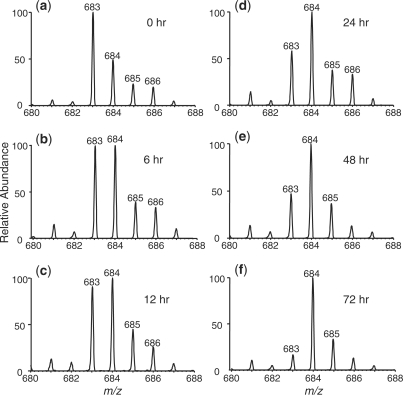
As depicted in Figure 2, we observed a time-dependent decrease in signal intensity for the p(mdCg)pdG with a concomitant increase in signal intensity for the pTgpdG. Since MS/MS provides better specificity, we decided to assess the kinetics of deamination based on MS/MS. In this regard, we normalized the area of peaks found in the SIC for the m/z 684 →541 transition, which is characteristic of pTgpdG, against that of peak found in the SIC for monitoring the fragmentation of the ion of m/z 330 (which is the [M – H]– ion of pdA). This normalization can facilitate the correction of difference in analyte loss during various steps of sample manipulation and the variation in instrument response. Plotting the normalized ratio versus the deamination time gives straight lines (Figure 3), supporting that the deamination follows a first-order reaction kinetics. The slope of the line facilitates the determination of the rate constant, which allows for the calculation of the half-life for the deamination of 5-methylcytosine glycol in duplex DNA (Figure 3 and the results are summarized in Table 2). It turned out that the rate constants for the deamination of the (5S,6S) and (5R,6R) diastereomers in duplex DNA at 37°C were 0.0186 and 0.0253 h–1, respectively, and the half-lives for their deamination were calculated to be 37.4 and 27.4 h, respectively.
Figure 2.
High-resolution ‘zoom-scan’ ESI-MS for monitoring the kinetics of deamination of 5-methylcytosine glycol in a 27-mer duplex DNA. Shown are the regions of the mass spectra for the [M – H]– ions of the dinucleotide p(mdCg)pdG (m/z 683) and its deaminated counterpart, pTgpdG (m/z 684) at 0 (a), 6 (b), 12 (c), 24 (d), 48 (e), and 72 h (f) after the initiation of the deamination reaction, and the spectra were averaged from the two major peaks found in Figure 1a and c. The 27-mer duplex sequence and deamination reaction conditions were described in ‘Materials and Methods’ section, and the initial extent of deamination was estimated to be ∼20%.
Figure 3.
Determination of the first-order rate constants for the deamination of 5-methylcytosine glycol in a 27-mer duplex DNA. Plotted are the ratios of peak area found in the SIC for the m/z 684 →541 transition (for pTgpdG) over that found in the SIC for monitoring the fragmentation of the ion of m/z 330 (for pdA) versus the deamination time. The solid square and triangle represent the results based on the formation of pTgpdG containing the (5R,6S) (the 19.5-min peak in Figure 1d) and (5S,6R) (the 20.1-min peak in Figure 1d) diastereomers of Tg (Scheme 1), respectively. The deamination reaction conditions were described in ‘Materials and Methods’ section.
Table 2.
Kinetic parameters for the deamination of 5-methylcytosine and 5-methylcytosine glycol at 37°C
We also assessed the kinetics of deamination mdCg in duplex DNA by measuring the decrease in signal intensity for p(mdCg)pdG, and the rate constant for the deamination of the major and minor diastereomers of mdCg were found to be 0.0177 and 0.0147 h–1, respectively (Figure S5). It remains unclear what accounts for the discrepancy in rate constants determined from the rise in signal for pTgpdG and the drop in signal for p(mdCg)pdG. However, we speculate that this might be attributed, in part, to the presence of co-eluting isobaric impurities with p(mdCg)pdG and/or the lack of quantitative release of mdCg from duplex DNA. We have shown that the nuclease P1-mediated release of pTgpdG from ODN is nearly quantitative and the product-ion spectra of the pTgpdG emanating from the deamination reaction are very similar as those of the standard compounds (vide supra). In addition, the increase in signal of pTgpdG has to arise from the deamination of mdCg in duplex DNA. Therefore, we reason that the rate constants determined from the signal increase in pTgpdG are reliable.
LC-MS/MS quantification of pTgpdG and pTgp(8-oxodG) lesions induced in duplex DNA by Cu(II)/H2O2/ascorbate
To quantify the total amount of Tg formed from the deamination of mdCg, we treated a self-complementary duplex ODNs with different concentrations of Cu(II)/H2O2/ascorbate (Table 1), digested the resulting ODNs with nuclease P1 to release the lesions as dinucleotide and analyzed the digestion mixtures by LC-MS/MS with stable isotope-labeled lesions as internal standard.
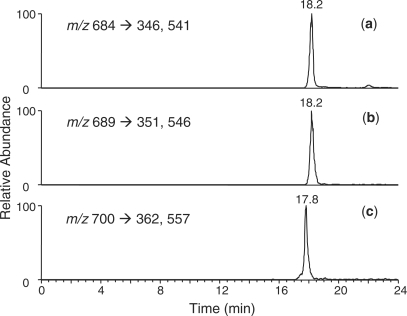
Prior to LC-MS/MS analysis, the digestion mixture was incubated at 37°C to allow for the extensive deamination of mdCg to Tg. In this context, it has been observed that the deamination of mdCg leads to its complete conversion to Tg without other degradation product at neutral pH, where the half-life is around 20 h for mononucleoside (5).
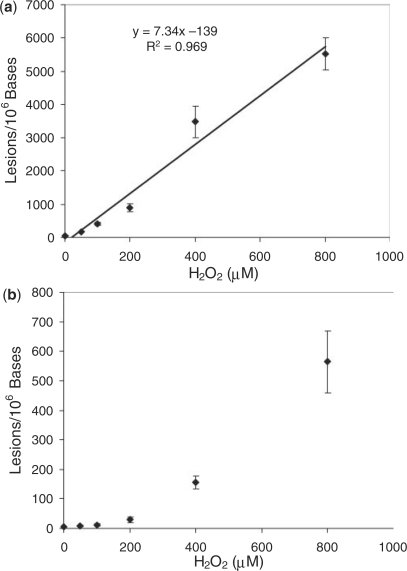
A fast chromatographic gradient was applied to allow for the two diastereomers to co-elute (Figure 4). The SIC for the m/z 684 →346, 541 transitions, which monitor the losses of thymidine glycol 5′-phosphate and the C5H5NO4 component, respectively (Figure S6), showed a fraction eluting at the same retention time as the labeled pTgpdG at 18.2 min. Similarly, the SIC for the m/z 700 →362, 557 transitions showed a peak at 17.8 min, which supported the formation of the pTgp(8-oxodG) tandem lesion. It is worth noting that the elimination of C5H5NO4 is a characteristic neutral loss from thymine glycol-containing dinucleotides and ODNs (Figure S6) (18). The quantification results showed that the pTgpdG exhibited a linear dose-responsive increase when the concentration of H2O2 was up to 800 µM; however, the yield of pTgp(8-oxodG) exhibits a nonlinear response over the dose range of Fenton-type reagents used (Figure 5, and calibration curves for LC-MS/MS quantification are shown in Figure S7). The yields of pTgp(8-oxodG) at low-dose range increase slowly with the rise in the concentrations of Cu(II)/H2O2/ascrobate. However, when DNA was treated with the Fenton-type reagent at the three higher doses, the yield of pTgp(8-oxodG) increased sharply. At the highest dose, the yield of pTgp(8-oxodG) is ∼10% of that of pTgpdG. The exact reason for the disproportionally high yield at higher doses remains ambiguous, though it is possible that the formation of the tandem lesion requires two oxidation events; the formation of mdCg at the 5-methyl-2′-deoxycytidine site may promote the oxidation of its neighboring 2′-deoxyguanosine, and vice versa.
Figure 4.
SICs for monitoring the m/z 684 →346,541 [(a), for unlabeled pTgpdG], m/z 689 →351, 546 [(b), for labeled pTgpdG] and m/z 700 →362,557 [(c), for unlabeled pTgp(8-oxodG)] transitions obtained from the LC-MS/MS analyses of the nuclease P1 digestion mixture of Fenton-reagent-treated ODNs under reaction condition C (Table 1).
Figure 5.
Cu(II)/H2O2/ascorbate-induced formation of pTgpdG (a) and pTgp(8-oxodG) (b) lesions in ODN d(AXGTAXGTAXGTAXGT) (‘X’ represents 5-methylcytosine). The values represent the means ± SD from three independent oxidation and quantification experiments.
Hydroxyl radical can be added to C5 = C6 double bond of mC to form the glycol lesions, and it can also abstract a hydrogen atom from the methyl group of mC to generate an allyl radical. The allyl radical can result in the formation of two other single-nucleobase lesion, i.e. 5-HmdC and 5-FodC (Scheme 1), as well as two intrastrand cross-links, i.e. G[8-5m]mC and mC[5m-8]G (11,23,24). To compare the damage products from different mC radicals, we also measured yields of 5-HmdC in the same ODN sequence. It turned out that the yield of Tg is comparable to that of 5-HmdC under the same experimental conditions, but was 5–6-fold lower than that of 8-oxodG (Figures 5a and Figure S8, and calibration curves are depicted in Figure S9). It is worth noting that the quantification results for 5-HmdC and 8-oxodG are consistent with the data that we obtained earlier for ODNs in different sequence contexts (11).
DISCUSSION
After the saturation of the C5 = C6 double bond, cytosine derivatives are susceptible to hydrolytic deamination. It was observed recently that the half-life of Cg is greatly enhanced in DNA, which is increased by 34-fold from 50 min in free nucleoside to 28 h in calf thymus DNA (10). A more recent study indicated that the half-life of Cg is 6.5 h in poly(dG-dC) (25). Even though it is shorter than what was previously reported, the half-life of Cg in duplex structure is still approximately eight times longer than Cg in free nucleoside. On the other hand, we observed that the deamination of the (5S,6S) diastereomer of mdCg in duplex DNA is merely 2-fold slower than the same compound in mononucleoside, whereas the placement of the (5R,6R) diastereomer in duplex DNA only increases the lifetime of the lesion by approximately 20% (Table 2). The striking difference in the effect of duplex structure on the deamination of Cg and 5-methylcytosine glycol suggests that the presence of a methyl group on the C5 carbon in 5-methylcytosine glycol, along with the saturation of the C5-C6 bond, may facilitate the lesion to adopt an extrahelical conformation in duplex DNA, thereby enabling the facile deamination of the lesion. Along this line, a previous NMR structural study demonstrated that the structurally related thymidine glycol is largely extrahelical (26). It is of note that, in isolated nucleoside, the (5S,6S) diastereomer is deaminated at a slightly slower rate than the (5R,6R) diastereomer, whereas the opposite is true when the two diastereomers are in duplex DNA. The difference might be due to the different steric hindrance experienced by the two diastereomers in duplex DNA, which may affect the nucleophilic attack of mdCg by a water molecule and its subsequent deamination to Tg. Future structural studies of duplex DNA housing the two diastereomers of mdCg or Tg will offer important insights about the difference of the two diastereomers of mdCg toward hydrolytic deamination.
As the presence of Tg inhibits the cleavage of its 3′ phosphodiester linkage by nuclease P1, which results in the liberation of the lesion as a dinucleotide bearing the 3′-flanking nucleotide (18–20), we quantified the Tg lesions formed at mCpG site as pTgpdG dinucleotide using negative-ion mass spectrometry. This also allows us to quantify the Tg-(8-oxodG) tandem lesion formed at mCpG site. In our study we used synthetic ODN containing an isotope-labeled 5′-pTgpdG*-3′ sequence (‘dG*’ represents the uniformly 15N-labeled 2′-deoxyguanosine) as internal standard, this method can effectively eliminate the error introduced by the potential lack of quantitative release of the lesion as a dinucleotide by nuclease P1 and spontaneous dephosphorylation during various stages of sample manipulation.
Previous experimental data obtained for poly(dG-mdC) upon exposure to γ rays (13) and theoretical calculations at the mononucleoside level (12) indicated that the hydroxyl radical can attack 5-methylcytosine at the C5 = C6 double bond and the 5-methyl group, with preference being on attack at the double bond to yield 5-methylcytosine glycol. The latter can deaminate to afford thymine glycol. However, the yield of thymine glycol in ODNs treated with Fenton-type reagents amounts to about five to six lesions per 1000 nucleosides at the highest dose, and is in a similar range as the other two single-nucleobase lesions arising from the 5-methyl radical of mC, i.e. 5-hydroxymethylcytosine (Figure 5a, Figure S8, and Table S1) and 5-formylcytosine (11). This result may indicate that the theoretical calculation at the mononucleoside level does not reveal completely the reactivity of the nucleoside in duplex DNA, where the methyl group in 5-methylcytosine, locating in the major groove, can be more accessible to hydroxyl radical attack than the C5 = C6 double bond.
The efficient formation of mdCg and its facile deamination to Tg, together with the production of 5-HmdC and 5-FodC, may account for the prevalent C →T transition mutation found at CpG site in human p53 gene, a hallmark for many types of human tumors (27). In this context, several mechanisms have been proposed to account for the ubiquitously observed C →T transition mutation at CpG site (1). Two of them involve the deamination processes. One is the direct deamination of 5-methylcytosine to thymine. In support of this mechanism, Shen et al. (28) found that the rate for the deamination of 5-methylcytosine in duplex DNA at 37°C is 3.3*108 h–1, which is twice as fast as the corresponding deamination of cytosine. The other involves the formation of mdCg and its subsequent deamination to Tg, and we showed that this latter deamination is about 7 orders of magnitude faster than the spontaneous deamination of 5-methylcytosine in duplex DNA (Table 2). For the two deamination pathways to be equally important in contributing to the C →T transition mutation at CpG site, mdCg has to form at a frequency of one lesion per 107 5-methyl-2′-deoxycytidine. Our quantification results showed that the rate for the formation of mdCg is about 5–6-fold lower than that of 8-oxodG, and a recent careful study showed that the basal level of 8-oxodG in human cells is approximately 2.2 lesions per 107 2′-deoxyguanosine (29). Thus, the two deamination pathways may make a similar extent of contribution to the frequently observed C →T mutation at CpG site. It is worth noting that the above analysis is under the assumption that the product arising from the spontaneous deamination of 5-methylcytosine, i.e. a G : T mismatch, and the product generated from the deamination of mdCg, i.e. a G : Tg pair, are repaired in cells at a similar efficiency. Further studies are required to assess whether this is indeed the case.
It is also important to discuss how the presence of an additional methyl group on the C5 carbon affects the ROS-induced damage to cytosine residues in DNA. The results presented in this article, together with our recently published data (11), revealed that the total yield of the single-nucleobase lesions, Tg, 5-HmdC and 5-FodC, emanating from mCpG in duplex DNA upon exposure to Cu(II)/H2O2/ascorbate is several fold higher than the major single-nucleobase lesion, 5-hydroxy-2′-deoxycytidine (5-OHdC), formed at unmethylated cytosine at CpG sites. A mutation frequency of 0.05% was reported for 5-OHdC after the lesion-bearing single-stranded M13 genome was propagated in Escherichia coli cells (30), and a mutation frequency of 0.03–0.28% was found for 5-FodC after the damage-harboring double-stranded shuttle vector was replicated in simian COS-7 cells (31). The mutagenic properties of 5-HmdC and mdCg have not yet been examined; however, the deaminated product of mdCg (i.e. Tg) is known to direct the predominant incorporation of adenine nucleotide during DNA replication (7–9). Together, the enhanced formation of oxidatively generated lesion from methylated cytosine, combined with the facile deamination of mdCg and the high mutagenic potential of the deaminated product, may render 5-methylcytosine more susceptible to produce C →T transition mutation than its unmethylated counterpart.
We also observed that the yield for the formation of the tandem lesion, i.e. Tg-(8-oxodG), is only about 10-fold less than that found for Tg that is 5′ neighboring to an unmodified dG (Figure 5). Therefore, the high yield for the formation of this tandem lesion at mCpG sites and the inefficient removal of 8-oxodG from Tg-(8-oxodG) site by human 8-oxoguanine DNA glycosylase (hOGG1) (32) may account for the high frequency of mCG →TT mutation observed after the CpG-methylated and Cu(II)/H2O2/ascorbate-treated pSP189 shuttle vector has been replicated in NER-deficient XPA cells (15). Along this line, both NER and long-patch BER repair pathway have been proposed for the repair of another type of tandem lesions composed of a thymine glycol and a neighboring 5′ 2-deoxyribonolactone (33).
Copper plays a pivotal role in maintaining the structure and integrity of chromatin (34,35). Our results suggest that transition metal ion, copper or iron (11), can induce significant amount of DNA damage, thereby posing a great challenge for people suffering from Wilson's disease and primary hemochromatosis, which cause abnormal accumulation of copper and iron in various organs (36,37). The tandem and/or cluster lesions, whose biochemical effects are largely unknown, constitute a major family of DNA damage products produced by Fenton-type reagents and ionizing radiation (38). The mutagenic properties of this particular type of tandem lesions are currently investigated in our laboratory.
Methylation pattern of cytosine residue at CpG sites is crucial for maintaining the epigenetic signal in human cells (39). The ROS-induced nucleobase modification at CpG sequence may also result in disturbed gene regulation and heritable epigenetic changes in chromatin (40).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (R01 CA 101864). Funding for open access charge: The National Institutes of Health (R01 CA 101864).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors want to thank Dr. Yuesong Wang for synthesizing the Tg-containing ODNs.
REFERENCES
- 1.Pfeifer GP. p53 mutational spectra and the role of methylated CpG sequences. Mutat. Res. 2000;450:155–166. doi: 10.1016/s0027-5107(00)00022-1. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. DNA lesions generated in vivo by reactive oxygen species, their accumulation and repair. NATO ASI Ser. A. 1999;302:251–257. [Google Scholar]
- 3.Dizdaroglu M. Mechanisms of oxidative DNA damage; lesions and their measurement. NATO ASI Ser. A. 1999;302:67–87. [Google Scholar]
- 4.Henle ES, Linn S. Formation, prevention, and repair of DNA damage by iron hydrogen peroxide. J. Biol. Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 5.Bienvenu C, Cadet J. Synthesis and kinetic study of the deamination of the cis diastereomers of 5,6-dihydroxy-5,6-dihydro-5-methyl-2′-deoxycytidine. J. Org. Chem. 1996;61:2632–2637. doi: 10.1021/jo951900e. [DOI] [PubMed] [Google Scholar]
- 6.McNulty JM, Jerkovic B, Bolton PH, Basu AK. Replication inhibition and miscoding properties of DNA templates containing a site-specific cis-thymine glycol or urea residue. Chem. Res. Toxicol. 1998;11:666–673. doi: 10.1021/tx970225w. [DOI] [PubMed] [Google Scholar]
- 7.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Translesion synthesis by human DNA polymerase eta across thymine glycol lesions. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 8.Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. Human DNA polymerase kappa bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J. Biol. Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- 9.Johnson RE, Yu SL, Prakash S, Prakash L. Yeast DNA polymerase zeta (ζ) is essential for error-free replication past thymine glycol. Genes Dev. 2003;17:77–87. doi: 10.1101/gad.1048303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tremblay S, Douki T, Cadet J, Wagner JR. 2′-deoxycytidine glycols, a missing link in the free radical-mediated oxidation of DNA. J. Biol. Chem. 1999;274:20833–20838. doi: 10.1074/jbc.274.30.20833. [DOI] [PubMed] [Google Scholar]
- 11.Cao HC, Wang YS. Quantification of oxidative single-base and intrastrand cross-link lesions in unmethylated and CpG-methylated DNA induced by Fenton-type reagents. Nucleic Acids Res. 2007;35:4833–4844. doi: 10.1093/nar/gkm497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grand A, Morell C, Labet V, Cadet J, Eriksson LA. H-center dot atom and (OH)-O-center dot radical reactions with 5-methylcytosine. J. Phys. Chem. A. 2007;111:8968–8972. doi: 10.1021/jp0737799. [DOI] [PubMed] [Google Scholar]
- 13.Zuo SJ, Boorstein RJ, Teebor GW. Oxidative damage to 5-methylcytosine in DNA. Nucleic Acids Res. 1995;23:3239–3243. doi: 10.1093/nar/23.16.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong HZ, Cao HC, Wang YS, Wang YS. Identification and quantification of a guanine-thymine intrastrand cross-link lesion induced by Cu(II)/H2O2/ascorbate. Chem. Res. Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee DH, O'Connor TR, Pfeifer GP. Oxidative DNA damage induced by copper and hydrogen peroxide promotes CG -> TT tandem mutations at methylated CpG dinucleotides in nucleotide excision repair-deficient cells. Nucleic Acids Res. 2002;30:3566–3573. doi: 10.1093/nar/gkf478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Wang Y. Synthesis and thermodynamic studies of oligodeoxyribonucleotides containing tandem lesions of thymidine glycol and 8-oxo-2′-deoxyguanosine. Chem. Res. Toxicol. 2006;19:837–843. doi: 10.1021/tx060032l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwai S. Synthesis of thymine glycol containing oligonucleotides from a building block with the oxidized base. Angew. Chem. Int. Ed. 2000;39:3874–3876. doi: 10.1002/1521-3773(20001103)39:21<3874::AID-ANIE3874>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y. HPLC isolation and mass spectrometric characterization of two isomers of thymine glycols in oligodeoxynucleotides. Chem. Res. Toxicol. 2002;15:671–676. doi: 10.1021/tx0155855. [DOI] [PubMed] [Google Scholar]
- 19.Bailey DT, DeFedericis HCC, Greene KF, Iijima H, Budzinski EE, Patrzyc HB, Dawidzik JB, Box HC. A novel approach to DNA damage assessments: measurement of the thymine glycol lesion. Radiat. Res. 2006;165:438–444. doi: 10.1667/rr3534.1. [DOI] [PubMed] [Google Scholar]
- 20.Weinfeld M, Soderlind KJM, Buchko GW. Influence of nucleic-acid base aromaticity on substrate reactivity with enzymes acting on single-stranded-DNA. Nucleic Acids Res. 1993;21:621–626. doi: 10.1093/nar/21.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lustig MJ, Cadet J, Boorstein RJ, Teebor GW. Synthesis of the diastereomers of thymidine glycol, determination of concentrations and rates of interconversion of their cis-trans epimers at equilibrium and demonstration of differential alkali lability within DNA. Nucleic Acids Res. 1992;20:4839–4845. doi: 10.1093/nar/20.18.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown KL, Adams T, Jasti VP, Basu AK, Stone MP. Interconversion of the cis-5R,6S- and trans-5R,6R-thymine glycol lesions in duplex DNA. J. Am. Chem. Soc. 2008;130:11701–11710. doi: 10.1021/ja8016544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q, Wang Y. Independent generation of 5-(2′-deoxycytidinyl)methyl radical and the formation of a novel crosslink lesion between 5-methylcytosine and guanine. J. Am. Chem. Soc. 2003;125:12795–12802. doi: 10.1021/ja034866r. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, Wang Y. Generation of 5-(2′-deoxycytidyl)methyl radical and the formation of intrastrand cross-link lesions in oligodeoxyribonucleotides. Nucleic Acids Res. 2005;33:1593–1603. doi: 10.1093/nar/gki301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremblay S, Wagner JR. Dehydration, deamination and enzymatic repair of cytosine glycols from oxidized poly(dG-dC) and poly(dI-dC) Nucleic Acids Res. 2008;36:284–293. doi: 10.1093/nar/gkm1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kung HC, Bolton PH. Structure of a duplex DNA containing a thymine glycol residue in solution. J. Biol. Chem. 1997;272:9227–9236. doi: 10.1074/jbc.272.14.9227. [DOI] [PubMed] [Google Scholar]
- 27.Rideout WM, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 28.Shen JC, Rideout WM, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22:972–976. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mangal D, Vudathala D, Park JH, Lee SH, Penning TM, Blair IA. Analysis of 7,8-dihydro-8-oxo-2′-deoxyguanosine in cellular DNA during oxidative stress. Chem. Res. Toxicol. 2009;22:788–797. doi: 10.1021/tx800343c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreutzer DA, Essigmann JM. Oxidized, deaminated cytosines are a source of C -> T transitions. in vivo. Proc. Natl Acad. Sci. USA. 1998;95:3578–3582. doi: 10.1073/pnas.95.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamiya H, Tsuchiya H, Karino N, Ueno Y, Matsuda A, Harashima H. Mutagenicity of 5-formylcytosine, an oxidation product of 5-methylcytosine, in DNA in mammalian cells. J. Biochem. 2002;132:551–555. doi: 10.1093/oxfordjournals.jbchem.a003256. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Wang Y, Wang Y. In vitro replication and repair studies of tandem lesions containing neighboring thymidine glycol and 8-oxo-7,8-dihydro-2′-deoxyguanosine. Chem. Res. Toxicol. 2009;22:574–583. doi: 10.1021/tx8003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM. DNA tandem lesion repair by strand displacement synthesis and nucleotide excision repair. Biochemistry. 2008;47:4306–4316. doi: 10.1021/bi7021427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agarwal K, Sharma A, Talukder G. Effects of copper on mammalian-cell components. Chem. Biol. Interact. 1989;69:1–16. doi: 10.1016/0009-2797(89)90094-x. [DOI] [PubMed] [Google Scholar]
- 35.Lewis CD, Laemmli UK. Higher-order metaphase chromosome structure – evidence for metalloprotein interactions. Cell. 1982;29:171–181. doi: 10.1016/0092-8674(82)90101-5. [DOI] [PubMed] [Google Scholar]
- 36.Ala A, Walker AP, Ashkan K, Dooley JS, Schilsky ML. Wilson's disease. Lancet. 2007;369:397–408. doi: 10.1016/S0140-6736(07)60196-2. [DOI] [PubMed] [Google Scholar]
- 37.Aaseth J, Flaten TP, Andersen O. Hereditary iron and copper deposition: diagnostics, pathogenesis and therapeutics. Scand. J. Gastroenterol. 2007;42:673–681. doi: 10.1080/00365520601075662. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y. Bulky DNA lesions induced by reactive oxygen species. Chem. Res. Toxicol. 2008;21:276–281. doi: 10.1021/tx700411g. [DOI] [PubMed] [Google Scholar]
- 39.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.